Abstract
BACKGROUND
Semaphorin 4F (S4F) has roles in embryological axon guidance and is expressed in adults. S4F is involved in cancer-induced neurogenesis.
METHODS
Prostate cells were transfected with S4F retrovirus. Cells and controls were used for a BrdU incorporation assay (proliferation) and in vitro scratch and matrigel transwell chamber invasion assay (migration). Monoclonal antibodies were developed using baculovirus expressed recombinant GST-S4F and used to immunostain tissue microarrays. Slides were imaged using deconvolution and analyzed using tissue segmentation. Data was correlated with clinico-pathological parameters, other biomarkers and survival analysis performed. Heterogeneity of S4F expression was analyzed with unsupervised clustering algorithms.
RESULTS
Proliferation rates measured by BrdU incorporation were higher in all S4F transfected cells. S4F over-expression was associated with increased motility of the cancer cells. S4F expression was over expressed in HGPIN/PCa than normal epithelium. S4F expression correlated with seminal vesicle invasion. Patients with high values of S4F in PCa cytoplasm are at significantly higher risk of biochemical recurrence, by univariate and multivariate analysis. S4F cytoplasmic expression in PCa cells also correlates with nerve density in PCa and perineural invasion diameter. Correlations were identified with NFkB and inversely with apoptosis in PNI.
CONCLUSION
This data demonstrates that S4F is significantly involved in human PCa progression. S4F is a key regulator of the interactions between nerves in the tumor microenvironment and cancer cells. Because of the importance of cancer nerve interaction in the biology of cancer and its clinical implication, S4F can be considered a major therapeutic target.
BACKGROUND
Nerves and cancer cells interact at many levels. Invasion of the nerve sheath by cancer cells, termed perineural invasion (PNI), is a key feature of human prostate cancer (PCa). Perineural invasion (PNI) is the process by which cancer cells wrap around nerves and the best described interaction between cancer and nerves. PNI is also a key route for PCa metastasis. Our PNI model (1) demonstrated specific interactions between PCa cells and nerves, which lead to co-stimulation of growth with reduced rate of apoptosis and an increased rate of proliferation through caveolin 1 and NFkB based mechanisms (2, 3). This phenomenon was validated in human tissues.
We have also recently described a novel biologic phenomeonon, cancer-related axonogenesis and neurogenesis (4). Our studies show that axon density is increased in cancer areas as well as in preneoplastic lesions compared to controls. Two and 3-dimensional reconstructions of entire prostates confirmed axonogenesis in human tumors. Finally, two in vitro models confirmed that cancer cells, particularly when interacting with nerves in PNI, induce neurite outgrowth in PCa. Axonogenesis is correlated with features of aggressive PCa and with recurrence in PCa. In addition, the number of neurons in the ganglia of patients with cancer was significantly higher than controls. This was the first description of cancer-related axonogenesis and neurogenesis (4). Accordingly, it is becoming more apparent that the biology regulated by nerves in cancer tissues is critical for the development of PCa cancer. Little is known about specific mechanisms of the interactions between nerves and cancer cells.
One of the members of the semaphorin family, semaphorin 4F, has been functionally coupled to cancer-induced axonogenesis (4). S4F is over-expressed only in PCa cells in the PNI model, not in nerves. Over-expression of S4F by PCa cells induces axonogenesis in a N1E115 axonogenesis assay, and S4F inhibition by siRNA reduces this effect (4). Also, siRNA in the PNI in vitro model on naïve DU145 cells reduces axonogenesis from the DRG at 48 hours.
In this study we will demonstrate that S4F is not only involved in axonogenesis, but that through potential autocrine and paracrine mechanisms it affects the proliferation and migration of cancer cells as well as the establishment of PNI. More importantly we will demonstrate using state of the art methodology, that S4F is critical in the interaction of nerves and cancer cells in human disease, and defines an aggressive phenotype of human PCa.
MATERIALS AND METHODS
Generation of Semaphorin 4F retrovirus
S4F was cloned as described previously(4). The retroviral expression system was developed in Dr. Garry Nolan’s lab. Retroviral vector pBMN-I-GFP was purchased from Addgene and retroviral packaging cell line Phoenix-A was obtained from ATCC Safe Deposit. To generate pBMN-I-GFP-4F, S4F cDNA was first inserted into pBMN-I-GFP EcoRI site, then a HA tag with N-terminal S4F cDNA obtained by RT-PCR was inserted into BamHI site (S4F N-terminal has a BamHI site): Forward primer: 5′-CCGGATCCATGTACCCATACGACGTCCCAGACTACGCTCCAAAGATGCCGGCCTCTG (contain an BamHI site); reverse primer: 5′-CCAACATAAAGTGTGTGG. Max Efficiency stbl2 Competent cells (Invitrogen) was used for produce pBMN-I-GFP-4F plasmid. pBMN-I-GFP-4F was then transfected into Phoenix-A cells by Calcium Phosphate Transfection kit (Invitrogen) according to Dr. Nolan’s lab protocol (http://www.stanford.edu/group/nolan/). Retrovirus containing media was harvested post-transfection 48 h and 72 h, filtered with 0.45 μm filter and stored in −80 °C.
Retroviral infection and expression of S4F
To study the autocrine effects of S4F on the PCa cells we transfected S4F into DU145, PC-3, LNCaP and PNT1A cells. These were retrovirally transfected using an expression system developed in Dr. Garry Nolan’s lab. Du145 and LnCap cells were planted in T25 flask at a density of 30–50%. The next day, cell growth media was removed, 1ml viral supernatant was mixed with 1 ml fresh cell growth media contain 8 μg/ml polybrene (final concentration) and added to cells. Three hours later, cells were washed with PBS, and 5 ml fresh growth media was added to cells. After 48 h, cells were separated for scratch, proliferation and invasion assays. S4F expression was confirmed by Western blot (LnCap cells) using anti HA antibody (Abcam) or quantitative PCR (Du145 and LnCap).
Proliferation assay
DU145, PC-3, LnCaP and PNT1A cells with or without virus infection were seeded in 4-well chamber slide (Lab-Tek II, Fisher Sci.,) at a density of 2–3 × 104, 1.5–3 × 104 to 2–3 × 104, 1.5–3 × 104 cells per well. Three days later, cell proliferation was assessed by the BrdU incorporation assay (Roche, Germany), which was performed according to manufacturer’s instruction.
In vitro scratch assay
DU145 and PC-3 cells were infected with S4F or vector retrovirus, then DU145-V, DU145-4F or PC3-V and PC3-4F cells were seeded in 60 mm plates. After cells were confluent, a “scratch” was created by using a p200 pipette tip. To remove the debris and smooth the edge of the scratch, cells were washed once with PBS and replaced with fresh growth medium. To obtain the same field during the image acquisition, markings were made as reference points close to the scratch. Photos were taken at 0, 24h (DU145-V, DU145-4F) or 0, 24h, 48h (PC3-V, PC3-4F) under microscope.
Invasion assay
DU145, PC-3, LNCaP and PNT1A cells with or without virus infection were resuspended in serum-free media and seeded into the upper Matrigel chamber of a transwell system (Fisher Sci.,) at a density of 1.5–3 × 104 cells per well. The bottom wells were filled with complete media containing 30 μg/mL of laminin. After incubating for 72 h, the cells that had invaded the Matrigel membrane were fixed with 4% formaldehyde and stained with 4,6-diamidino-2-phenylindole (DAPI). The cells were then counted under a fluorescence microscope.
Generation of monoclonal S4F antibodies
The monoclonal S4F antibodies 986 and 988 and the baculovirus expressed recombinant GST-S4F ex protein were generated in the Dan L. Duncan Cancer Center Baculovirus/Monoclonal Antibody Shared Resource at Baylor College of Medicine. We assume that SEMA-4F and SEMA-W (an isoform of SEMA-4F) have the same transmembrane domain (Proc. Natl. Acad. Sci., Vol. 96, pp2491–2496, 1999), then the extra-cellular domain of S4F (S4F-ex) cDNA was obtained by PCR: forward primer: 5′-CGGAATTCAGATGCCGGCCTCTGCTG (contain a EcoR I site); reverse primer: 5′-GCGAATTCTCAAGCATCTCGCTGGCTGCC (contain a EcoR I site). The S4F-ex cDNA was inserted into the pAcSecG2T (BD Biosciences) baculovirus transfer vector (pAcSecG2T-S4F ex), and then pAcSecG2T-S4F ex and BD BaculoGold Linearized Baculovirus DNA (BD Biosciences) were co-transfected into Sf9 insect cells to obtain the recombinant baculovirus expressing GST-S4F ex. Recombinant GST-S4F ex protein was produced by infecting High Five cells with the recombinant baculovirus at a MOI of 1.0 for 48 hours. The purified GST-S4F ex fusion protein was used to immunize BALB/c mice, and the fusion was performed with spleen cells from one mouse and the FOX-NY mouse myeloma to produce the S4F hybridomas/monoclonal antibodies. The monoclonal S4F antibodies 986 and 988 were produced as culture supernatants in spinner vessels, precipitated by ammonium sulfate, and purified using a standard Protein G Sepharose purification protocol. Antibodies have been tested successfully for use in western blots, immunohistochemistry in paraffin embedded tissues and as neutralizing for axonogenesis. Antibody validation was performed. LnCap human prostate cancer cells were infected with Vector (V) or S4F lentivirus, and cell lysate was extracted for Western blot analysis using the anti-S4F monoclonal antibody produced in our laboratory. Bands were identified in the expected distribution.
S4F in Human Tissues
PCa progression microarray
Two mm cores of areas of normal cancer, high-grade prostatic intraepithelial neoplasia (HGPIN) and cancer were cored from a total of 100 patients. They were included in 6 blocks and will be referred to as the progression TMA. This set permits the study of HGPIN markers and comparison with normal prostate and cancer. The TMAs were constructed by using a manual tissue arrayer (Beecher Instruments, Silver Spring, MD).
Outcomes Tissue microarray
For this array whole-mount slides were reviewed and mapped. The index tumor, defined as the largest and/or highest GS was identified on the slide, and areas representative of the highest GS were circled. Areas of tumor were circled and a 2 mm core were obtained from these areas and transferred to a recipient paraffin block.
Clinical and pathologic characteristics
The initial cohort consisted of 1120 patients who underwent radical prostatectomy (RP) at Baylor College of Medicine (Houston, TX)–affiliated hospitals between 1983 and 1998. We qualified 640 cases for building TMAs based on the following criteria: (1) patients did not receive preoperative treatment, (2) patients were operated on between 1983 and 1998, and (3) sufficient PCa tissue is available for building TMA. The full cohort patient characteristics have been published before (5–7). A total of 240 patients had analyzable S4F data for this study. Thirteen patients had lymph node metastasis, 90 extra capsular extension, 27 seminal vesicle invasion, 31 positive surgical margin, 28 patients had biochemical recurrence and 12 died of PCa. Tissue recruitment was in accordance with institutional review board approval.
Immunohistochemistry
Immunohistochemical staining of S4F on TMA slides was conducted by using an automated immunostainer (DAKO, Carpinteria, CA). Briefly, sections were deparaffinized in xylene, rehydrated through decreasing concentrations of alcohol ending in phosphate-buffered saline, subjected to steam heat in 10 mmol/L citrate buffer (pH 6.0) for 40 minutes in a vegetable steamer, then allowed to cool off at room temperature for an additional 10 minutes. After endogenous peroxidase activity was quenched in 3% hydrogen peroxide solution in distilled water, sections were incubated with rabbit polyclonal antibody against S4F (1:40, overnight at 48C; cat no. 9462). Sections were washed and the bound antibody was detected by using a DAKO Envision Plus kit (DAKO) with diaminobenzidine as chromogen. Finally, sections were counterstained with hematoxylin, dehydrated, and mounted. Negative controls were sections immunostained as above, but normal rabbit serum was used instead of primary antibody.
Image procurement and interpretation
An automated slide scanner (Bacus Laboratories, Lombard, IL) was used to digitize all the stained PCa progression TMA slides to produce an image of every dot and also inform the dot coordinates on the slide. Each image was interpreted for immunoreactivity by using a 0 to 3+ semiquantitation scoring system for both the intensity of stain and percentage of positive cells (percent labeling frequency). For the intensity, the grading scale ranged from no detectable signal (0) to strong signal seen at low power (3); 2 corresponds to moderate signal seen at low to intermediate power, and 1 corresponds to weak signal seen only at intermediate to high power. Labeling frequency was scored as 0 (0%), 1 (1% – 33%), 2 (34%–66%), or 3 (67%–100%). To represent the intensity of hot spots, the highest intensity value was used. The staining index was obtained by multiplying the score of intensity with that of percentage. In the case of nuclear and cytoplasmic expression, both nuclear and cytoplasmic staining signals were interpreted and recorded separately.
Deconvolution Imaging and NuanceTM
Using our newly developed S4F antibody we have combined deconvolution imaging (Nuance™) and image segmentation technology (inForm™) to quantitate biomarker expression more reliably. The NuanceTM™ multispectral deconvolution imaging system (CRi) was used to image the outcomes tissue microarray. The system utilizes an optimized high-throughput tunable filter and its spectral range is 420 to 720nm. Each final image is composed of numerous component images that specifically target small portions of the spectrum. The final result is an image that does not look different from an image captured with a regular RGB camera, but can be analyzed with greater detail because it carries significantly superior amounts of discriminating information.
The inFormTM advanced image analysis software permits tissue segmentation. It is based on learn-by-example automated image processing with object recognition and data analysis tools. inFormTM was successfully trained to find tissue types (cancer vs. stroma). Subsequently it was used to automatically assess IHC staining levels, on a cell-by-cell and sub-cellular basis, for per-cell phenotyping.
Statistical analysis
The semiquantitative differences of S4F expression between normal, preneoplastic and PCa were compared between normal prostate and cancer specimens by using matched pair analysis (Wilcoxon rank tests). The S4F quantitaive data from the outcomes cohort obtained using combined deconvolution imaging (NuanceTM) and image segmentation technology (inFormTM) required a different analysis. Data is provided on a cell per cell basis (nuclear vs. cytoplasmic), per tumor compartment, per patient. This created a statistical challenge that required new analytical methodology. The intensity span was divided into 10 intensity bins. Each bin was then populated with the percentage of cells that fell within such intensity span. Each bin was then weighted for the intensity of stain and the percentage of cells expressing the stain multiplied against the weight for each bin. Finally, a weighted average is obtained from each patient that more truthfully reflects the total protein concentration per tumor. Four values were obtained, epithelial nuclear and cytoplasmic, and stromal nuclear and cytoplasmic.
For survival analysis, the endpoint was the biochemical recurrence of the cancer, defined as serum PSA level higher than 0.4 ng/mL or clinical evidence of progression. Time to recurrence was defined as the interval between the date of surgery and the date of identification of biochemical recurrence. The predictive value of S4F for recurrence-free survival was evaluated using the Kaplan-Meier actuarial analysis and the log-rank test. Kaplan-Meier survival curves were constructed for patients with low and high levels of S4F expression. Actual cutoff value of S4F expression for division into high expression and low expression groups was selected using minimum P-value method”. The Cox univariate and multivariate proportional hazard models were used to determine the hazard ratios. Cox multivariate analysis was applied to assess the prognostic value of S4F in presence of known predictors such as clinical stage, preoperative PSA (Pre-PSA), extracapsular extension (ECE), seminal vesicle invasion (SVI), margins, and Gleason grade. The hazard ratio and its 95% confidence interval were recorded for each marker. S4F expression was also tested for correlations with some other biomarkers in our database. All analyses were performed with statistical software (SPSS 11.0, SPSS, Chicago, IL).
RESULTS
Semaphorin Effects on PCa Proliferation
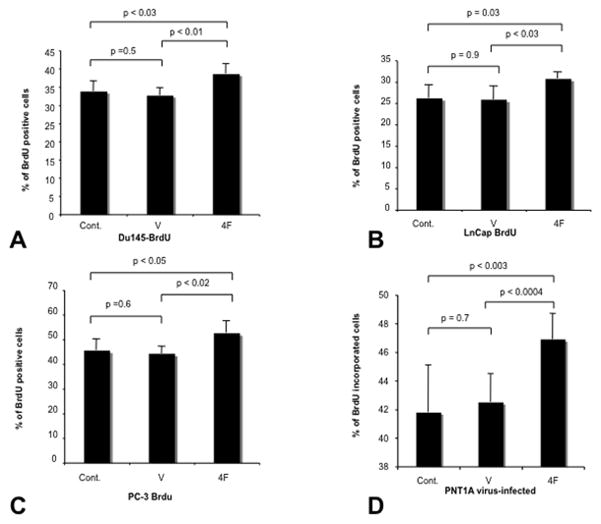
To study the autocrine effects of S4F on the PCa cells we over expressed S4F using a retrovirus into DU145, PC-3, LNCaP and PNT1A cells. Empty vectors and naïve cells were used as controls. Proliferation rates measured by BrdU incorporation were consistently higher in S4F transfected cells. The percentage of BrdU positive cells were increased with S4F over expression in all 4 cell lines examined compared to naïve and empty vector controls (DU145: 33.8%±2.8 and DU145 ev: 32.7%±2.1 vs. DU145 S4F: 38.6%±2.8; p<0.05) (Figure 1A); (LNCAP: 26.2%±3.1 and LNCAP ev: 25.9±3.1 vs. LNCAP S4F: 30.8±1.5; p<0.05) (Figure 1B); (PC3: 45.6%±4.5 and PC3 ev: 44.3±2.9 vs. PC3 S4F: 52.6±4.9; p<0.05) (Figure 1C); (PNT1A: 41.8%±3.3 and PNT1A ev: 42.5%±2 vs. PNT1A S4F: 46.9±1.8; p<0.05) (Figure 1D). PCa proliferates at a rate of approximately 5% in human tumors, as measured by Ki67, such that any increase might be clinically significant.
Figure 1. Proliferation is increased by S4F expression.
Proliferation rates as measured by BrdU incorporation were higher in S4F transfected cells consistently. The percentage of BrdU positive cells were increased with S4F over expression in all 4 cell lines examined compared to naïve and empty vector controls DU145 (Figure 1A); LNCAP (Figure 1B); PC3 (Figure 1C); PNT1A (Figure 1D).
Semaphorin Effects on PCa Migration and Invasion
Motility was tested using invasion assay, scratch assays and the PNI model. In all 3 assays S4F over-expression was associated with increased motility of the cancer cells. In the scratch assay S4F transfected cells completely covered the exposed area within 24 Hrs with PC3 cells and 48 hours with DU145 cells (Figure 2A) and PC3 cells (Figure 2B), while empty vector controls failed to do so at the same time points Invasion assays confirmed that cancer cell motility was also increased as the number of migrating cells increased in S4F+cells than controls (DU145: 77±2.1 and DU145 ev: 75.3±6 vs. DU145 S4F: 90.7±11.9; p<0.05) (Figure 3A); (LNCAP: 75±8.2 and LNCAP ev 77±6.3: vs. LNCAP S4F: 85.7±7.1; p<0.05) (Figure 3B); (PC3: 77.7±7.3 and PC3 ev 78.8±8.7: vs. PC3 S4F: 93.2±6.8; p<0.05) (Figure 3C); (PNT1A: 71.3±8.1 and PNT1A ev: 74.7±7.3 vs. PNT1A S4F: 88±9.3; p<0.05) (Figure 3D).
Figure 2. Motility is increased by S4F expression.
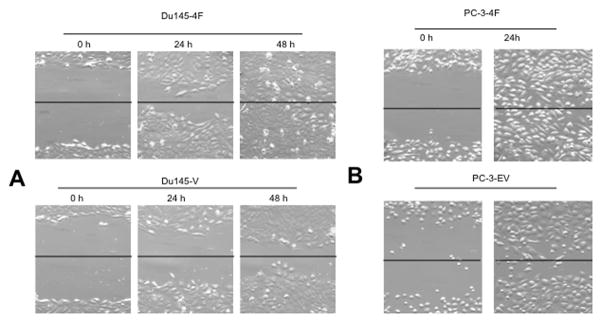
In the scratch assay S4F transfected cells completely covered the exposed area within 24 Hrs with PC3 cells and 48 hours with DU145 cells, while empty vector controls failed to do so at the same time points (Figure 2A). The same was observed with PC3 cells (Figure 2B).
Figure 3. Invasion is increased by S4F expression.
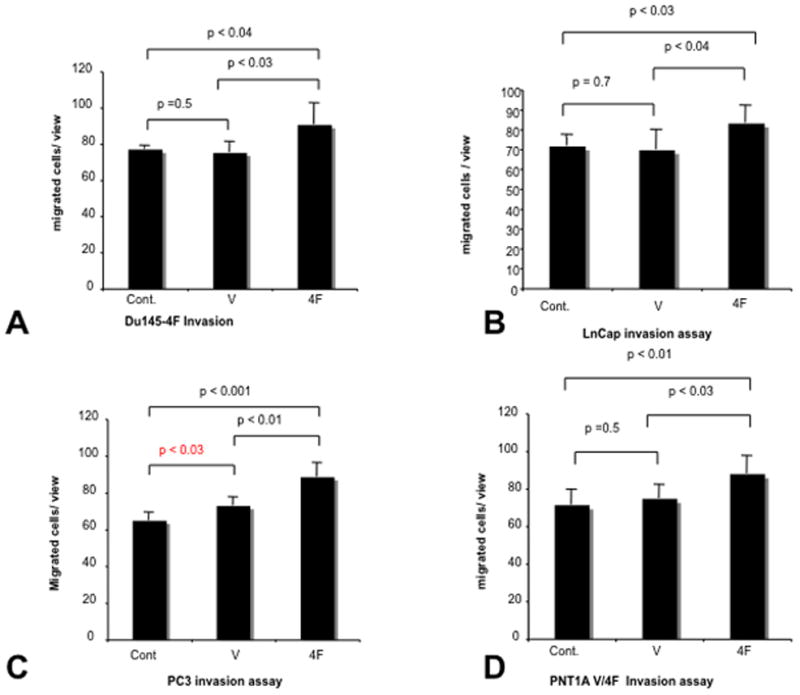
Invasion assays confirmed that cancer cell motility was also increased as the number of migrating cells increased in S4F+cells than controls DU145 (Figure 3A); LNCAP (Figure 3B); PC3 (Figure 3C); PNT1A (Figure 3D).
The effects are consistent in all cell lines studied. This increased in migration is corroborated by human PCa data (see below). Increased S4F expression in PCa, either cytoplasmic of nuclear is associated with increased seminal vesicle invasion, a measure of expansion of PCa outside of the prostate, and an ominous predictive marker for aggressive disease (S4F PCa cyto rho=0.184, p=0.004; S4F PCa nucl, rho= 0.162, p=0.011).
We also have an preliminary indication that S4F+ cells migrated directionally towards the dorsal root ganglia than empty vector controls, indicating that S4F might be involved in formation of PNI (data not presented). Therefore S4F effects in PCa might not only be related to increased axonogenesis, but also through an autocrine loop that increases growth, migration and PNI establishment.
Semaphorin 4F in Human PCa Progression
S4F was initially identified through gene array studies comparing cancer cells in the PNI in vitro model with and without DRG/nerves. Using visual semiquantitation in the progression human TMA, S4F expression was localized in the cytoplasm as well as nuclei of epithelial components of normal tissue (Figure 4A), HGPIN (Figure 4B), and PCa (Figure 4C). On analysis, S4F was expressed more frequently in cytoplasmic location in HGPIN and PCa than normal (4.17 and 4.27 vs. 0.12; p<0.05 (Figure 4D). Stromal S4F expression was also identified.
Figure 4. Immunohistochemistry of S4F in human PCa tissues.
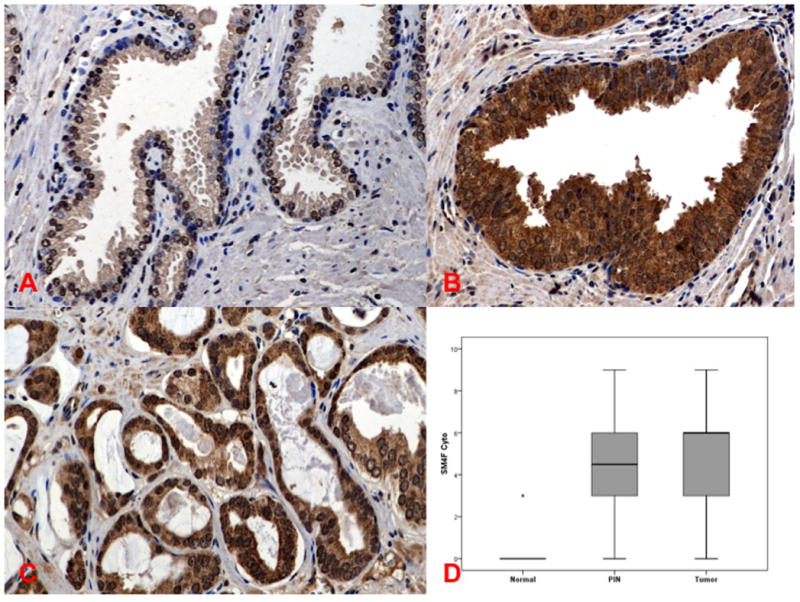
Semaphorin 4F in Human PCa Progression. S4F expression was localized in the cytoplasm as well as nuclei of epithelial components of normal tissue (Figure 4A), HGPIN (Figure 4B), and PCa (Figure 4C). S4F was expressed more frequently in cytoplasmic location in HGPIN and PCa than normal (Figure 4D). Stromal S4F expression can also be identified.
Identification of S4F in the PCa and Stromal Compartments
For this study we used the progression array. After extensive testing with the deconvolution imaging method we have established that hematoxylin and eosin gives the best discrimination of structure for image segmentation. Hence, instead of using hematoxylin as a background stain for the immunohistochemical stains, we use a combination of hematoxylin and eosin. The resulting image is difficult to interpret with the naked eye as can be seen in Figure 5A. The inForm™ software was then trained to recognize and distinguish stroma, and cancerous epithelium. Figure 5B shows the segmentation of the PCa from the stroma. Every nucleus within each compartment is then recognized and the biomarker quantified within each cell and sub cellular compartment (nuclear vs. cytoplasm). Figure 5C shows each individual nuclei in the PCa, circled in green, surrounded by their individual cytoplasm. Figure 5D shows the same for the stromal component. S4F expression levels for individual cell data per compartment was then quantified and used for subsequent correlation and survival studies.
Figure 5. Identification of S4F in the PCa and Stromal Compartments.
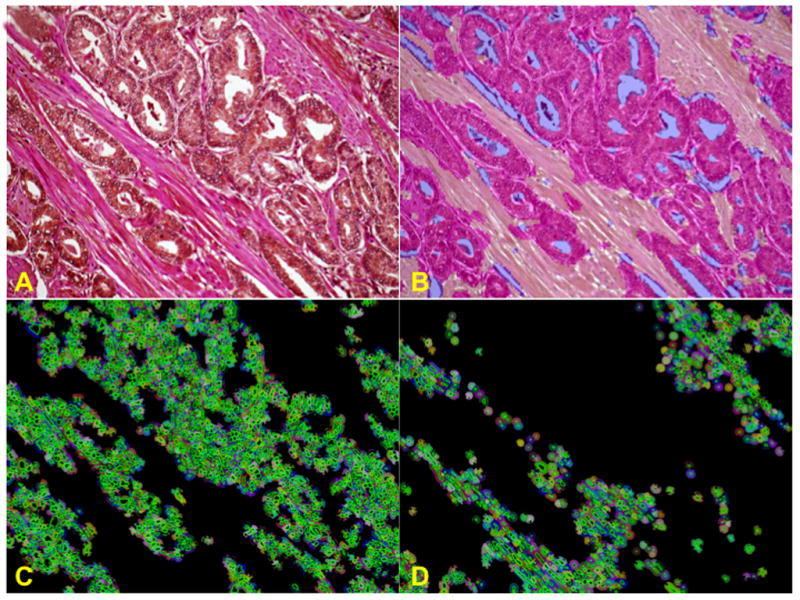
A cohort tissue microarray was stained, imaged and analyzed using image deconvolution (Nuance) and segmentation (InForm). This permits separation of compartments within the tumor (PCa vs. stroma). Individual cells are then recognized within the PCa and stroma and analyzed for nuclear vs. cytoplasmic stain. Figure 5A shows S4F immunohistochemistry with DAB and Hematoxylin & eosin as background stain. Image is difficult to interpret with the naked eye. Figure 5B shows the segmentation of the PCa from the stroma. Figure 5C shows each individual nuclei only in the PCa, circled in green, surrounded by their individual cytoplasm. Figure 5D shows the same exclusively for cells in the stromal component. S4F expression levels for individual cell data per compartment was then quantified and used for subsequent correlation and survival studies.
Heterogeneity of S4F Expression in Human PCA
The progression tissue microarray cohort was stained, imaged and analyzed using image deconvolution (Nuance) and segmentation (InForm). This permits separation of compartments within the tumor (PCa vs. stroma). Individual cells are then recognized within the PCa and stroma and analyzed for nuclear vs. cytoplasmic stain. Data are provided on a cell per cell basis. S4F was expressed more frequently in PCa than stromal cells (median values; PCa cyto 2.68 vs. stroma cyto 0.96; PCa nucl 2.67 vs. stroma nucl 1.00).
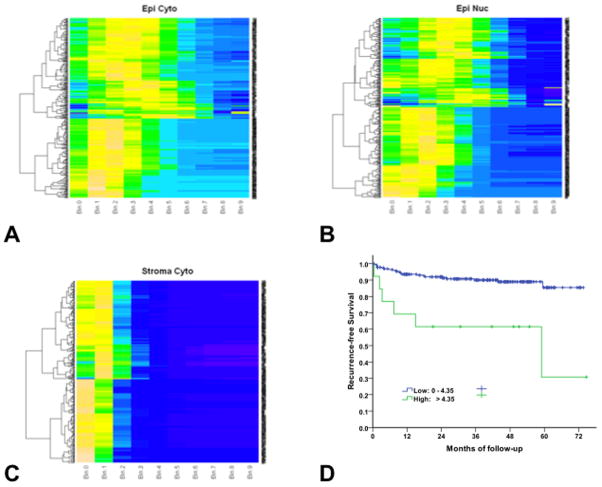
To better visualize and understand the heterogeneity of expression of this biomarker within patients we used an unsupervised clustering algorithm. Fig. 6 shows the unsupervised clustering of S4F PCa cytoplasmic expression (Figure 6A), nuclear expression (Figure 6B) and stromal cytoplasmic expression (Figure 6C).
Figure 6.
Figures 6 A, B and C, show unsupervised clustering of the expression of S4F in the nucleus and stroma of the cancer and stroma. Each patient corresponds to a row, and the expression bins are columns. Color-coding shows the percentage of cells present in each expression bin, with blue representing low % of expressing cells in the bin, and orange high % of expressing cells. S4F PCa cytoplasmic expression (Figure 6A), PCa nuclear expression (Figure 6B) and stromal cytoplasmic expression (Figure 6C). Figure 6D shows a Kaplan-Meier survival analysis of PCa patients with high or low S4F expression. The recurrence-free survival was significant higher in patients with low level of S4F (S4F <4.35; 227 cases) than that in those with high level of S4F (S4F ≥4.35; 13 cases) (p=0.0002).
Each patient corresponds to a row, and the expression bins are columns. Color-coding shows the percentage of cells present in each expression bin, with blue representing low % of expressing cells in the bin, and orange high % of expressing cells. Results suggest that cell clusters with high S4F expressing in the cytoplasm is a major discriminator (Figure 6A), corroborated by the survival data to follow. In contrast, the major clustering factor for nuclear expression of S4F in PCa cells is the presence or absence of expression (Figure 6B). This is an entirely new way of understanding biomarker expression in human cancer and will permit exploration of the value of heterogeneity in expression. New tools are needed to fully understand the biological and predictive information obtained within this type of data.
Correlations
The expression levels in the cytoplasm and nucleus of PCa cells were not correlated to each other. The same is true for the nuclear and cytoplasmic expression in the stromal cells. However, a correlation was identified between the cytoplasmic expression in PCa and nuclear expression in stromal cells (rho=0.644; p<0.001).
S4F cytoplasmic expression in PCa cells also correlates with nerve density in PCa (rho=0.135; p=0.04) and perineural invasion (PNI) diameter (rho=0.237; p=0.004). The higher the levels of expression within the cytoplasm of the PCa cells, the higher the nerve density and the diameter of perineural invasion. This data corroborates involvement of this molecule in PCa induced axonogenesis and perineural invasion. While we did not identify a correlation between S4F expression and proliferation indices in PCa as measured by Ki67, inverse correlations between S4F expression and apoptosis as measured by TUNEL, were identified, particularly with cells in the perineural space (S4F PCa cytoplasmic rho=−0.362, p=0.027; S4F PCa nucleus rho=−0.334, p=0.043).
S4F expression results were compared with previous biomarker data available at BCM in the same cohort of patients. Significant correlations were identified with nuclear expression of NFkB (S4F PCa cytoplasmic rho=0.225, p=0.009; S4F PCa nucleus rho=0.263, p=0.002; S4F stroma nucleus rho=0.220, p=0.01). This data confirms that S4F is involved in PCa induced axonogenesis in human tissues, as well as in the development of PNI invasion and its subsequent survival advantage for the PCa cells in PNI. The correlations between S4F expression and NFkB are consistent with PNI mechanisms identified in prior publications (3).
Semaphorin 4F as a Biomarker of Aggressive PCa
To test the predictive potential of S4F we obtained a weighted average from each patient. This data point more accurately reflects the total protein concentration per tumor. “S4F expression in the cytoplasm of PCa cells is marginally significant for biochemical recurrence as a continuous variable when using the 95 % confidence interval (HR 1.5; p=0.056).” After extensive search for an optimal cutoff by using minimum P value method, a value of 4.35 was found to be the best for categorizing patients as high expression group (S4F ≥4.35; 13 cases) or low expression group (S4F <4.35; 227 cases). The difference in recurrence-free survival between patients with low level of S4F and those with high level of S4F is clearly illustrated with Kaplan-Meier survival curves (Figure 6D). Univariate analysis showed that patients with high level of S4F had 5-fold increase in estimated risk of biochemical recurrence within follow-up time compared with those with low level of S4F [HR (hazard ratio)=5.43, 95%CI (95% confidence interval) (2.19–13.40); P = 0.0002; (Table 1)]. More importantly, S4F was an independent indicator for biochemical recurrence; that is, when clinical and pathologic conditions were assumed identical, patients with high level of S4F expression had more than seven times of risk for biochemical recurrence within follow-up time compared with those with low level of S4F [HR=7.55, 95%CI (2.35–24.31); P=0.0007; (Table 1)].
Table 1.
Univariate analysis showed that patients with high level of S4F had <5-fold increase of risk for biochemical recurrence compared with those with low level of S4F [HR=5.426 (hazard ratio, 5.426 (95% confidence interval, 2.19–13.40); P = 0.0002]. S4F was an independent indicator for biochemical recurrence. [hazard ratio, 7.551 (95% confidence interval, 2.345–24.312); P = 0.000].
| Univariate | HR | 95.0% CI for HR | p-value | |
|---|---|---|---|---|
| Lower | Upper | |||
| S4F Epi Cyto (groups) | 5.43 | 2.20 | 13.40 | .0002 |
| Multivariate | HR | 95.0% CI for HR
|
p-value | |
|---|---|---|---|---|
| Lower | Upper | |||
|
| ||||
| Ln (PSA) | 2.03 | 1.14 | 3.60 | .0160 |
| LN | 21.59 | 6.84 | 68.17 | <0.0001 |
| ECE | 1.67 | .53 | 5.25 | .3811 |
| SVI | 1.25 | .47 | 3.38 | .6534 |
| MARGINS | 5.22 | 1.91 | 14.29 | .0013 |
| Gleason Grade | 4.70 | 1.88 | 11.72 | .0009 |
| S4F Epi Cyto (groups) | 7.55 | 2.35 | 24.31 | .0007 |
In this model, Gleason, Pre-PSA, lymph node status, and margins were also independently significant. However, ECE and SVI were dropped out of the model when assessed against S4F. Nuclear PCa expression and stromal expression of S4F were not significant predictors.
This data, in aggregate are confirmatory of the role of S4F in human disease, as a critical regulator of cancer nerve interactions and as a contributing factor in aggressive phenotypes.
CONCLUSION
S4F has been proposed as a molecule involved in PCa induced axonogenesis. New data presented here corroborates involvement in axonogenesis in human PCa. In this article we demonstrate novel biological functions for S4F. We identified that S4F over expression induces PCa proliferation, migration and is possibly involved in the establishment of PNI. Hence, S4F is a critical regulator of the interaction between cancer and nerves and the progression to aggressive PCa. Using state of the art quantitative methodology we corroborate the in vitro data in human PCa and demonstrate that S4F is a potential biomarker for PCa aggressive disease, measured by biochemical recurrence.
The nature of the interaction between tumor and host is a determinant of aggressive human PCa. It is now accepted that PCa requires the stimulatory effects of the host response to acquire an aggressive phenotype. The host response is a multicellular reaction. The most frequently component is reactive stroma, the myofibroblastic response to tumors. Angiogenesis and immune response are also accepted components of the host response. The function of nerves in the host response mechanisms been recently brought to the forefront. Our group has described a novel phenomenon, cancer induced axonogenesis and neurogenesis. This phenomenon is regulated, at least in part by a member of the semaphorin family, S4F.
Semaphorins are highly conserved molecules; classes III to VII are found in all mammals and are characterized by the presence of an acid N-terminal Sema domain (8). Classes IV–VII are transmembrane or membrane-anchored. The known function is embryological axon guidance as collapsing factors and mediators of axon repulsion (9). Semaphorins also have roles in cardiovascular development and the immune response (10). M-sema H, correlates with the metastatic ability of mouse tumor cell lines (11) and semaphorin E as a non-MDR drug resistance gene of human cancers (12). Semaphorin 4F has been shown to be involved in branch tubulogenesis in the developing breast (13).
Due to the effects of other semaphorins on cancer cells we propose that the complex interactions between cancer cells and nerves results in elevated S4F expression by the cancer cells and that S4F over expression by the cancer results not only in axonogenesis, but also in elevated cancer cell proliferation, elevated cancer cell migration and elevated PNI owing to increased axon density and altered cancer cell biology.
There are several potential sources of S4F. Of note is that S4F has not been detected in neurons or nerves. S4F was originally identified in gene array studies of the PNI in vitro model in cancer cells interacting with neurons than controls without neuronal input. The expression of S4F by PCa cells was validated in human tumors. S4F was also identified in gene array studies of human PCa reactive stroma versus normal host stroma. This expression was also detected in human samples. Hence S4F could act in an autocrine fashion, derived from the PCa cells and on the PCa cells; and/or in a paracrine fashion, from the reactive stromal cells, on the PCa cells. The data presented in this study suggests that the S4F derived from the PCa cells has greater biologic significance, as measured by the correlations with nerve density, PNI, with seminal vesicle invasion and inversely with apoptosis, and the survival studies.
We have also identified that S4F has effects on the proliferation and migration of non neoplastic PCa cells (PNT1A). Although we cannot corroborate this data in human tissues at this point, it suggests that S4F can be involved in the homeostasis of non neoplastic prostate, either directly on the epithelial cells, or through the maintenance of the supporting neural network. Furthermore, S4F as well as other semaphorins have been involved in ductal morphogenesis of the breast. It is possible that S4F has similar functions in prostate organogenesis. Future studies will address this issue.
Another significant aspect of this article is the use of novel and state of the art technology to test the predictive potential of biomarkers. True quantitation of protein expression has remained an elusive goal. The combination of image deconvolution and image segmentation gives us the ability to study the expression of S4F, and other proteins, selectively within different compartments of the tumor, in essence dissecting the effects on the cancer cells and the host response. Our combination of novel technology and the resources available permit this discrimination. The use of extensive tissue resources with significant follow, and a database with many other quantified biological correlates, permits a better understanding of the biologic and clinical significance of S4F. In this case, we have identified that S4F has an effect on both the cancer cells and the neural microenvironment, serving as a key regulator of the interaction between cancer and nerves. We were also able to dissect how S4F compartmentalization in the nucleus or the cytoplasm of PCa or stromal cells can affect the development of an aggressive phenotype of PCa. Another significant advantage is the ability to study the effects of heterogeneity of expression and the significance of clusters of high expressing cells. Furthermore the methodology used to reach these conclusions is not only state of the art, but will form the basis for reproducibility of S4F as a biomarker in validation studies.
We conclude through the use of human tissue derived data that S4F is over expressed in preneoplastic and neoplastic cells than normal epithelial cell and that S4F is associated with aggressive PCa. S4F is associated with increased nerve density in PCa and with PNI diameter, the ultimate measurement of interaction between cancer cells and nerves. Moreover, S4F expression in the cytoplasm of PCa cells is independently predictive of biochemical recurrence.
Accordingly, targeting the mechanisms related to the S4F-regulated axonogenesis and/or neurogenesis and its effects on the PCa cells themselves might inhibit the formation and progression of PCa. The natural progression of nerve-cancer interaction-based therapies now requires targeting molecules that regulate this interaction. We believe that S4F is the most significant molecule in the interaction between cancer and nerves. Understanding the biological mechanisms of S4F action is essential to determining whether these molecules and affected pathways might become key targets for novel therapeutic approaches. Future studies of molecules needed for S4F effect will provide understanding of novel biology and provide potential therapeutic targets that affect not only the tumor but also its microenvironment. In addition, we believe that S4F effects on cancer and environment are not only found in PCa and can potentially be a generalizable mechanism for a number of other cancers.
STATEMENT OF TRANSLATIONAL RELEVANCE.
Interaction between nerves and cancer are key to cancer progression. Our studies have shown that perineural invasion delivers pro growth conditions resulting in cancer cell decreased apoptosis and increased proliferation. Prostate cancer also induces axonogenesis/neurogenesis, and this process is regulated in part by semaphorin 4F. In this article we demonstrate that semaphorin 4F is the key critical regulator for neuro-epithelial interactions, from cancer induced neurogenesis to perineural invasion. Semaphorin 4F induces cancer growth and migration. Our data also indicates that semaphorin 4F regulatory biology also is present in human samples and predictive of progression. As such, targeting the critical regulator of the interaction that defines prostate cancer aggressiveness is key to detaining prostate cancer progression.
Acknowledgments
This work was supported by National Institutes of Health grant TMEN U54CA126568-01, and a Prostate Cancer Foundation Creativity Award.
Footnotes
Conflicts of Interest
The Authors have no conflicts to report that they believe could be construed as resulting in an actual, potential, or perceived conflict of interest with regard to the manuscript submitted for review.
BIBLIOGRAPHY
- 1.Ayala GE, Wheeler TM, Shine HD, Schmelz M, Frolov A, Chakraborty S, et al. In vitro dorsal root ganglia and human prostate cell line interaction: redefining perineural invasion in prostate cancer. Prostate. 2001 Nov 1;49(3):213–23. doi: 10.1002/pros.1137. [DOI] [PubMed] [Google Scholar]
- 2.Ayala GE, Dai H, Tahir SA, Li R, Timme T, Ittmann M, et al. Stromal antiapoptotic paracrine loop in perineural invasion of prostatic carcinoma. Cancer Res. 2006 May 15;66(10):5159–64. doi: 10.1158/0008-5472.CAN-05-1847. [DOI] [PubMed] [Google Scholar]
- 3.Ayala GE, Dai H, Ittmann M, Li R, Powell M, Frolov A, et al. Growth and survival mechanisms associated with perineural invasion in prostate cancer. Cancer Res. 2004 Sep 1;64(17):6082–90. doi: 10.1158/0008-5472.CAN-04-0838. [DOI] [PubMed] [Google Scholar]
- 4.Ayala GE, Dai H, Powell M, Li R, Ding Y, Wheeler TM, et al. Cancer-related axonogenesis and neurogenesis in prostate cancer. Clin Cancer Res. 2008 Dec 1;14(23):7593–603. doi: 10.1158/1078-0432.CCR-08-1164. [DOI] [PubMed] [Google Scholar]
- 5.Ayala G, Thompson T, Yang G, Frolov A, Li R, Scardino P, et al. High levels of phosphorylated form of Akt-1 in prostate cancer and non-neoplastic prostate tissues are strong predictors of biochemical recurrence. Clin Cancer Res. 2004 Oct 1;10(19):6572–8. doi: 10.1158/1078-0432.CCR-04-0477. Epub 2004/10/12.eng. [DOI] [PubMed] [Google Scholar]
- 6.Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003 Oct 15;9(13):4792–801. [PubMed] [Google Scholar]
- 7.Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003 Oct 1;63(19):6244–51. [PubMed] [Google Scholar]
- 8.Tamagnone L, Comoglio PM. To move or not to move? Semaphorin signalling in cell migration. EMBO Rep. 2004 Apr;5(4):356–61. doi: 10.1038/sj.embor.7400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Wit J, Verhaagen J. Role of semaphorins in the adult nervous system. Prog Neurobiol. 2003 Oct;71(2–3):249–67. doi: 10.1016/j.pneurobio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Kumanogoh A. Involvement of immune semaphorins in the immune system. Tanpakushitsu Kakusan Koso. 2002 Dec;47(16 Suppl):2254–60. [PubMed] [Google Scholar]
- 11.Christensen CR, Klingelhofer J, Tarabykina S, Hulgaard EF, Kramerov D, Lukanidin E. Transcription of a novel mouse semaphorin gene, M-semaH, correlates with the metastatic ability of mouse tumor cell lines. Cancer Res. 1998 Mar 15;58(6):1238–44. [PubMed] [Google Scholar]
- 12.Yamada T, Endo R, Gotoh M, Hirohashi S. Identification of semaphorin E as a non-MDR drug resistance gene of human cancers. Proc Natl Acad Sci U S A. 1997 Dec 23;94(26):14713–8. doi: 10.1073/pnas.94.26.14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris JS, Stein T, Pringle MA, Davies CR, Weber-Hall S, Ferrier RK, et al. Involvement of axonal guidance proteins and their signaling partners in the developing mouse mammary gland. J Cell Physiol. 2006 Jan;206(1):16–24. doi: 10.1002/jcp.20427. [DOI] [PubMed] [Google Scholar]