Abstract
Ubiquitination is countered by a group of enzymes collectively called deubiquitinases (DUBs) - about 100 of them can be found in the human genome. One of the most interesting aspects of these enzymes is the ability of some members to selectively recognize specific linkage types between ubiquitin in polyubiquitin chains and their endo and exo specificity. The structural basis of exo-specific deubiquitination catalyzed by a DUB is poorly understood. UCH37, a cysteine DUB conserved from fungi to humans, is a proteasome-associated factor that regulates the proteasome by sequentially cleaving polyubiquitin chains from their distal ends, i.e., by exo-specific deubiquitination. In addition to the catalytic domain, the DUB features a functionally uncharacterized UCH37-like domain (ULD), presumed to keep the enzyme in an inhibited state in its proteasome-free form. Herein we report the crystal structure of two constructs of UCH37 from Trichinella spiralis in complex with a ubiquitin-based suicide inhibitor, ubiquitin vinyl methyl ester (UbVME). These structures show that the ULD makes direct contact with ubiquitin stabilizing a highly unusual intra-molecular salt bridge between Lys48 and Glu51 of ubiquitin, an interaction that would be favored only with the distal ubiquitin but not with the internal ones in a Lys48-linked polyubiquitin chain. An inspection of 39 DUB-ubiquitin structures in the protein data bank reveals the uniqueness of the salt bridge in ubiquitin bound to UCH37, an interaction that disappears when the ULD is deleted, as revealed in the structure of the catalytic domain alone bound to UbVME. The structural data are consistent with previously reported mutational data on the mammalian enzyme, which, together with the fact that the ULD residues that bind to ubiquitin are conserved, points to a similar mechanism behind the exo specificity of the human enzyme. To the best of our knowledge, these data provide the only structural example so far of how the exo specificity of a DUB can be determined by its non-catalytic domain. Thus, our data show that, contrary to its proposed inhibitory role, the ULD actually contributes to substrate recognition and could be a major determinant of proteasome-associated function of UCH37. Moreover, our structures show that the unproductively oriented catalytic cysteine in the free enzyme is aligned correctly when ubiquitin binds, suggesting a mechanism for ubiquitin selectivity.
Introduction
The ubiquitin proteasome system (UPS), present in all eukaryotes, is responsible for the majority of controlled degradation and recycling of proteins within the cell (1-5). Poly-ubiquitinated, and to some extent mono-ubiquitinated, proteins are recognized and degraded by the 26S proteasome, a 2.5 MDa self-compartmentalizing proteolytic complex (6-13). It is composed of two major units: the 20S core particle (CP) consisting of 28 subunits, and the 19S regulatory particle (RP) containing 19 subunits in yeast. The proteolytic active sites are housed within the luminal chamber of the barrel-shaped CP, capped on both ends by the RP, which contains ubiquitin receptors and enzymes that prepare substrates for degradation. Entry of substrates into the CP is regulated by the RP, primarily by opening and closing of the substrate translocation channel. Before the substrate is translocated into the narrow channel leading to the lumen of the CP, it is obligatorily deubiquitinated with the help of the RP-resident JAMM metalloprotease Rpn11 (14-16), and unfolded by Rpt subunits that sit within the base subcomplex of the RP (7, 9, 14). However, additional regulation is performed by proteasome-associated deubiquitinating enzymes, whose underlying mechanism is still poorly understood(7, 17).
Attachment of ubiquitin to a lysine residue(s) on target proteins is catalyzed by the sequential action of three enzymatic systems: E1 (ubiquitin-activating), E2 (ubiquitin-conjugating) and E3 (ubiquitin ligating) enzymes (18, 19). Usually, ubiquitination of a target protein results in the attachment of a polyubiquitin chain in which successive ubiquitin moieties are attached to one of the seven lysines, or the N-terminal amino group of the preceding monomer, to generate a homopolymeric structure (18, 20). Polyubiquitin chains of distinct topology are thus generated depending on which amino group of ubiquitin is used for chain extension (lysines 6, 11, 27, 29, 33, 48, 63 or the amino group of Met 1). A polyubiquitin chain of a specific topology is meant for a specific type of functional outcome (20-25). For example, a Lys48 (K48)-linked chain usually serves as the signal for proteasomal degradation whereas K63 chains signal other types of functions such as endocytosis, DNA repair, and NF-κB signaling(24, 26).
Ubiquitination works as a reversible post-translational modification, like phosphorylation. Deubiquitinating enzymes, or DUBs, can hydrolytically remove ubiquitin from protein adducts, thereby opposing the action of ubiquitin conjugating machinery (27-33). Consequently, DUBs have been found to play important regulatory roles in numerous ubiquitin-dependent cellular processes(32-35). Mechanistically speaking, these enzymes can be categorized into two main groups: cysteine proteases and zinc metalloproteases. The zinc metalloproteases consist of only one family, the JAB1/MPN/MOV34 metalloenzymes (JAMMs). The cysteine proteases are further broken down into four families based on the structure of their catalytic domain: ubiquitin carboxyl-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), ovarian tumor proteases (OTUs), and Machado-Josephin domain proteases (MJDs) (32).
UCH37 (also known as UCHL5) is a 37-kDa DUB of the UCH family and is one of the two proteasome-associated DUBs, the other one being USP14 (Ubp6 in yeast), known to regulate protein degradation by the mammalian proteasome (36-40). These associated DUBs, along with Rpn11, a constitutive member of the RP, carry out deubiquitination at the proteasome. However, the activities of the three enzymes are distinct. Rpn11 is responsible for en-block removal of polyubiquitin chains prior to (or concurrently with) unfolding and translocation of the substrate into the CP, an activity that appears to be coupled to substrate degradation (15-17). USP14 and UCH37 on the other hand are known to have chain-trimming functions (17, 37, 41). The importance of these associated DUBs to proteasome function was revealed through pharmacological inhibition of these enzymes. A small-molecule inhibitor of USP14 appears to accelerate proteasomal degradation of certain substrates, whereas UCH37 inhibition can stall proteolysis, consistent with distinct functional roles played by the two enzymes (42-44).
UCH37 was first identified as the PA700 isopeptidase, the cysteine DUB tightly associated with the RP, also known as PA700 (38, 45, 46). Like other UCH family members, it contains a conserved catalytic triad of a cysteine, a histidine, and an aspartate. UCH37 has a canonical UCH domain with 45% similarity to UCHL1 and 49% similarity to UCHL3, its single-domain family members (47-50). It also has an additional C-terminal tail domain responsible for its interaction with the Rpn13 subunit of the RP (51-54). Proteasome-bound UCH37 is thought to behave as an “editor”, relieving poorly ubiquitinated substrates from degradation by sequentially dismantling their K48-linked polyubiquitin chains from the very distal end, removing one ubiquitin at a time (37, 38, 45). Such a type of chain-disassembling activity can be termed as an exo cleavage activity in contrast to the endo activity, which leads to dismantling of chains by cleavage between internal ubiquitins. Although it has respectable UbAMC (ubiquitin aminomethylcoumarin) hydrolysis activity independently, UCH37 has been shown to require association with the proteasome in order to cleave diubiquitin (and polyubiquitin) chains (37). Additionally, its UbAMC hydrolysis activity is enhanced upon binding with Rpn13 (37, 54). Interestingly, UCH37 also associates in the nucleus with the human Ino80 chromatin-remodeling complex, where it is held in an inactive state compared to the free enzyme (55). It thus serves as an example of a DUB whose catalytic activity is both positively and negatively regulated by binding to specific protein partners, making it an attractive target for structural studies. Crystal structures have been solved for both the catalytic domain and full-length human UCH37 (56-58); however, the mechanism of its catalytic regulation upon binding to associated protein factors is not known. Any mechanistic understanding of its regulation must require structural information of UCH37 and its catalytic domain bound to ubiquitin, which has yet to be reported.
TsUCH37 is a recently-characterized lower organism homolog of UCH37 from Trichinella spiralis (Ts), an infectious helminth found nearly worldwide. TsUCH37 was identified by White et. al. by incubation of whole-cell lysate of Ts larvae with the HA-UbVME probe (HA, the hemagglutinin epitope, fused with the N-terminus of ubiquitin vinyl methyl ester), an epitope-tagged irreversible inhibitor of cysteine DUBs (59). Its structural and functional homology with human UCH37 was then confirmed by sequence analysis, co-immunoprecipitation with proteasomal subunits, and UbAMC hydrolysis assays. TsUCH37 is 45% identical to its human homolog and was shown to pull down TsADRM1, the corresponding Rpn13 homolog, by co-immunoprecipitation(59). The sequence and functional conservation between the Ts and human enzymes implies a similar chain-editing role of the former at the proteasome. In order to understand the mechanisms associated with UCH37, we have crystallized two constructs of TsUCH37 bound to ubiquitin vinyl methyl ester. The structures illuminate the mode of ubiquitin recognition in the enzyme by revealing binding interactions with the catalytic domain, which are conserved among UCH enzymes, and interactions unique to UCH37, notably ubiquitin binding by the ULD, providing further explanation of the proteasome-associated exo-specific deubiquitination activity of the DUB.
Materials and Methods
Cloning, Expression, and Purification
TsUCH37cat
TsUCH37cat (residues 1-226) was subcloned from the full-length construct (residues 1-309) in pET28a(+) into pGEX-6P-1 (GE Biosciences) by using BamHI and XhoI restriction sites. The protein was expressed in E. coli Rosetta cells (Novagen) grown at 37°C in LB media containing 100 μg/L ampicillin to an OD600 of 1.0, and then induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 18°C for 16 hrs. Harvested cells were resuspended in lysis buffer (1x phosphate buffered saline, 400 mM KCl) and lysed with French press. The lysate was then purified on a glutathione S-transferase (GST) column (GE Biosciences) followed by cleavage of the GST tag by PreScission Protease (GE Biosciences) per the manufacturer’s instructions. It was further purified by size exclusion chromatography on a Superdex 75 column (GE Biosciences). Intein-fused ubiquitin1-75 in pTXB1 was expressed in E. coli Rosetta cells and purified on chitin beads (New England Biosciences). Ubiquitin vinyl methyl ester (UbVME) was synthesized by overnight incubation of Ub1-75-MESNa (MESNA: sodium mercaptosulfonate) with glycine vinyl methyl ester, and then purified on a MonoS cation exchange column (GE Biosciences). Glycine vinyl methyl ester was synthesized by a modified, previously-published procedure (60). TsUCH37cat was reacted with UbVME for four hours, followed by purification on a MonoQ anion exchange column (GE Biosciences) to separate any unreacted TsUCH37cat. Selenomethionine TsUCH37cat protein (SeMet TsUCH37cat) was grown in M9 minimal media supplemented with selenomethionine, reacted with UbVME, and purified as above.
TsUCH37ΔC46
TsUCH37FL was subcloned previously into pET28a(+) with an N-terminally fused His tag (Novagen). TsUCH37FL was expressed in E. coli Rosetta cells, grown at 37°C in LB media containing 10 μg/L kanamycin to an OD600 of 0.8, and then induced with 0.5 mM IPTG at 18°C for 16 hrs. Harvested cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.6, 200 mM NaCl, 3 mM β-mercaptoethanol) and lysed by French press. His-tagged TsUCH37FL was purified by immobilized metal affinity chromatography (IMAC) and eluted with lysis buffer including 500 mM imidazole. Eluted proteins were further purified by size exclusion chromatography (SEC) on a Superdex 75 column (GE Biosciences) in 50 mM HEPES, pH 7.6, 3 mM dithiothreitol (DTT). SDS PAGE on the fractions indicated a cleavage of the full-length protein, so the construct described is actually a proteolytic cleavage product of the full-length protein. The crystal structure (described below) lacks density for the last 46 amino acids from the C-terminus; therefore, this construct will hereafter be described as TsUCH37ΔC46. Fractions containing the target protein were pooled, concentrated, and reacted with UbVME. UbVME was synthesized and reacted with purified TsUCH37ΔC46 as was done with TsUCH37cat. In order to separate unreacted TsUCH37ΔC46, the complex was further purified by SEC on a Superdex 75 column (GE Biosciences).
Crystallization and Structure Determination
TsUCH37cat-UbVME
TsUCH37cat-UbVME complex was concentrated to 3 mg/mL in 50mM Tris pH 7.6, 200mM NaCl, 1mM DTT. Crystals were grown in two days at room temperature by hanging drop vapor diffusion in 3 M ammonium sulfate, 0.1 M bicine pH 9.0 with 2 mM L-glutathione (mixture of oxidized and reduced) additive. Crystals were cryoprotected in 2.5 M sodium malonate and flash frozen in liquid nitrogen(61). Diffraction data was collected on a Mar300 CCD detector (Mar USA) at the 23-ID-B beamline at Argonne National Laboratory. Data up to 1.7 Å were collected on SeMet TsUCH37cat - UbVME crystals at the selenium peak (0.979 Å) for SAD (single-wavelength anomalous dispersion) phasing. Data were processed by HKL2000(62).
The initial model was obtained by Se-SAD phasing in the Phenix AutoSol wizard(63). Its sequence was built in using the Phenix AutoBuild wizard, as well as manual model building in Coot (63, 64). Structure refinement was done in Phenix using TLS refinement (with the entire asymmetric unit taken as one TLS group), as well as optimized weighting for stereochemical restraints(63). The data was run through Phenix Xtriage, which confirmed the chosen space group, C2, and did not detect evidence of crystal twinning(63). The completeness of the crystallographic data for TsUCH37cat-UbVME was less than ideal (see Table 2), however, this did not hinder structure solution or the generation of the structural model presented herein and can be accounted for by poor completeness in the highest resolution shells.
Table 2.
Crystallographic and Refinement Statistics
| SeMet TsUCH37cat –UbVME | TsUCH37ΔC46 –UbVME | |
|---|---|---|
| Data collection | ||
| Space group | C1 2 1 | R3 |
| Cell dimensions | ||
| a, b, c (Å) | 171.2, 55.8, 73.9 | 147.4, 147.4, 40.5 |
| α β γ (°) | 90, 113.4, 90 | 90, 90, 120 |
| Wavelength (Å) | 0.979 | 1.033 |
| Resolution (Å) | 50.00-1.70 | 50-2.0 |
| (1.73-1.70)a | (2.03-2.00) | |
| Rsym or Rmergeb (%) | 8.7 (50.0) | 8.5 (83.8) |
| I / σI | 15.9(3.0) | 4.9 (4.1) |
| Completeness (%) | 88.5 (42.0) | 100.0 (100.0) |
| Redundancy | 6.8 (3.5) | 5.8 (5.7) |
|
| ||
| Refinement | ||
| Resolution (Å) | 27.9-1.7 | 38.6-2.0 |
| No. unique reflections | 62326 / 3126 | 22270 / 2002 |
| Rworkc / Rfreed | 17.4 / 21.1 | 19.3 / 24.0 |
| No. atoms | ||
| Proteinee | 4650 | 2428 |
| Ligand | 24 | 8 |
| Water | 437 | 100 |
| Average B-factors (Å2) | ||
| Protein | 36.2 | 43.4 |
| Ligand | 36.5 | 32.4 |
| Water | 44.0 | 44.3 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.013 | 0.009 |
| Bond angles (°) | 1.48 | 1.07 |
| Ramachandran plot | ||
| Favored (%) | 98.1 | 97.7 |
| Allowed (%) | 1.6 | 1.0 |
| Outliers (%) | 0.4 | 1.3 |
Numbers in parentheses refer to data in the highest resolution shell.
Rmerge =Σ|Ih - <Ih>|/Σ Ih , where Ih is the observed intensity and <Ih> is the average intensity.
Rwork = Σ ||Fobs|-k|Fcal||/ Σ|Fobs|
Rfree is the same as Robs for a selected subset (5% and 9%, respectively) of the reflections that was not included in prior refinement calculations.
Ordered residues: SeMet TsUCH37cat-UbVME structure (Pro-3 to Gly141 and Lys153 to Asp224 in Chain C; Pro-3 to Gly141 and Gln152 to Gln225 in Chain A), and TsUCH37ΔC46-UbVME structure (Gly4 to Lys57, Thr72 to Gly141, and Glu157 to Ala263)
TsUCH37ΔC46-UbVME
TsUCH37ΔC46-UbVME complex was concentrated to 5 mg/mL in 50 mM HEPES, pH 7.6, 2 mM DTT. Crystals were grown in 60 days at room temperature in 0.2 M ammonium chloride, pH 5.8 and 18% PEG3350. Crystals were cryoprotected in ethylene glycol and flash frozen in liquid nitrogen. Diffraction data was collected on a Mar300 CCD detector (Mar USA) at the 23-ID-B beamline at Argonne National Laboratory. Data up to 2.0 Å were collected on TsUCH37ΔC46-UbVME crystals at 1.033 Å. Data were processed by HKL2000(62).
The initial model was obtained by molecular replacement using the Phenix AutoMR wizard, with a monomer of the TsUCH37cat –UbVME structure as the search model(63). Manual model building was done in Coot and structure refinement was done initially in Refmac using TLS refinement, then using simulated annealing and individual B-factor refinement in Phenix(63, 64). The data was run through Phenix Xtriage, which confirmed the chosen space group, R3, and did not detect evidence of crystal twinning(63).
UbAMC Hydrolysis Assay
TsUCH37cat was diluted in reaction buffer (50mM Tris pH 7.6, 0.5 mM EDTA, 0.1% bovine serum albumin, 5 mM DTT) to a final reaction concentration of 7 nM and pre-incubated at 30°C for five minutes prior to addition of the UbAMC substrate (Boston Biochem). UbAMC cleavage was measured on a Tecan fluorescence plate reader (Männedorf, Switzerland) with 380 nm excitation and 465 nm emission wavelengths at 30°C. Data was fit to Michaelis-Menten kinetics in SigmaPlot (Systat Software, San Jose, CA).
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted the Beckman-Coulter XLA analytical ultracentrifuge. The sample was extensively dialyzed against 50 mM Tris-HCl, 200 mM NaCl and 1 mM DTT pH 7.4. The TsUCH37cat and TsUCH37cat-UbVME concentration ranged from 10-32 μM. The samples were centrifuged at 50,000 rpm using a two-sector 1.2 cm path-length carbon-filled epon centerpiece. The experiments were run on an An-50 Ti rotor at 20°C. The density and relative viscosity of the buffers were calculated using SEDNTERP v. 1.09 (http://www.rasmb.bbri.org/rasmb/windows/sednterp-philo) 1.0079 g mL−1 and 0.01036 Poise. The partial specific volume (vbar) of the protein was also calculated from the protein sequence using SEDNTERP (0.7340 ml g−1 for TsUCH37cat and 0.7317 ml g−1 for TsUCH37cat-UbVME). The samples were monitored at 280 nm with a 4-min delay and 150 scans. The c(s) distributions were analyzed using SEDFIT v. 13.0b(65).
Molecular Dynamics Simulations
A model of full-length TsUCH37 was generated by the SwissModel homology modeling server using the structure of the full-length human protein as a template(66). Missing ULD residues produced by the model were appended onto the TsUCH37ΔC46-UbVME structure in Coot and a single round of refinement was done in Phenix, to produce a final model referred to hereafter as “the system”(63, 64). The system was solvated in a box of TIP3P water with the minimum distance between any solute atom and the boundary of the box set to 10 Å. The system was neutralized with 15 Na+ ions, which were automatically positioned by the tleap program. MD simulations were performed using Amber 10 with the Amber force field ff03(67). Periodic boundary conditions were applied and the full electrostatic energy was calculated using particle mesh Ewald (PME) method(68). The simulation consisted of three sequential steps: energy minimization for 5000 steps (2500 steps of steepest-descent followed by 2500 steps of conjugate gradient minimization), equilibration for 100 ps of solvent with the protein restraint with a force constant of 5 kcal/(mol·Å), and final MD simulation for 2 ns. All simulations were carried out at 300 K with constant volume. A time step of 2 fs was used and the SHAKE was applied to constrain the bonds involving hydrogen atoms(69).
Results
TsUCH37, like its mammalian counterpart, contains a catalytic UCH domain, and an additional polypeptide chain following it called the C-terminal tail comprising the conserved UCH37-like domain (ULD) followed by a putative KEKE motif (Fig. 1b) (37, 51, 70, 71). The ULD in human UCH37 is thought to have an inhibitory role, presumably by folding onto the catalytic domain thereby occluding ubiquitin binding (37). However, how ubiquitin binds to UCH37 has not been structurally characterized. To gain insight into how ubiquitin is recognized by TsUCH37, we aimed at crystallizing both the catalytic domain of TsUCH37 bound to ubiquitin vinyl methyl ester (UbVME), referred to here as TsUCH37cat-UbVME, as well as the UbVME complex of the full-length protein. UbVME is a suicide substrate of cysteine DUBs, which react with the former via nucleophilic attack of the catalytic cysteine at the vinyl group of the VME moiety, resulting in an irreversible modification whereby a covalent bond is formed between the catalytic cysteine and the VME portion of the inhibitor (Fig. 1a) (36, 48, 60). This covalent adduct is thought to mimic the acyl-enzyme intermediate formed during deubiquitination reactions catalyzed by the DUB (drawings II and IV in Fig. 1a). Taking diubiquitin as the substrate, the distal ubiquitin moiety is the acyl component of the acyl-enzyme intermediate, with the proximal ubiquitin acting as the leaving group during isopeptide bond hydrolysis (in diubiquitin, a lysine residue of one ubiquitin, called the proximal ubiquitin, is linked via isopeptide bond to the C-terminal carboxylate group of another ubiquitin, called the distal ubiquitin) (III in Fig. 1a).
Figure 1.
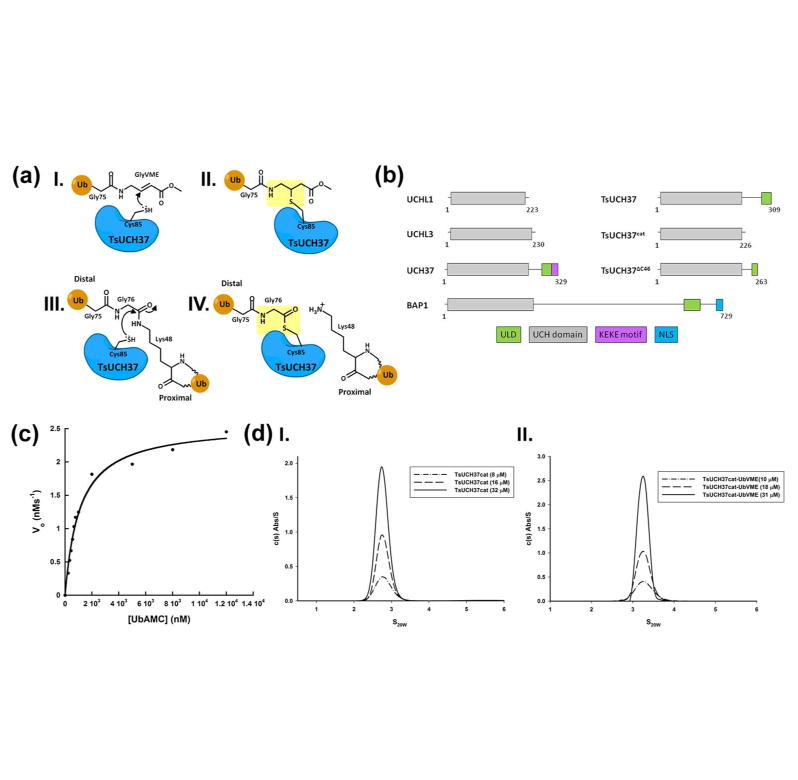
(a) Schematic structures representing inhibition of UCH37 by UbVME (I and II); Definition of proximal and distal ubiquitin in a diubiquitin substrate (III); Schematic structure of the acyl-enzyme intermediate formed during deubiquitination catalyzed by a cysteine DUB (IV). The UbVME adduct (II) mimics the acyl-enzyme intermediate (IV), as shown in yellow shading. (b) Domain diagrams of TsUCH37 constructs compared to other UCH family members with UCH domains boxed in grey and additional domains boxed and labeled as shown. (c) Kinetic assay of UbAMC hydrolysis by TsUCH37cat. (d) Analytical ultracentrifugation profiles of TsUCH37cat (left) and TsUCH37cat-UbVME complex (right), indicating that both are monomeric in solution.
TsUCH37cat-UbVME crystallized in the C2 space group with two molecules of the complex in the asymmetric unit. Our attempts to crystallize the full-length version, however, has met with limited success so far, the full-length protein being susceptible to proteolysis as indicated by at least two closely migrating bands in SDS-PAGE (data not shown). While attempting to purify the full-length construct, we managed to retrieve a truncated version of the protein lacking 46 amino acids from the C-terminal end of the protein (see Materials and Methods). This truncated protein was purified by Ni affinity chromatography, reacted with UbVME and the complex was purified using ion-exchange chromatography. This complex, hereafter referred to as TsUCH37ΔC46-UbVME (TsUCH37 missing the last 46 residues), crystallized in the R3 space group with one complex in the asymmetric unit.
The catalytic activity of TsUCH37cat was measured by UbAMC hydrolysis assay (Fig. 1c), which yielded Michaelis-Menten parameters as shown in Table 1. Compared to the catalytic domain of human UCH37, TsUCH37cat has approximately 20-fold less KM, indicating a higher affinity for this substrate than the human protein, but a 100-fold lower kcat, a substantially lower turnover number. Consequently, TsUCH37cat is nearly 5-fold less efficient than the UCH domain of human UCH37.
Table 1.
Kinetic Parameters for TsUCH37cat
| Enzyme | KM (nM) | kcat (s-1) | kcat/KM × 105 (M-1 s-1) | |
|---|---|---|---|---|
| TsUCH37cat | 1085 | 0.37 | 3.4 | |
| UCH37N240a | 21493 | 34 | 16 | |
| UCHL3a | 77.1 | 19 | 2414 | |
| UCHL1a | 47.0 | 0.03 | 7.4 | |
Kinetic parameters previously determined, from Boudreaux et. al. 2012(75).
Crystals of the TsUCH37cat-UbVME complex diffracted to 1.7 Å. The structure was solved by single-wavelength anomalous dispersion (SAD) using anomalous scattering from selenium (TsUCH37cat was labeled with selenium). Manual model building using Coot, followed by multiple rounds of refinement using Phenix, produced a final model with an R factor of 17.4% and Rfree of 21% (see Table 2 for crystallographic and refinement parameters)(63, 64). The final refined model corresponding to the asymmetric unit consists of two copies of the TsUCH37cat-UbVME complex, composed of TsUCH37cat, residues 1-226, covalently connected via a thioether bond linking the catalytic cysteine with the VME group of UbVME (residues 1-75 of ubiquitin attached to GlyVME as the 76th residue, which is modeled as methyl 4-amino butanoate). The refined model was of high stereochemical quality with less than 0.2% of residues in the disallowed region of the Ramachandran plot and scoring in the upper 98% according to Molprobity evaluation (72). The structure of TsUCH37ΔC46-UbVME (2.0 Å resolution) was solved by molecular replacement using the TsUCH37cat-UbVME structure as the search model (Table 2). The final refined model with good stereochemical quality (less than 0.2 % of residues in the disallowed region of Ramachandran plot and Molprobity score of 63%) has 5-263 amino acids of the protein and one UbVME linked via a thioether bond to the catalytic cysteine. The structures of the UCH domain in the two constructs are very similar, except for two loop regions (see below), with Cα root-mean-square deviations (rmsd) of 0.32 Å between the two (the loop regions were excluded from the calculation of rmsd). When discussing the structure of the UCH domain alone or its interaction with UbVME, we will therefore use the structure of the TsUCH37cat-UbVME complex since its resolution is higher, while specifically mentioning any structural feature that is different in the UCH domain of TsUCH37ΔC46-UbVME.
Initial analysis of the structure revealed that the two copies in the asymmetric unit of TsUCH37cat-UbVME crystals are linked by a disulfide bond between Cys71 of the two TsUCH37cat chains (Fig. 2a). It is possible that the disulfide bond forms because the protein exists as a dimer in solution bringing the cysteines into proximity, or, is a result of crystallographic packing. In order to determine if this disulfide is a crystallographic artifact or a biologically-relevant association, we determined the oligomerization state of complexed and uncomplexed (apo) TsUCH37cat by sedimentation velocity analytical ultracentrifugation (AUC). We found that both the complex and the apo protein exist as monomers in solution with sedimentation coefficients (S20,w) of 3.3 and 2.8, respectively (Fig. 1d), indicating that this disulfide is likely a result of crystal packing. TsUCH37 is expected to be predominantly localized to cytosol, a reducing environment, and therefore should not rely on disulfide-mediated dimerization for catalytic activity. Moreover, the observation that TsUCH37ΔC46-UbVME is a monomer in the asymmetric unit, and that the segment of residues 57-71, which is used as a part of the dimer interface in the crystals of the TsUCH37cat-UbVME complex, is disordered in the TsUCH37ΔC46-UbVME structure (Figs. 2 and 3) supports the notion that the dimer observed in the TsUCH37cat-UbVME structure is a crystallographic dimer and may not exist in solution. The two copies of the complex in the dimer observed in the crystals of TsUCH37cat-UbVME have very similar structure with rmsd of 0.39 Å between Cα atoms. We will therefore focus on one of them in discussions presented below.
Figure 2. Crystal structures of TsUCH37 constructs bound to UbVME.
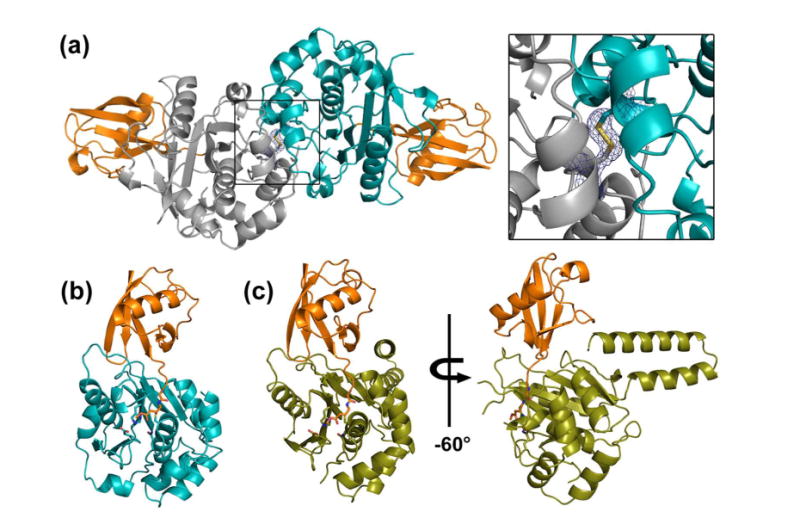
(a) Dimeric structure of TsUCH37cat bound to UbVME (orange) in crystals. Monomers are shown in teal (chain A) and grey (chain C). Inset shows the disulfide bridge that links the two subunits via Cys71. The electron density is rendered from the 2Fo-Fc map contoured at 1σ. (b) Monomer of TsUCH37cat -UbVME structure. (c) Structure of TsUCH37ΔC46-UbVME, with TsUCH37ΔC46 shown in olive and UbVME in orange.
Figure 3. Secondary structures of TsUCH37 constructs.
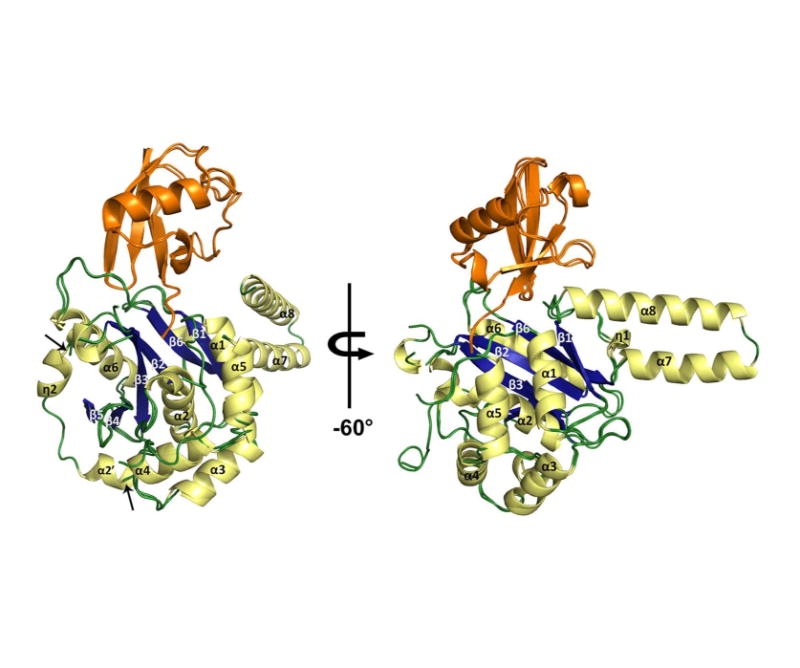
TsUCH37ΔC46-UbVME and TsUCH37cat-UbVME are superposed with α-helices and 310-helices shown in pale yellow, β-sheets in blue, loops in green, and UbVME in orange. Arrows indicate where the TsUCH37ΔC46-UbVME structure lacks density, compared to TsUCH37cat-UbVME, from residues 57-71.
Overall Structure of the UCH domain of TsUCH37
The overall structure of the TsUCH37 catalytic domain is similar to that of other structurally-characterized UCH enzymes (49, 50, 73). It has the classical alpha-beta-alpha fold, in which a central 6-stranded β-sheet is surrounded by six alpha helices, five on one side (α1, α2, α3, α4, and α5) and one on the other (α6) (Fig. 3). The overall architecture of TsUCH37cat can be seen as bilobal, with the one of the lobes comprising helices α1, α2, α3, α4, and α5, and the other comprising the β-sheets and α6. The active site is located at the interface of the two lobes, with Cys85 from α2 in one lobe and His161 from β3 in the other forming the catalytic Cys-His pair. An adjacent loop provides the third member of the triad, Asp176. Most of the secondary structural elements seen in TsUCH37cat are conserved in UCHL1 and UCHL3, with the only noticeable difference being the conformation of a segment following β2, residues 57-71. This segment is a helix in UCHL1 and UCHL3 and is in somewhat of an extended loop-like conformation in human UCH37 (hUCH37), but is fairly ordered; in the structure of the various constructs of human UCH37 solved so far, this loop has been found to be in a similar conformation regardless of crystallographic packing (Fig. S1)(56-58, 74). In contrast, this segment appears to be flexible in TsUCH37 and is visualized only in the TsUCH37cat-UbVME structure, in which it forms the dimer interface between the two subunits in the asymmetric unit. In TsUCH37ΔC46-UbVME, a crystallographic monomer, this loop is disordered (Fig. 3). Although the possibility that its binding can influence the loop dynamics cannot be ruled out, it is unlikely that UbVME has anything to do with the dynamic behavior of the loop since it does not bind to it. We therefore propose that the loop is intrinsically flexible in TsUCH37, but can become ordered under certain circumstances, such as under the constraints of crystallographic packing.
It is possible that the corresponding loop segment in hUCH37 is somewhat dynamic as well, but it appears to be significantly more flexible in TsUCH37. The significance of this difference in dynamics between the two proteins is not clear at the moment. Intriguingly, the loop’s dynamic behavior appears to have an effect on the conformation of a tryptophan residue (Trp55) adjacent to the active site (Fig. S2). This tryptophan is conserved among Schizosaccharomyces pombe (Sp), Ts and human UCH37 (Fig. S3). In TsUCH37ΔC46-UbVME, Trp55 makes contact with the OMe group of the suicide inhibitor, which in the actual substrate (a ubiquitinated protein or the diubiquitin motif of a polyubiquitin chain) would be replaced by the hydrocarbon portion of the isopeptide-linked lysine side chain (Fig. 1a). The same residue in TsUCH37cat-UbVME shows a different orientation with respect to the OMe group and appears to have adopted a more open position for interaction with the isopeptide unit (Fig. S2). Therefore, Trp55 may not only provide important contacts with the isopeptide link to hold it in place near the active site, it could also confer certain plasticity to the active site of UCH37, which may be useful for an induced-fit type of engagement with the substrate.
As stated before, in the TsUCH37ΔC46-UbVME structure, we are able to visualize 40 additional amino acids after the UCH domain, the first 40 amino acids (residues 223 to 263) of the ULD in TsUCH37. The polypeptide chain, after emerging from the C-terminus of the UCH domain, adopts a helical structure of 6 turns (α7), takes a U-turn and then continues as a helix (α8). α7 and α8 are arranged as a helix-turn-helix motif with a number of inter-helical contacts, and this motif adopts a similar orientation with respect to the UCH domain as observed in hUCH37 (Figs. 3 and 4b) (57). The only difference in this motif between TsUCH37 and hUCH37 is that it is somewhat shorter in the former. The ULD in TsUCH37 appears to have a proteolytically susceptible region after Ala263, perhaps immediately following it, producing the C-terminal truncation we are observing here. When we model the missing part of the ULD, using the structure of hUCH37 as a template (see Material Methods), it is apparent that α8 could have continued on after the cleavage site (Fig. 4b) almost as a long helix all the way up to residue 285, except for an interruption at Arg268 where four successive residues, including the arginine, adopt non-helical dihedral angles producing a kink (a kink featuring equivalent residues is also seen in the template structure). As expected from the hUCH37 structure, the model shows that after the interruption, the helix would terminate at or near the amino acid 285 (Fig. 4), where the polypeptide chain reverses its direction as a turn segment that appears to cap the c-terminus of the helix. The putative KEKE motif was not modeled because it is absent in the template structure. Interestingly, the structure of TsUCH37ΔC46-UbVME reveals side chains from α8 making contact with ubiquitin, specifically with its Lys48 residue, an interaction that may explain the distal-end specificity displayed by UCH37 (discussed in more details below). Also, the side chains from the modeled part of the ULD, missing in our structure, appear to present themselves for additional contacts with ubiquitin. Indeed the two most conserved residues in the ULD, Glu265 and Asn272, are facing ubiquitin and lie within contact distances (Fig. 4c). Thus, it is possible that they may actually bind to ubiquitin. Alternatively, in contrast to what is predicted by the model these residues may be used for making contact with Rpn13, explaining why they are conserved.
Figure 4. ULD-ubiquitin interactions.
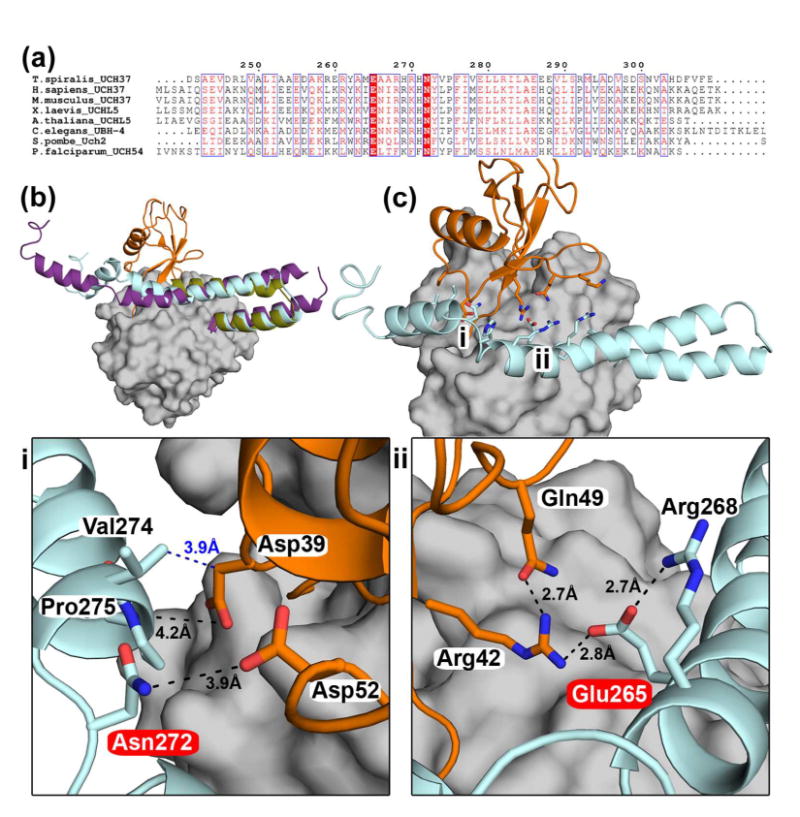
(a) Sequence alignment of the ULD of UCH37 highlighting conserved residues in UCH37 homologs. Glu265 and Asn272 (according to Ts numbering) are absolutely conserved, highlighted in red. (b) Superposition of TsUCH37ΔC46 -UbVME (the ULD in olive, UbVME in orange), human UCH37 (the ULD in purple, PDB ID 3IHR), and TsUCH37 with the entire ULD modeled (cyan) based on the structure of the ULD in human UCH37. The model was generated using SwissModel and MD simulation (please see Materials and Methods). This model is taken from a snapshot collected at 1.3 ns during an MD simulation run of 2 ns. (c) The structure of TsUCH37-ubiquitin complex with the entire ULD modeled as shown in (b), showing that the conserved residues of the ULD could make additional contacts with ubiquitin. The regions marked i and ii are expanded in the panels below. The UCH domain is surface-rendered in grey.
Active-site geometry
The catalytic triad in this cysteine protease assumes a canonical arrangement in the ubiquitin bound complex. The distance between the catalytic cysteine and histidine is 3.9 Å (Nδ-Sγ distance) in both structures and that between the histidine and the aspartate is 2.8 Å (Nɛ-Oδ) in TsUCH37cat-UbVME and 2.9 Å in TsUCH37ΔC46-UbVME. The distance between the CɛH group of the catalytic histidine and the side-chain carbonyl oxygen of the oxy-anion stabilizing glutamine (Gln79) is 3.3 Å in TsUCH37cat-UbVME and 3.1 Å in TsUCH37ΔC46-UbVME, suggesting a significant CH•••O interaction between them, an interaction seen in other cysteine proteases as well (75). We were unable to crystallize the apo form of either TsUCH37cat or TsUCH37ΔC46. In their place, we use the structure of apo human UCH37 to gain insight into structural changes in the active site region that may occur upon ubiquitin binding(57, 58). Comparison with the structures of the human UCH37 reveals that the catalytic cysteine has changed its orientation going from apo to ubiquitin-bound form, adopting a more productive orientation in the latter, an orientation in which the catalytic cysteine’s side chain faces the catalytic cleft (Fig. 5g). This analysis suggests that UCH37 exists in an unproductive form in absence of ubiquitin, with the catalytic thiol facing the interior of the protein rather than the open space in the catalytic cleft (58), but is induced to adopt a more productive form upon its binding. Thus, UCH37 may offer yet another example of a UCH DUB which undergoes substrate-induced reorganization to a more productive form(48, 76).
Figure 5. Ubiquitin recognition by TsUCH37.
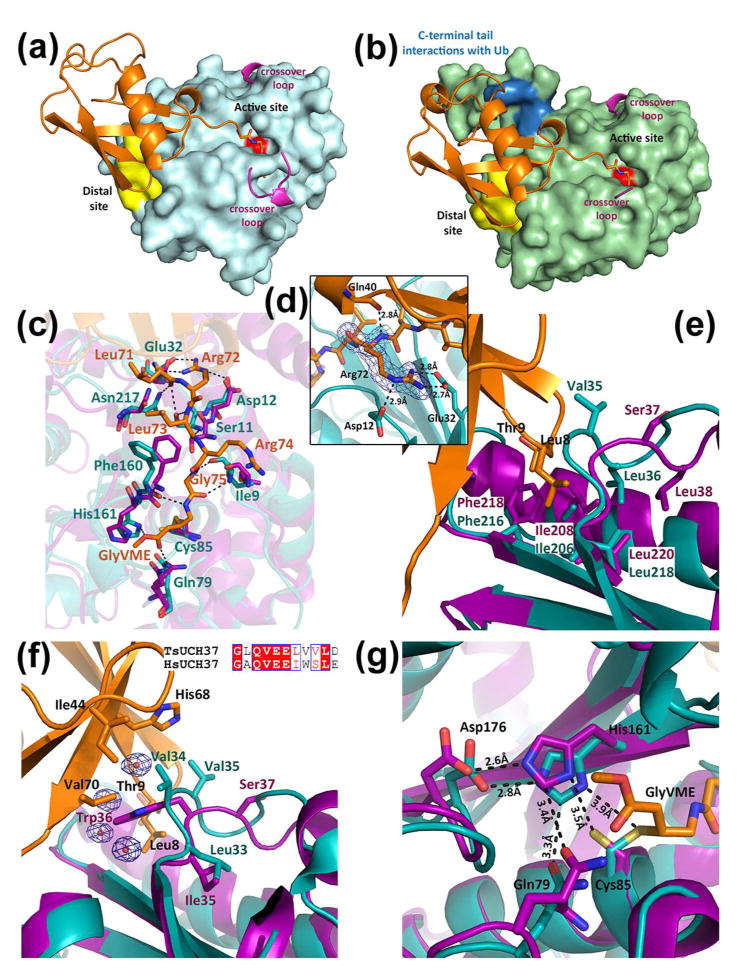
(a) Surface rendering of TsUCH37cat shown in cyan with ubiquitin binding regions highlighted. The distal site is shown in yellow, the active-site cysteine is shown in red, and resolved portions of the crossover loop are shown in pink. (b) Surface rendering of TsUCH37ΔC46 shown in green with ubiquitin binding regions highlighted as in (a), except with additional C-terminal tail ubiquitin binding residues highlighted in blue. (c) Interactions near the active site cleft with the C-terminal hexapeptide tail of ubiquitin. UbVME residues are shown in orange, TsUCH37 residues are shown in teal, and human UCH37 residues in purple. (d) Interactions of Arg72 of ubiquitin with surrounding residues of TsUCH37cat. Density from the 2Fo-Fc map is contoured at 1σ, shown in blue mesh. (e) UCH37 distal site binding residues, with TsUCH37 in teal and human UCH37 in purple. (f) Ile44 patch interacting residues, with UbVME in orange, TsUCH37 in teal, human UCH37 in purple. Waters involved in binding are also shown, enveloped with density from 2Fo-Fc map contoured at 1σ. Sequence alignment of this region in TsUCH37 compared to human UCH37 is shown as an inset. (g) Active site of TsUCH37 (in teal), showing the catalytic residues, compared to human UCH37 (in purple), with UbVME in orange.
Crossover loop flexibility
A common structural feature present in all UCH enzymes is the crossover loop, which in TsUCH37 spans residues 141 to 157 (connecting α5 with β3). It straddles the active site cleft as a flexible loop and is known to provide steric constraint, limiting the size of the leaving group at the C-terminus of ubiquitin (74, 77). Accordingly, UCH enzymes, such as UCHL1 and UCHL3, can only cleave small leaving groups from the C-terminus of ubiquitin, not large proteins or another ubiquitin (47). However, UCH37 is known to cleave diubiquitin (and polyubiquitin chains), but only when it is associated with the RP, being activated upon binding to its protein co-factor, Rpn13 (37). All previously solved structures of UCH enzymes bound to ubiquitin have shown a resolved crossover loop, which makes contact with at least one residue from the C-terminal tail of ubiquitin. In the apo form of UCHL3, the closest homolog of UCH37, the loop is disordered but becomes ordered when ubiquitin is bound (48, 50). The ubiquitin-bound structures of PfUCHL3 and the yeast ubiquitin hydrolase Yuh1 show an ordered crossover loop making contacts with side chains on the C-terminal tail of ubiquitin(73, 78). In contrast, the structures of the TsUCH37-UbVME constructs present the only examples so far of a UCH DUB in which the crossover loop is still disordered even after ubiquitin is bound, indicating that the loop is flexible and does not contribute to ubiquitin binding. A small network of van der Waals interactions and hydrogen bonds seem to stabilize part of the crossover loop (residues 152 to 157) in a short helical conformation in the structure of TsUCH37cat-UbVME, but the same segment in the TsUCH37ΔC46-UbVME structure is disordered and hence not visible, supporting dynamic sampling of conformations by this loop. The observation that the crossover loop is flexible despite the bound ubiquitin may be related to its activation by its proteasome co-factor Rpn13 (37). By not engaging with ubiquitin, the loop is available to freely interact with the co-factor, which may stabilize it in a conformation that leaves the active site maximally open to accommodate the isopeptide bond between two ubiquitins or between ubiquitin and an acceptor protein.
Interactions with Ubiquitin
The interaction of UbVME with the TsUCH37cat UCH domain buries a total of 2355 Å2 of solvent accessible surface area, a value comparable to such buried area in other UCH domain ubiquitin complexes (buried accessible surface area in TsUCH37ΔC46-UbVME is 2479 Å2) (48, 76). The interaction is predominantly localized at two areas on TsUCH37, the active site cleft and the distal site (Figs. 5a and 5b) The active-site cleft engages the C-terminal hexapeptide segment, Leu71ArgLeuArgGly-Gly76VME, of UbVME with numerous intermolecular contacts that include van der Waals forces, hydrogen bonding, electrostatic and water-mediated interactions (Fig. 5c). This segment sits in the active-site cleft with an extended conformation to maximize interactions with both backbone and side chain atoms of nearby residues of the enzyme. As seen in other UCH structures, the narrowest part of the active site cleft surrounds the terminal Gly-Gly motif, with the last Gly (GlyVME in this case) being placed immediately adjacent to the Sγ atom of the catalytic cysteine, precisely located for nucleophilic attack on the scissile peptide bond (Figs. 5a and 5b). It is interesting to note that Arg72 of UbVME is engaged in at least three major interactions (Fig. 5d), suggesting that it contributes significantly to stabilizing the enzyme-substrate complex. The interactions with Arg72 imply that TsUCH37 will find NEDD8 (neural precursor cell expressed, developmentally down-regulated 8, a structurally similar ubiquitin like protein modifier that shares 60% sequence identity with ubiquitin) as a poorer substrate since this arginine is replaced by alanine in NEDD8. Indeed TsUCH37 does not cleave NEDD8-AMC (see Figure S4). Many of the active site interactions observed in the ubiquitin-bound structures of UCHL1, UCHL3, PfUCHL3 and Yuh1 are conserved in both TsUCH37 structures. Additionally, those residues surrounding the C-terminal hexapeptide tail of ubiquitin are highly conserved between the Ts and human protein (Fig. 5c).
The interactions at the active site cleft appear to be necessary for precise cleavage at the terminal glycine residue of ubiquitin, while the distal site provides additional interaction to stabilize the enzyme-substrate complex (Figs. 5e and 5f). The distal site engages the N-terminal β-hairpin of ubiquitin, which docks by utilizing interactions primarily involving the two-residue β-turn segment, Leu8-Thr9 of ubiquitin. These interactions are mostly hydrophobic in nature, involving van der Waals contact of Leu8-Thr9 with Val35, Leu36, Ile206, Phe216 and Leu218, residues that constitute the surface-exposed hydrophobic crevice that is the distal site. Leu36, Ile206, Phe216 and Leu218 are conserved among Sp, Ts, Pf and hUCH37 (Fig. S3), suggesting the importance of distal-site binding in the enzyme-substrate recognition.
Ile44 of ubiquitin, a residue widely used in recognition by ubiquitin-binding proteins including DUBs, is seen making van der Waals contacts with Val34 on a greasy loop in TsUCH37, residues 34-36 (residues Val35 and Leu36 extend into the distal-site pocket) (Fig. 5f). A similar motif is used in other UCH enzymes to bind to Ile44 of ubiquitin. Val34 of TsUCH37 also makes contacts with His68 and Val70, which, together with Ile44 and Leu8 from the N-terminal β-hairpin turn, form the so-called Ile44-patch on ubiquitin. Thus, the binding potential of the Ile44-patch on ubiquitin appears to be fully satisfied in structures of the two complexes presented here, with each residue in the patch making at least one contact with the enzyme. The structural data presented here is supported by previously reported mutational analysis on the PA700 isopeptidase. Replacing Ile44 and Leu8 from the Ile44 patch to alanine in the distal ubiquitin of a diubiquitin substrate results in significantly impaired catalysis with no detectable hydrolysis product (45). Val34 and Val35 are replaced by tryptophan and serine, respectively, going from Ts to hUCH37 (Fig. 5f) (Val34 provides additional contacts with Val70 of UbVME). These residues also show variability among other UCH family members. Subtle differences in the Ile44-patch-binding residues could be one of the contributing factors in the difference in KM between human and Ts UCH37, especially as most of the residues at the active site are conserved between the two.
There appear to be no striking conformational changes between the ubiquitin-bound form of TsUCH37 and the apo hUCH37 except for the aforementioned reorientation of the catalytic cysteine. However, we cannot rule out that significant conformational changes might have occurred as a result of ubiquitin binding in the Ts enzyme since we could not crystallize its apo form.
Ubiquitin binding by the ULD
As mentioned earlier, the ULD of hUCH37 was thought to have an inhibitory role, presumably by folding onto the catalytic domain and obstructing substrate binding (37). In contrast, the structure of TsUCH37ΔC46-UbVME provides crystallographic evidence that the ULD can actually contribute to ubiquitin binding, and therefore can play a productive role in catalysis. Arg261 and Tyr262 on α8 of the ULD approach ubiquitin to engage in van der Waals contact with of three of its side-chains, Lys48 (with Arg261), Gln49 and Arg72 (both with Tyr262) (Fig. 6). Most notably, Arg261 is oriented in such a way to engage in close van der Waals contact with the hydrocarbon portion of the Lys48 side chain, forcing it to adopt an unusual conformation that allows an intra-molecular salt-bridge interaction with Glu51. This interaction is not observed in any of the 39 other ubiquitin-bound DUB structures currently found on the RCSB protein data bank, catalogued in Table 3; the Lys48-Glu51 distance is greater than 5.8 Å in all. Figure 6b shows the orientation of the same lysine in TsUCH37cat-UbVME. Clearly, the orientation is different in this structure and the intra-molecular salt-bridge in ubiquitin is absent, suggesting that Arg261 of the ULD plays a role in inducing the unusual orientation of Lys48 of ubiquitin. Arg261 is conserved among Sp, Ts and human UCH37 (Fig. 7), but is replaced with leucine in PfUCH37 (also known as PfUCH54). Tyr262 is conserved in human and Ts but is substituted with tryptophan in Sp and PfUCH37. Inspection of the structure reveals that the van der Waals contact with Lys48 is still feasible with leucine in place of arginine and tryptophan can conservatively replace tyrosine as well. Thus, it is likely that ULD binding with Lys48 and subsequent formation of the intra-molecular salt-bridge we are observing here is a conserved feature of UCH37 in general.
Figure 6. ULD of TsUCH37 binding to ubiquitin.
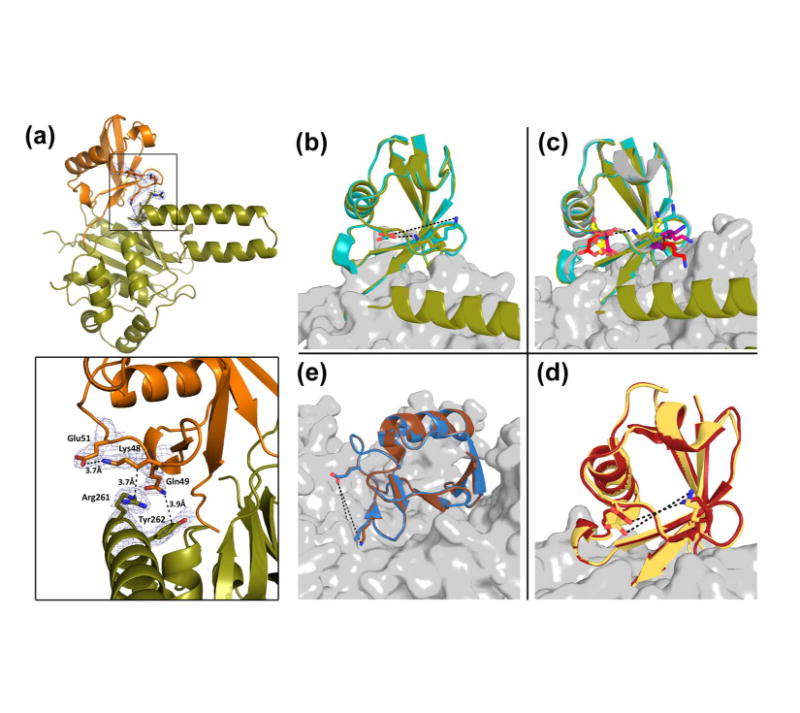
(a) TsUCH37ΔC46 (in olive) ULD residues interacting with UbVME (in orange). Inset shows Arg261 and Tyr262 interactions with UbVME, as well as the intra-molecular salt bridge formed between Lys48 and Glu51 of ubiquitin. Density rendered from 2Fo-Fc map contoured to 0.7σ. (b-e) Comparison of the Lys48-Glu51 distance in ubiquitin observed in other DUB-ubiquitin structures. (b) Lys48 and Glu51 form a 3.7Å salt bridge in TsUCH37ΔC46 – UbVME structure (olive), but not in TsUCH37cat-UbVME structure (9.9 Å). (c) The same distance in all other UCH-ubiquitin structures are ≥9Å: UCHL3-UbVME in yellow (PDB ID 1XD3), UCHL1-UbVME in red (PDB ID 3KW5), Yuh1-Ubal in pink (PDB ID 1CMX), PfUCHL3-UbVME in purple (PDB ID 2WDT), TsUCH37cat-UbVME in teal. (d) The same distance in OTU-ubiquitin structures: Otu1-ubiquitin in dark red (PDB ID 3BY4) is 8.7 Å, and in DUBA-Ubal in pale yellow (PDB ID 3TMP) is 6.0 Å. (e) The same distance in HAUSP/USP7-Ubal (PDB ID 1NBF in blue) is 10Å and in USP14-Ubal (PDB ID 2AYO in brown) is 10.9Å.
Table 3.
Lys48-Glu51 distances for all DUB-ubiquitin complexes
| UCH Family | ||
| PDB ID | DUB-ubiquitin complex | Lys48-Glu51 distance (Å) |
| 1XD3 | UCHL3-UbVME | 9.1, 11.5* |
| 1CMX | YUH1-Ubal | 10.8 |
| 2WDT | PfUCHL3-UbVME | 7.3, 9.6 |
| 3IFW | UCHL1 S18Y-UbVME | 8.7 |
| 3KVF | UCHL1 I93M-UbVME | 12.3 |
| 3KW5 | UCHL1-UbVME | 13.1 |
| USP Family | ||
| PDB ID | DUB-ubiquitin complex | Lys48-Glu51 distance (Å) |
| 3TMP | DUBA-Ubal | 8.3 |
| 2Y5B | USP21-linear diUbal | 7.8, 10.8, 6.6 |
| 1NBF | HAUSP-Ubal | 10.0, 10.7 |
| 2AYO | USP14-Ubal | 10.9 |
| 3MHS | SAGA complex (UBP8)-Ubal | 9.2 |
| 2HD5 | USP2, Ub | 9.0 |
| 2G45 | IsoT, Ub | 8.6, 8.9 |
| 2J7Q | M48 USP-UbVME | 9.9, 10.8 |
| 3V6E | USP2, Ub variant | 7.0 |
| 3V6C | USP2, Ub variant | 7.4 |
| 3IHP | USP5, Ub covalent | 9.6, 6.3 |
| 3MTN | USP21, ubiquitin-based USP21-specific inhibitor | 8.5 |
| 3IT3 | USP21, Ub covalent | 7.4, 7.5, 7.4, 7.4 |
| 3N3K | USP8, covalent Ub-like variant | 10.4 |
| 3NHE | USP2a, Ub | 7.4 |
| 2IBI | USP2, Ub covalent | 7.6 |
| OTU Family | ||
| PDB ID | DUB-ubiquitin complex | Lys48-Glu51 distance (Å) |
| 4IUM | arterivirus papain-like protease 2, Ub | 7.4 |
| 3ZNH | CCHF viral, Ub-propargyl | 9.5 |
| 4I6L | OTUB1, Ub | 9.1 |
| 3PT2 | Viral OTU, Ub | 10.7 |
| 4HXD | Nairovirus viral OTU, Ub | 8.3, 10.1 |
| 3BY4 | OTU, Ub | 6.0 |
| 3PRM | CCHF viral OTU, Ub | 9.5, 10.0 |
| 3PRP | CCHF viral OTU, Ub | 10.3, 10.9 |
| 3C0R | OTU, Ub | 8.4 |
| 4DHZ | h/ceOTUB1-ubiquitin aldehyde-UBC13∼Ub | 9.2, 8.8 |
| 4DHJ | ceOTUB1 ubiquitin aldehyde UBC13∼Ub complex | 11.4, 8.5, 6.9, 11.8, 9.8, 10.1 |
| 4DDI | OTUB1/UbcH5b∼Ub/Ub | 7.5, 12.6, 12.6, 7.5, 12.6, 7.5 |
| 4DDG | OTUB1/UbcH5b∼Ub/Ub | 11.3, 7.1, 8.0 |
| 3PHW | OTU Domain of CCHF Virus, Ub | 7.6, 7.2, 9.0, 5.8 |
| MJD Family | ||
| PDB ID | DUB-ubiquitin complex | Lys48-Glu51 distance (Å) |
| 3O65 | Ataxin-3-like, Ub | 9.6, 11.4, 12.8, 11.5 |
| 2JRI | Ataxin 3, Ub | 12.3, 12.9 |
| JAMM Family | ||
| PDB ID | DUB-ubiquitin complex | Lys48-Glu51 distance (Å) |
| 2ZNV | AMSH-LP, Lys63-linked diubiquitin | 9.0, 8.3, 11.9, 11.3 |
Multiple distance entries refer to that in the other subunits of the crystallographic asymmetric unit.
Figure 7.
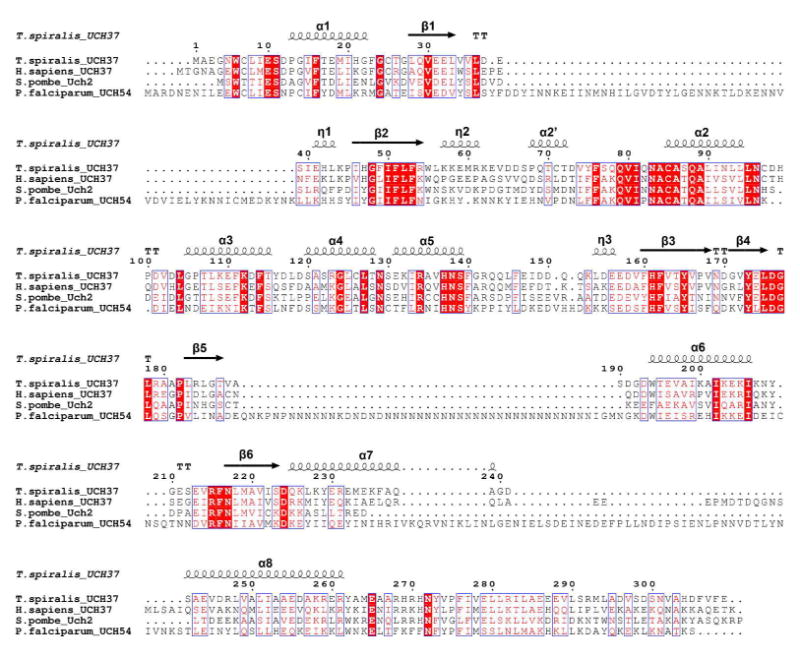
Sequence alignment of TsUCH37 and other homologs: human UCH37, S. pombe Uch2, and P. falciparum UCH54. Secondary structures for the two TsUCH37 structures are annotated above (e.g. α1 = alpha helix 1, β1 = beta sheet 1, η1 = 310 helix 1). α2’ and η2 are not resolved in the TsUCH37ΔC46 –UbVME structure, and helices α7 and α8 are not present in the TsUCH37cat-UbVME construct.
UCH37, as a part of PA700, is known to selectively cleave polyubiquitin chains from the very distal end, sequentially removing one ubiquitin at a time (38). The structural basis of this exo cleavage specificity is not yet known. The unique orientation of Lys48 stabilized by Arg261 leading to the intra-molecular salt-bridge may explain this selectivity. We propose that although a similar type of interaction between Arg261 and ubiquitin’s Lys48 is possible with an internal ubiquitin, the intra-molecular salt-bridge will be absent in this case since the amino group of the lysine is acylated and hence not charged. Thus, it is the lack of an additional interaction with an internal ubiquitin that makes binding to Lys48 of the terminal ubiquitin more favored, hence the exo selectivity.
Discussion
UCH37 is a proteasome-associated UCH DUB known to have polyubiquitin chain-editing function. It preferentially cleaves the chain from its very distal tip (38). Such a function might rescue certain substrates from being committed to further downstream action of the proteasome (38). It is also possible that certain substrates carry inappropriate polyubiquitin tags that are not optimal for their degradation. The chain-editing function might be essential for releasing these substrates to clear up ubiquitin receptors for binding to productive substrates. A regulator of proteasome function, it is itself regulated by binding to the proteasome: UCH37 is activated upon binding to Rpn13, a subunit of PA700 (the 19S proteasome or RP), the mechanism of which is not understood. We report here the structure of two constructs of UCH37 from the infectious helminth Trichinella spiralis (Ts) bound to the suicide inhibitor UbVME. This work constitutes the first structural analysis of a ubiquitin-pathway protein in the organism showing how ubiquitin is recognized by this UCH family DUB in Ts. The structures reveal striking conservation of the ubiquitin binding mode among UCH DUBs, from lower eukaryotes to human (Fig. S5). It also shows important structural differences between other UCH DUBs, such as UCHL1 and UCHL3, some of which could be used for specialized function of UCH37. While revealing interesting differences, the Ts structures provide a number of details that may also hold true for the human enzyme, advancing our understanding of UCH37 in general.
The active-site cysteine may undergo ubiquitin-mediated reorientation to a more productive form (Fig. 5g), making UCH37 yet another example of a UCH DUB that shows regulation of activity by ubiquitin, a feature that may provide selectivity to this group of cysteine proteases. Structures of the two constructs reveal invariant parts of the enzyme, likely less dynamic parts, while also revealing parts that are more dynamic in nature, such as the segment from residues 57-71 and Trp55. Future studies would reveal the role of such dynamic parts in catalysis or regulation thereof.
Importantly, the structure of the construct with the additional 40 amino acids after the UCH domain reveals that the ULD could contribute to ubiquitin binding (Figs. 4 and 6), an unexpected finding since it was thought to be inhibitory in the human enzyme (37). The interaction of Arg261 on the ULD appears to engage Lys48 of the distal ubiquitin in a way that would be energetically most favored with the very terminal ubiquitin in a polyubiquitin chain, possibly explaining the exo specificity displayed by mammalian UCH37. This structural data is consistent with previously reported mutational analysis probing substrate specificity of the PA700 isopeptidase: mutation of Lys48 to cysteine on the distal ubiquitin of a diubiquitin substrate results in severely impaired catalysis (45). Apart from the broad agreement with the aforementioned experimental work, this observation of the intra-molecular Lys48-Glu51 salt bridge in the distal ubiquitin, apparently induced by Arg261, is purely crystallographic at this point, although it seems unlikely that lattice forces have anything to do with it. Even if the opposite is true, the fact that such interactions are physiologically relevant cannot be ignored. The lack of an intra-molecular Lys48-Glu51 salt bridge in any other ubiquitin-bound DUB structures to date (Table 3) makes this unusual interaction more intriguing, and worth additional pursuit. This observation therefore lays the structural groundwork for future mutational analysis aimed at validating their existence in solution and their role in substrate specificity.
It is interesting to note that a salt-bridge interaction, albeit an intermolecular one, involving Lys48 of ubiquitin and an acidic side chain of the enzyme is also seen in the structure of USP7 bound to ubiquitin aldehyde (the Lys48 side chain of the distal ubiquitin is interacting with Asp305 and Glu308 of USP7)(79). Such bifurcated salt-bridges will perhaps contribute substantially to the enzyme binding the distal ubiquitin in a K48-linked chain, based on which one may predict that USP7 will also exhibit exo specificity. This needs to be examined. Preferential cleavage from the very distal tip of a Lys48-linked polyubiquitin chain may be a feature common to DUBs that work on chains of this topology. Lys48-linked chains are known to adopt a compact structure (80). However, the terminal ubiquitin, being less packed than the internal ones (packed from both sides), is more likely to fray and be susceptible to DUB cleavage for stereochemical reasons. Certain DUBs may have evolved a mechanism to grab onto those fraying ends and start disassembling chains from there. There may be other DUBs that prefer internal ubiquitins, or the terminal ones on the other extreme end of the chain, such as Isopeptidase-T (USP5) (81), and there may be some with no preference at all. The structure of AMSH-LP (a Lys63-linked chain specific DUB) in complex with Lys63-linked diubiquitin shows that Lys63 on the distal ubiquitin is not engaged by the enzyme, suggesting it is unlikely to show any preference between the terminal and internal cleavage sites (82). This is consistent with the structure of a Lys63-linked chain, which adopts a more extended conformation in crystals and perhaps in solution as well (83-85). Future structural studies should reveal more details explaining exo and endo specificity seen in certain DUBs.
The structural analysis, combined with MD simulation, shows the contribution of the ULD in ubiquitin binding. In theory, certain residues in TsUCH37’s ULD, missing in our structure, also appear to be correctly positioned for contacting ubiquitin. Notably, the modeling study provides a possible explanation of why Glu265 and Asn272 are so strictly conserved in UCH37 from different organisms, with virtually no exception. Contributing to ubiquitin binding, as suggested by our modeling studies, may be one of the functional constraints underlying the conservation of the amino acids, although one cannot rule out if binding to other proteins such as Rpn13 may be involved. It should be noted that Bap1, a UCH DUB mutated in several cancers, also features a ULD (70, 71, 86). Like UCH37, Bap1 becomes activated upon binding to a larger protein complex, demonstrated with the Drosophila orthologue, Calypso, binding to the polycomb repressor DUB complex (87). Interestingly, the putative ubiquitin-binding residues of the ULD of UCH37 are also conserved in Bap1 (data not shown), suggesting a role in ubiquitin binding for Bap1’s ULD as well (in some Bap1 orthologues the glutamate corresponding to TsUCH37’s Glu265 is replaced with an aspartate). However, human Bap1 has a linker of approximately 300 amino acids separating the UCH domain and its ULD. It will be interesting to see how the ULD positions itself to bind ubiquitin, if it does. Of more interest is to know if the ULD has independent ability to bind to ubiquitin.
ACCESSION NUMBERS
Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 4I6N and 4IG7.
Supplementary Material
Acknowledgments
The authors acknowledge Venugopalan Nagarajan and Craig Ogata at beam line 23-ID-B of the Advanced Photon Source for assistance with data collection. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. We thank Emma DeWalt and Garth Simpson from the Department of Chemistry, Purdue University, for their assistance with SONICC imaging of initial crystals.
Funding:
Financial support from the National Institutes of Health (1R01RR026273) (C.D.) is gratefully acknowledged. The General Medicine and Cancer Institutes Collaborative Access Team (GM/CA CAT) of the Advanced Photon Source at Argonne National Laboratory has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104).
Abbreviations
- SDS PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- DTT
dithiothreitol
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- UbVME
ubiquitin vinyl methyl ester
- UbAMC
ubiquitin aminomethylcoumarin
- UCH37
ubiquitin carboxyl-terminal hydrolase 37
- DUB
deubiquitinating enzyme or deubiquitinase
Footnotes
Supporting Information Supporting figures and NEDD8 hydrolysis data (Fig. S4). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A, Schwartz AL. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology. 2002;35:3–6. doi: 10.1053/jhep.2002.30316. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 7.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 9.Matyskiela ME, Martin A. Design principles of a universal protein degradation machine. J Mol Biol. 2012;425:199–213. doi: 10.1016/j.jmb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 13.Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Guterman A, Glickman MH. Deubiquitinating enzymes are IN/(trinsic to proteasome function) Curr Protein Pept Sci. 2004;5:201–211. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- 15.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 16.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 17.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 200170:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 19.Schulman BA. Twists and turns in ubiquitin-like protein conjugation cascades. Protein Sci. 2011;20:1941–1954. doi: 10.1002/pro.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 21.Haglund K, Dikic I. Ubiquitylation and cell signaling. Embo J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fushman D, Wilkinson KD. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol Rep. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 25.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 201213:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 26.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 28.Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 29.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695. 2004:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. Faseb J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson KD. DUBs at a glance. J Cell Sci. 2009;122:2325–2329. doi: 10.1242/jcs.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 34.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsou WL, Sheedlo MJ, Morrow ME, Blount JR, McGregor KM, Das C, Todi SV. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS One. 2012;7:e43112. doi: 10.1371/journal.pone.0043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 38.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 39.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 40.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 44.D'Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44:1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem. 1997;272:28438–28446. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 46.Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C. Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol. 2004;344:697–706. doi: 10.1016/j.jmb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- 47.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 48.Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 49.Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. Embo J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. Embo J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishio K, Kim SW, Kawai K, Mizushima T, Yamane T, Hamazaki J, Murata S, Tanaka K, Morimoto Y. Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain. Biochem Biophys Res Commun. 2009;390:855–860. doi: 10.1016/j.bbrc.2009.10.062. [DOI] [PubMed] [Google Scholar]
- 57.Burgie SE, Bingman CA, Soni AB, Phillips GN., Jr Structural characterization of human Uch37. Proteins. 2011 doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maiti TK, Permaul M, Boudreaux DA, Mahanic C, Mauney S, Das C. Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme. Febs J. 2011;278:4917–4926. doi: 10.1111/j.1742-4658.2011.08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White RR, Miyata S, Papa E, Spooner E, Gounaris K, Selkirk ME, Artavanis-Tsakonas K. Characterisation of the Trichinella spiralis deubiquitinating enzyme, TsUCH37, an evolutionarily conserved proteasome interaction partner. PLoS Negl Trop Dis. 2011;5:e1340. doi: 10.1371/journal.pntd.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 61.Holyoak T, Fenn TD, Wilson MA, Moulin AG, Ringe D, Petsko GA. Malonate: a versatile cryoprotectant and stabilizing solution for salt-grown macromolecular crystals. Acta crystallographica Section D, Biological crystallography. 2003;59:2356–2358. doi: 10.1107/s0907444903021784. [DOI] [PubMed] [Google Scholar]
- 62.Otwinowski ZaM, W . Processing of X-ray Diffraction Data Collected in Oscillation Mode. Vol. 276. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- 63.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D: Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 65.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophysical journal. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 67.Case D, Darden T, Cheatham T, III, Simmerling C, Wang J, Duke R, Luo R, Walker R, Zhang W, Merz K. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 68.Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98:10089. [Google Scholar]
- 69.Ryckaert JP, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 70.Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O'Rourke K, Dixit VM, Wilson AC. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanchez-Pulido L, Kong L, Ponting CP. A common ancestry for BAP1 and Uch37 regulators. Bioinformatics. 2012;28:1953–1956. doi: 10.1093/bioinformatics/bts319. [DOI] [PubMed] [Google Scholar]
- 72.Chen V, Arendall W, Headd J, Keedy D, Immormino R, Kapral G, Murray L, Richardson J, Richardson D. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. Embo J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou ZR, Zhang YH, Liu S, Song AX, Hu HY. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
- 75.Boudreaux DA, Chaney J, Maiti TK, Das C. Contribution of active site glutamine to rate enhancement in ubiquitin C-terminal hydrolases. Febs J. 2012;279:1106–1118. doi: 10.1111/j.1742-4658.2012.08507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boudreaux DA, Maiti TK, Davies CW, Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc Natl Acad Sci U S A. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Popp MW, Artavanis-Tsakonas K, Ploegh HL. Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J Biol Chem. 2009;284:3593–3602. doi: 10.1074/jbc.M807172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Artavanis-Tsakonas K, Weihofen WA, Antos JM, Coleman BI, Comeaux CA, Duraisingh MT, Gaudet R, Ploegh HL. Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3. J Biol Chem. 2010;285:6857–6866. doi: 10.1074/jbc.M109.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 80.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 81.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 83.Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, Hayashi H, Murata S, Hiroaki H, Sato M, Tanaka K, Shirakawa M. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–875. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 86.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


