Abstract
Existing super-resolution fluorescence microscopes compromise acquisition speed to provide subdiffractive sample information. We report an analog implementation of structured illumination microscopy that enables 3D super-resolution imaging with 145 nm lateral and 350 nm axial resolution, at acquisition speeds up to 100 Hz. By performing image processing operations optically instead of digitally, we removed the need to capture, store, and combine multiple camera exposures, increasing data acquisition rates 10–100x over other super-resolution microscopes and acquiring and displaying super-resolution images in real-time. Low excitation intensities allow imaging over hundreds of 2D sections, and combined physical and computational sectioning allow similar depth penetration to confocal microscopy. We demonstrate the capability of our system by imaging fine, rapidly moving structures including motor-driven organelles in human lung fibroblasts and the cytoskeleton of flowing blood cells within developing zebrafish embryos.
Introduction
Modern fluorescence microscopy combines contrast, molecular specificity, speed, and biocompatibility to enable the visualization of live cellular processes. Unfortunately, diffraction limits the resolution of a widefield fluorescence microscope to ~250 nm laterally and 500–750 nm axially, and achieving even this ‘diffraction-limited’ performance is difficult in practice. Super-resolution imaging techniques1 overcome this difficulty, yet speed, imaging duration, and field of view are severely compromised in existing implementations, especially when compared to conventional microscopy. For example, single-molecule imaging2,3 or stimulated emission depletion microscopy (STED)4 enable sub-100 nm spatial resolution over cellular areas, but are limited to imaging speeds of ~0.01–1 Hz (with faster imaging trading field of view and image quality for speed5). Neither single-molecule imaging nor STED is currently practical for time-lapse volumetric imaging, due to the low acquisition speed and phototoxic illumination intensities (104 – 107 W/cm2 peak intensities, tens of mW average power).
In contrast to single-molecule imaging and STED, linear structured illumination microscopy (SIM)6,7 provides a more modest resolution increase (√2 better than the diffraction limit, 2-fold better after deconvolution) but requires 103 – 106 lower illumination intensities, provides much faster acquisition rates (up to 11 Hz in 2D8 and 0.2 Hz in 3D9), and computationally rejects out of focus light originating elsewhere than the focal plane. These advantages enable optically-sectioned super-resolution imaging over hundreds of 3D timepoints. Using a sparse lattice of excitation points in combination with pinholes in the emission path and appropriate image processing (ref.10–13, Supplementary Note 1) confers the further advantage that out-of-focus light may be physically rejected, extending the depth penetration of SIM and allowing live super-resolution imaging at depths ~50 μm from the coverslip surface (‘multifocal SIM’ or MSIM13). Regardless of the specific implementation, all previous efforts illuminate the sample with ~10–100 excitation patterns, acquire one camera exposure per pattern, and digitally combine the resulting images to produce a single 2D super-resolution image. The need to capture and combine many raw images per plane has fundamentally limited the speed of SIM relative to conventional microscopy.
We report an analog implementation of structured illumination microscopy that doubles the spatial resolution of a fluorescence microscope, with no tradeoff in speed, phototoxicity, or field of view. By eliminating the need to acquire and digitally combine multiple camera exposures, our method removes the only drawback of SIM compared to conventional fluorescence microscopy and permits super-resolution image acquisition and display in real time. Our technique enables multicolor volumetric imaging at rates comparable or faster than line-scanning or spinning disk confocal microscopy, permitting inspection of sub-mitochondrial detail otherwise obscured by diffraction or motion-blur. We further highlighted the advantage of our method relative to existing SIM implementations by performing noninvasive super-resolution imaging of interacting protein distributions at volumetric rates 15x faster than previously reported. Finally, we demonstrate super-resolution imaging at unprecedented speed by visualizing cytoskeletal detail within flowing blood cells in vivo (at 37 Hz) and capturing the millisecond-scale remodeling and growth of the endoplasmic reticulum (ER, at 100 Hz).
Results
Analog image processing for instantaneous super-resolution
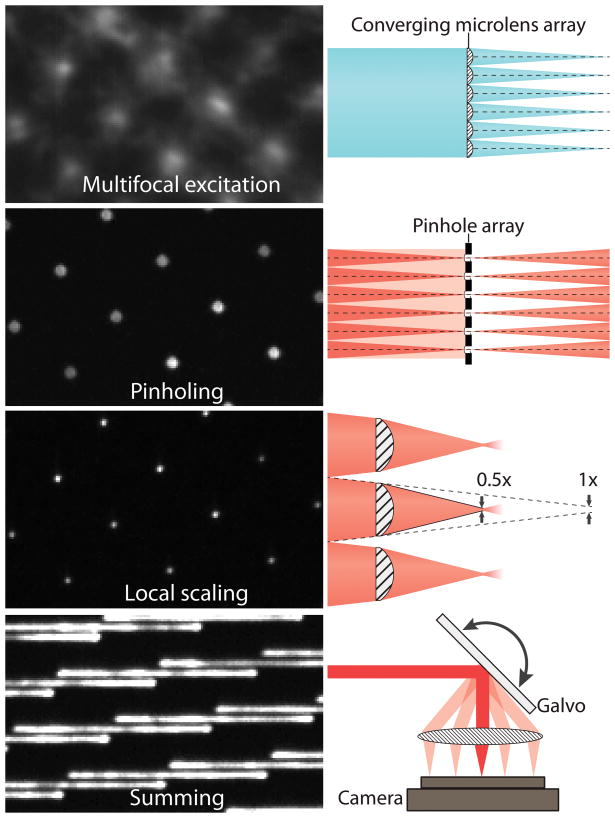
The key realization underlying our approach is that every step of the digital combination in MSIM (fully explained in Supplementary Note 1) may be performed optically, in an analog, effectively instantaneous fashion. MSIM data acquisition and processing may be conceptually divided into multiple steps: (i) sparse, multifocal illumination of the sample; (ii) recording the resulting fluorescence image with a camera; (iii) digital pinholing to reject out-of-focus emission; (iv) digital 2x contraction of each pinholed emission (without changing overall image dimensions); (v) repetition of steps (i–iv) at different positions of the multifocal excitation pattern until the entire field has been illuminated; (vi) digital summation of the resulting images to generate a super-resolution image with √2 improved resolution; (vii) deconvolution to recover the full 2x resolution improvement. Here we implement steps (i–vi) entirely in hardware (Fig. 1): using (I) a microlens array to generate a multifocal excitation pattern; (II) a matched pinhole array to reject out-of-focus emissions; (III) a matched microlens array to locally contract each pinholed emission; (IV) a galvanometric mirror to translate the excitation pattern and sum the fluorescence emissions resulting from (I–III) prior to image collection with a camera.
Fig. 1. Key steps in implementing instant structured illumination.
A converging microlens array is used to produce a multifocal excitation. After exciting the sample, out-of-focus fluorescence is rejected with a pinhole array that is matched to the microlens array. A 2x local contraction of each pinholed fluorescence emission is achieved with the aid of a second, matched microlens array. A galvo serves to raster multifocal excitation and sum multifocal emission, producing a super-resolution image during each camera exposure (for clarity, only a partial galvo scan is shown in this figure). Raw data corresponding to each of these steps: left. Cartoon representation: right. See also Supplementary Fig. 1.
Our instantaneous structured illumination microscope (‘instant SIM’, Supplementary Fig. 1–6) directly captures optically-sectioned images with √2 improvement in resolution in a single camera exposure, producing resolution doubled images after deconvolution. As in MSIM, imaging Alexafluor 488-labeled microtubules in a fixed, human osteosarcoma (U2OS) cell with instant SIM revealed an apparent microtubule width of 150 nm, doubling widefield resolution and allowing us to resolve microtubules with 110 nm center-to-center separation (Supplementary Fig. 7). We confirmed this result on 100 nm fluorescent beads (145 +/− 14 nm, N = 10), also obtaining the previously reported ~2x increase in axial resolution (356 +/− 37 nm) enabled by SIM (Table 1, Supplementary Figs. 8–9).
Table 1.
Apparent width of 100 nm subdiffractive beads, as measured in different microscopes.
| Instant SIM, widefield mode | Instant SIM raw | Instant SIM + decon | SDC raw | SDC + decon | LSC raw | LSC + decon | |
|---|---|---|---|---|---|---|---|
| NA | 1.45 | 1.45 | 1.45 | 1.49 | 1.49 | 1.46 | 1.46 |
| Lateral FWHM (nm) | 281 ± 11 | 213 ± 26 | 145 ± 14 | 260 ± 11 | 188 ± 14 | 369 ± 30 | 250 ± 38 |
| Axial FWHM (nm) | 680 ± 30 | 511 ± 24 | 356 ± 37 | 633 ± 36 | 429 ± 20 | 661 ± 31 | 401 ± 14 |
Abbreviations: NA, numerical aperture; SDC, spinning disk confocal; LSC, line-scanning confocal; FWHM, full width at half maximum. ‘Widefield mode’ measurements were conducted on the instant SIM, but with microlens arrays and pinholes removed. Beads appear sharpest in instant SIM, despite the lower NA. Means ± standard deviations were derived from ten beads. ‘Raw’ values were taken as parameters for theoretical Gaussian PSFs used when deconvolving datasets used in this paper.
Rapid and non-invasive 3D super-resolution imaging
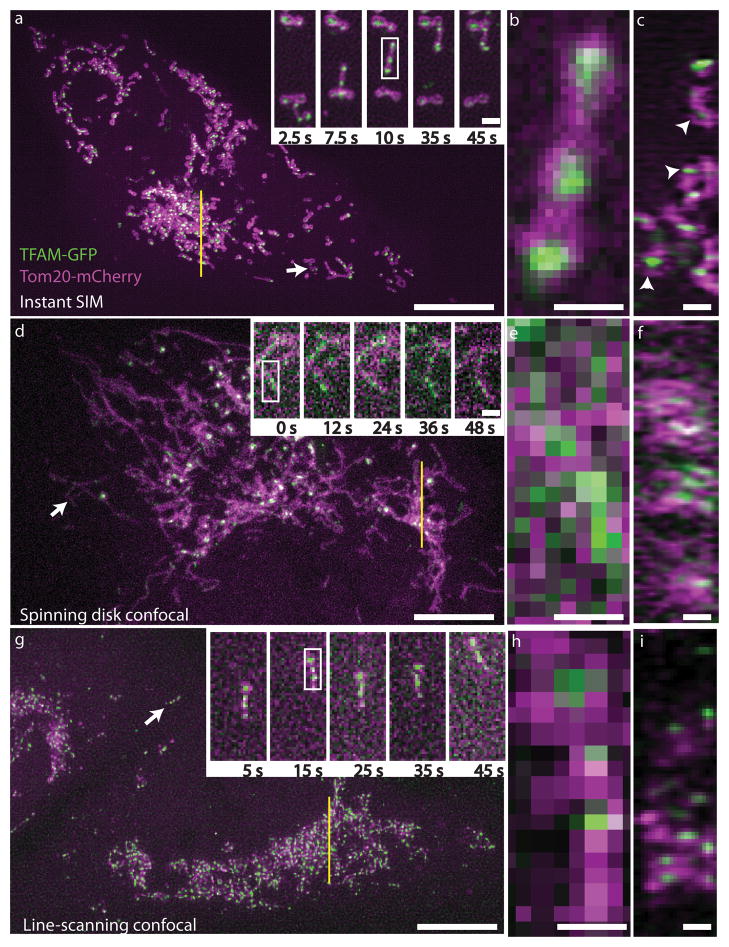
To demonstrate the capability of instant SIM for time-lapse interrogation of whole cellular volumes, we imaged a variety of dynamic protein distributions in transformed human lung fibroblasts. For example, we created a GFP chimera of TFAM, a mitochondrial DNA transcription factor that also acts to package mitochondrial DNA in nucleoids14 and imaged it along with Tom20-mCherry, a member of the translocase multimeric complex of outer mitochondrial membrane that functions to selectively transport proteins from cytosol into the mitochondrial intermembrane space (ref.15, Fig. 2a, Supplementary Video 1). The speed of instant SIM enabled us to collect each 2-color, 3 μm thick volume in 1.2 s (~15x faster than previous multicolor volumetric SIM on samples of equivalent thickness16), clearly resolving even fast-moving mitochondria that migrated at rates of 0.5 – 1 μm/s (Fig. 2b). Our resolution was sufficiently high to discern internal voids within most mitochondria (evident in the Tom20 channel, Supplementary Fig. 10). TFAM nucleoids (puncta sized at our resolution limit, in accordance with other super-resolution experiments on fixed cells17) were engulfed within mitochondrial voids (Fig. 2b), an observation confirmed in axial views of the sample (Fig. 2c). To our knowledge, STED is the only other super-resolution technique that has been used to reveal the absence of Tom20 from the inner mitochondrial space18,19, yet these experiments were performed on fixed cells, presumably due to STED’s slow acquisition speed or phototoxicity.
Fig. 2. Instant SIM enables high-speed multicolor super-resolution imaging.
Live MRL-TR transformed human lung fibroblasts expressing TFAM-GFP (green) and Tom20-mCherry (red) were imaged in instant SIM (a–c), spinning disk confocal (d–f), and a fast, line-scanning confocal microscope (g–h). a, d, g: Maximum intensity projections (XY) of 3 μm thick volumes are shown, with subregions (white arrows) at indicated timepoints shown in higher magnification insets. Note that the line-scanning confocal inset in g has been rotated 90 degrees relative to lower magnification view. Scalebars: 10 μm and 1 μm (inset). b, e, h: Higher magnification of mitochondria highlighted by white rectangles in a, d, g insets. Scalebars: 0.5 μm. c, f, i: Axial (ZY) view of ~270 nm thick slices (yellow lines) from a, d, g. White arrowheads indicate TFAM enclosed by Tom20 in instant SIM. Scalebars: 1 μm. See also Supplementary Videos 1–3 and Supplementary Fig. 8–12.
Functionally and conceptually, instant SIM is much like a spinning disk confocal microscope with higher resolution, so we compared our method to this popular, proven technology. For two-color volumes of equivalent thickness, our spinning disk system was ~10-fold slower than instant SIM (Supplementary Video 2), and both lateral and axial resolutions were worse than instant SIM (Fig. 2d–f, Table 1, and Supplementary Fig. 8–9). With (Fig. 2e) or without (Supplementary Fig. 11) deconvolution, the resolution of our spinning disk was insufficient to observe sub-mitochondrial voids (Supplementary Fig. 12) and blurred nucleoids enough that they often appeared as continuous, not discrete, entities (Fig. 2e–f). We also performed a comparison to a fast line-scanning confocal microscope that enabled imaging at similar volumetric acquisition rates to instant SIM (Fig. 2g–i, Supplementary Video 3). As with the spinning disk, with (Fig. 2h) or without (Supplementary Fig. 11) deconvolution mitochondrial voids were obscured (Supplementary Fig. 12). Furthermore, the loss in axial resolution relative to instant SIM was enough that the enclosure of TFAM by Tom20 was completely masked by diffraction (Fig. 2i, Table 1, Supplementary Figures 8–9).
The low intensities employed in instant SIM (~5–50 W/cm2) allowed us to volumetrically image suitably bright samples over tens of timepoints without obvious photobleaching or signs of photodamage such as membrane retraction or blebbing. For example, we visualized the dynamics of abundant H-Ras and Rab8A GTPases that are key players in growth factor signaling cascades and intracellular trafficking20, over 60 – 100 imaging volumes (Supplementary Videos 4–5). Instant SIM revealed the formation and movement of tubular endosomes, and also highly mobile small vesicles in proximity to the basal membrane (Supplementary Video 5). In light of emerging interest for interaction of peroxisomes with mitochondria21, we performed live imaging of GFP labeled peroxisomes in conjunction with Tom20-mCherry (Supplementary Video 6). Strikingly, our spatiotemporal resolution also allowed us to visualize rapid interactions of peroxisomes with mitochondrial spheroids (Supplementary Video 7). Finally, we used the ER-peroxisome escort protein GFP-SEC16B and imaged its interactions with mCherry labeled peroxisomes (Supplementary Video 8), observing ‘kiss and run’ interactions between these two partners, most likely representing transfer of material from ER to peroxisomes (Supplementary Video 9, ref.22). In all cases, our acquisition rate was at least 15x faster than other SIM implementations9,16.
Faster than video-rate super-resolution imaging
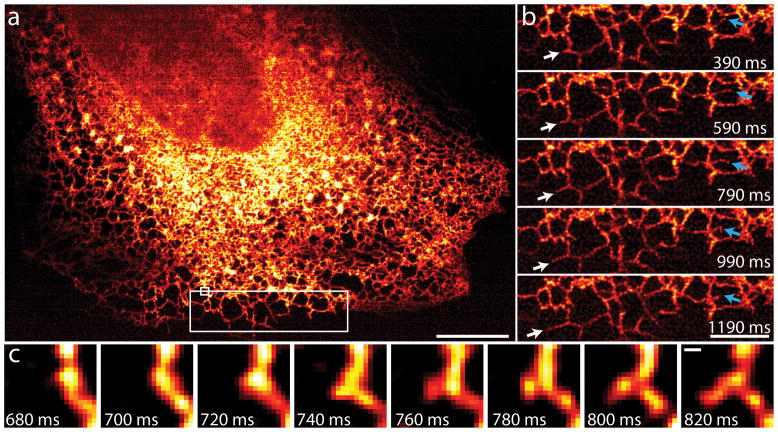
In order to fully exploit the speed of instant SIM, we recorded dynamics of the endoplasmic reticulum in a single 2D plane at 100 Hz, over hundreds of timepoints ((Fig. 3a, Supplementary Video 10). We chose the ER due to its highly plastic nature and rapid movement23. Our frame rate allowed easy observation and quantification of the rapid growth (~3.5 μm/s, white arrows in Fig. 3b) and remodeling (blue arrows) of individual ER tubules. We also observed the formation and growth of new ER tubules, events that occurred in < 140 ms (Fig. 3c) and that would have been blurred in any other SIM implementation, even at the very fastest 2D frame rates reported8. We note that our imaging rate is 50x faster than other methods that record ER dynamics at similar effective resolution24, despite the 40x larger imaging area we cover.
Fig. 3. Instant SIM reveals ER dynamics at 100 Hz.
A: First image in 200 timepoint series, showing ER labeled with GFP-Sec61A within MRL-TR transformed human lung fibroblasts. Data were acquired at the coverslip surface. Scalebar: 10 μm. b: Higher magnification view of the large white rectangle in a. White arrows mark growth of an ER tubule, blue arrows indicate remodeling of an ER tubule. Scalebar: 5 μm. c: Higher magnification view of the small white rectangle in a, indicating formation of a new tubule within 140 ms. Scale bar: 200 nm. See also Supplementary Video 10.
An advantage of instant SIM is that its combination of computational and physical optical sectioning enables super-resolution imaging at sample depths ~10x deeper than possible with other SIM implementations13. We used this feature to image blood flow 20–25 μm from the coverslip surface, within cranial vessels of 3 day old zebrafish embryos expressing a GFP transgene that labeled microtubules. Our imaging rate of 37 Hz was fast enough to ‘freeze’ individual blood cells, revealing subcellular detail despite their rapid movement (25 μm/s, Fig. 4, Supplementary Video 11). Some blood cells displayed ‘tails’ (yellow arrows, Fig. 4) composed of multiple microtubules (yellow arrowheads), in addition to intracellular microtubules that spanned the cell body (red arrowheads). In at least some cases, microtubule bundles are splayed apart, revealing what are likely to be individual microtubules, as suggested by their sparse labeling. We speculate that these cells with tails may be neutrophils, as the tails appeared visually similar to ‘sling’ structures that help neutrophils roll in the presence of high shear forces25, although the identity of these cells awaits confirmation.
Fig. 4. Imaging vascular flow with subcellular resolution at 37 Hz.

Top panel: maximum intensity projection across 100 2D frames (spanning 2673 ms), highlighting GFP labeled microtubules in blood cells located within cranial vessels, 20 μm into a 3 day old zebrafish embryo. Direction of motion is from left to right. Sharp cell boundaries indicate the absence of motion blur. Scale bar: 10 μm. Bottom panel: Spatial regions corresponding to the red boxes in the top panel at indicated times. Yellow arrows: ‘tail’ structure at the end of the same cell; yellow arrowheads: microtubules feeding into tail; magenta arrowheads: microtubules present inside the cell. Scalebars: 2 μm. See also Supplementary Video 11.
Discussion
Relative to previous forms of structured illumination microscopy that provide comparable resolution, optical sectioning, and depth penetration13, instant SIM is 100x faster. Taking into account processing time, the speedup factor exceeds ten thousand. Due to its near-instantaneous acquisition and display, instant SIM enables imaging at rates exceeding state-of-the art commercial confocal microscopes, but with better resolution. Relative to STED and single-molecule imaging techniques, instant SIM is orders of magnitude faster over comparable fields of view. Nevertheless, spatiotemporal resolution, light efficiency, and depth penetration could be further improved.
Our 3D acquisition speed is limited by the inertia of our sample stage, but optical refocusing strategies26 circumvent this problem and could enable higher volumetric frame rates. Our optical setup is reminiscent of a swept-field confocal system27 with a single additional optical element (the microlens array in the emission path). This implies that commercial hardware might be easily modified to provide instant SIM capability, yet the correspondingly large number of optics in the emission path reduces signal levels and thus the maximum possible acquisition speed. We note that the multifocal excitation, pinholing, and local contraction operations necessary in instant SIM might be performed in a spinning-disk confocal geometry with many fewer emission optics, improving speed and signal levels accordingly. Using multifocal excitation necessarily implies a lower degree of optical sectioning than excitation with a single focused beam, due to pinhole cross-talk. Indeed, at the pinhole spacing we chose, instant SIM provided sectioning similar to spinning disk confocal microscopy and inferior to point- or line-scanning confocal microscopy (Supplementary Fig. 13). Sectioning strength and depth penetration would be improved if pinholes were spaced further apart, or if multiphoton excitation were used. Finally, if fluorescence emission is made to depend nonlinearly on excitation intensity, spatial resolution could be further increased28,29. The combination of photostable, reversibly-switchable fluorescent proteins24,30 with an instant SIM optical design may thus enable video-rate super-resolution imaging in the sub-100 nm regime.
Online Methods
Instant SIM Instrumentation
All optics were bolted to a 4′ × 8′ × 8″ optical table (Newport) in order to minimize external vibrations. A rapid automated modular microscope (Applied Scientific Instrumentation, RAMM-FULL-INVAR) served as the microscope body that housed sample, objectives (Olympus, PlanApo, NA 1.45 TIRF, for single cells, or UPLSAPO 60XS, NA 1.3 silicone oil, for zebrafish embryos samples), and automated XY stage equipped with Z axis piezo (Applied Scientific Instrumentation, PZ2300) that moved the axial position of the sample relative to the objective. For exciting fluorescence, two lasers were used: a 1W, 488 nm laser and a 0.5 W, 561 nm laser (Coherent, Genesis MX488-1000 STM and Genesis MX561-500 STM). Lasers were combined via a dichroic mirror (DC, Semrock, LPD01-488RU-25) and passed through an acousto-optical tunable filter (AOTF, AA Optoelectronic, Quanta Tech, AOTFnC-400.650-TN) for shuttering control. The intensity of each laser was maximized by tuning the rotation of half wave plates (Thorlabs, WPH10M-488 and AHWP05M-600) placed in front of each laser. After the AOTF, the beams were expanded 8.9x with a beam expander (Edmund, f = 45 mm, 64–837 and Thorlabs, f = 400, AC254-400-A), and passed through a converging microlens array (Amus, f = 1.86 mm, 1 mm thick, 25 mm diameter, antireflection coated over 400–650 nm, APO-Q-P222-F1.86), compensator plate (CVI Melles-Griot, PW1-2025-UV) and 6 mm thick dichroic mirror (TDC, Iridian Spectral Technologies, 488–561 DM). The compensator plate was used to cancel astigmatism that would otherwise arise when the focused beamlets passed through the tilted dichroic mirror. The resulting multifocal illumination was reimaged with a 1:1 telescope, consisting of 2 scan lenses (Scan lens 1 and 2, Special Optics, f = 190 mm, 55-S190-60-VIS) placed in a 4f configuration. An additional demagnification of 116.7x was achieved by reimaging the resulting excitation to the sample plane with tube lens (Edmund, f = 350 mm, NT49-289-INK) and objectives (f = 3 mm), also placed in a 4f configuration, and aligned so that the rear focal plane of the tube lens coincided with the front focal plane of Scan lens 2. Rotations of a 2-sided galvanometric mirror (Galvo, Nutfield Technology, QS-12 Galvo-Based Single-Axis Scan Set, P-PWR15 (power supply), S-0152 (Connector Kit), 10-2564 (mounting block); and Sierra Precision Optics, SPO9086 Rev B X-Mirror, double-coated) placed midway between each scan lens served to translate the multifocal array at the sample plane, thus covering the imaging field.
Several of these optics were reused for emission, as fluorescence was collected along the same path, through objective, tube lens, scan lenses 1 and 2, and galvanometric mirror, before reflection from the 6 mm thick dichroic mirror. Since the galvanometric mirror introduced an equal and opposite rotation angle on the return path (‘descanning’), the multiple fluorescence foci produced at the focus of scan lens 1 were stationary. A pinhole array, with pinhole spacing equivalent to the spacing between microlenses in the converging microlens array (Photosciences, Chrome on 0.090″ thick quartz, 222 μm pinhole spacing, 40 μm pinhole diameter) placed at the front focal plane of scan lens 1 served to reject out-of-focus fluorescence emission present around each fluorescent beamlet. The resulting filtered beamlets were relayed to a secondary microlens array via a 1:1 imaging telescope (Relay lenses, Thorlabs AC508-300-A-ML) and focused through a second microlens array (Amus, f = 0.93 mm, 1 mm thick, 25 mm diameter, antireflection coated over 400–650 nm, APO-Q-P222-F0.93) that locally contracted each fluorescent focus 2x, while preserving the orientation of each foci (see further discussion below and Supplementary Fig. 3). The locally scaled multifocal array thus produced was reimaged to a scientific grade complementary metal-oxide semiconductor camera (PCO-TECH, pco.edge) via additional scan lenses (Scan Lenses 3 and 4, Special Optics, f = 190 mm, 55-S190-60-VIS) placed in a 4f configuration. By placing the galvanometric mirror at the midpoint between scan lenses, fluorescence was rescanned onto the camera, producing a super-resolution, sectioned image of the sample plane. A filter wheel (Sutter, FG-LB10-BIQ and FG-LB10-NW) and notch filters (Semrock, NF03-488E-25 and NF03-561E-25) placed immediately before the cameras served to reject excitation laser light. These optical elements are shown in Supplementary Fig. 1.
Our magnification of 116.7x resulted in an imaging pixel size of 55.5 nm. Excitation power varied between 5–50 W/cm2 depending on the particular sample.
Placement of the second microlens array
In order to achieve a 2x local contraction of each fluorescent focus in the descanned path while retaining the orientation of each focus, we placed the second microlens array one focal length before the image that would have been produced by Scan lens 3 had we not placed the second microlens array in the optical path. The justification for this choice can be derived mathematically starting from the Gaussian form of the thin lens equation:
Where o is the object position, i the image position, and f the focal length of the lens. The magnification M = −i/o
We desire that M = 0.5, i.e that the image of each fluorescent foci is a locally contracted but upright (non-inverted) version of the foci prior to passage through the microlens array. Substituting i = −0.5 o in the lensmaker equation implies that o = −f (the object is virtual, placed one focal length away from the microlens array on its downstream side) and i = − 0.5 f (the image is formed 0.5 f from the microlens array, on the same side as the object). A geometric proof is presented in Supplementary Fig. 3.
Brightfield Imaging
The instant SIM produces an optically sectioned fluorescence image at the camera, but we often found it useful to acquire widefield (non-sectioned) brightfield images for scouting purposes. We implemented brightfield imaging by illuminating the sample from above with a lamp (Danray Products LMP-24M), and directing the transmitted light to the camera with removable mirrors (Thorlabs, MFF001), bypassing most of the optics in the fluorescence path (Supplementary Fig. 2).
Alignment of Instant SIM Optics
Most optics in Supplementary Fig. 1 are straightforward to align. The most sensitive alignment involves finding the correct rotation angle of the emission path microlens array about the optical axis. The instant SIM ‘paints’ a 2D image with a 1D scan of a 2D grid of illumination points, so the angle between the grid axes and the scan direction is critical for uniform illumination of the image. Small rotations (barely mechanically perceptible) of the emission path microlens array from its optimal position can cause substantial striping artifacts in our images. In practice, we find the best alignment procedure is to image a thin, uniform fluorescent solution while carefully adjusting the rotation angle of the emission path microlenses and inspecting the resulting images for striping. The emission path pinholes and the excitation microlenses can then be (re)aligned to the emission path microlenses as required, since all three of these optics are mounted on removable magnetic stages (Thorlabs, KB3X3). One aligned, the system is stable and maintenance-free.
Instant SIM Instrument Control and Data Acquisition
Our previous MSIM implementation13 controlled data acquisition with a custom Python script, but relied on manufacturer-supplied software and manual adjustment of hardware for exploring samples and identifying regions of interest. This is simple to implement and allows high-performance acquisition, but requires well-trained users and often leads to substantial bleaching of the sample during the scouting process. The instant SIM is substantially faster, and a unified graphical interface was essential for taking full advantage of the device’s capability for real-time super-resolution while minimizing sample perturbation. The instant SIM hardware is computer-controlled using custom software written in the Python language (ref.31 and www.scipy.org), and is freely available at code.google.com/p/msim.
We implemented graphical controls using the built-in Python library Tkinter. Image data is displayed in real time, on-screen using the Python libray pyglet (http://www.pyglet.org/). Three-dimensional volume data and timelapses are saved to disk as TIF files, and automatically opened in ImageJ (http://rsb.info.nih.gov/ij/) as virtual stacks. We controlled the pco.edge camera using a manufacturer-supplied DLL and the built-in Python library ctypes. The Sutter filter wheel is controlled through a virtual serial port over USB, and the X/Y positions of the ASI stage and the Z-position of the ASI objective stepper motor are interrogated through a physical serial port. Both the virtual and physical serial ports are controlled using the Python library pyserial (http://pyserial.sourceforge.net/). The galvanometric mirror, AOTF, piezo stage, brightfield bypass mirrors, and trigger signals to the PCO camera are all controlled by external voltages. A 16-channel analog-out card (National Instruments, NI 9264) provides these voltage signals, and is controlled through a manufacturer-supplied DLL and the Python ctypes library.
The camera and the AOTF have low input impedance. The analog-out card specifications lead us to believe that it could drive enough current to work with this impedance, but in practice we found this false. We thus built a buffer amplifier using an op-amp, box, and power supply (Radio Shack) to provide enough current to drive the camera, two channels of the AOTF, and the AOTF blanking input.
High-speed operation of the instant SIM requires careful optimization of control voltages. Supplementary Fig. 4 shows an example of control voltages for a 5-timepoint, 2-color timelapse acquisition. Several features of these voltage signals are worth noting:
Even in the fully ‘off’ state, the AOTF still transmits a non-negligible fraction of the excitation laser, so between acquisitions, we leave the galvo mirror ‘parked’ in a highly deflected position to protect the sample from unnecessary phototoxicity. The ‘unparking’ of the galvo takes the longest time, so this occurs first.
We operate the pco.edge camera in ‘rolling shutter’ mode, which provides lower noise and higher speed relative to ‘global shutter’ mode. However, in rolling shutter mode, a lag exists between the external trigger signal and the moment when the entire camera chip is exposing. This lag is shorter than the galvo ‘unparking’ time, so it occurs next.
The galvanometric mirror vibrates if it scans too slowly, causing a substantial striping artifact in image data. For this reason, we only tune galvo scan speed over a narrow range, and implement longer exposures by accumulating several sweeps of the galvo. In Supplementary Fig. 4, the galvo executes three sweeps per exposure. Between exposures, the galvo waits in a deflected position for whatever operation is the limiting factor. In this case, the rolling shutter mode time lag is the limiting factor between timepoints.
The 488 and 561 lasers alternately illuminate the sample, providing fast 2-color 2D timelapse operation. All our experiments (as illustrated in Supplementary Fig. 4) used maximally permissive notch emission filters that eliminate 488 nm and 561 nm excitation light while transmitting all other wavelengths. This combination of filters works well for probes which may be distinguished by excitation alone, such as GFP and mCherry at similar probe density. A sample brightly tagged with td-Tomato and weakly tagged with GFP might show substantial crosstalk between channels, and require switching between two bandpass filters for each timepoint. The filter wheel we used has reasonably fast switching time (~10–50 ms), but switching between filters would still slow acquisition substantially.
Supplementary Fig. 5 shows an example of control voltages for a 4-slice, 2-color Z-stack. Voltages are similar to the 2D 2-color timelapse, with a few differences:
The Z axis piezo stage control voltage is no longer constant. The piezo can be controlled through the serial port or an external voltage, but we found the external voltage control faster and more precise. The manufacturer specifies that a 5 volt signal will move the piezo to the same position as a serial port command of ‘0’, but we found the required ‘0’ signal to be 5.08 volts. It is unclear whether this difference is due to the Z axis piezo or our analog-out card.
The limiting factor between timepoints alternates between the rolling shutter mode time lag, and the time period we allow for axial motions of the Z piezo stage. We allow a ~10 ms delay for stage motion. Moving the piezo stage this rapidly requires careful optimization of the voltage waveform, as we found that fast movements of the piezo would otherwise cause substantial vibration with a resonant period of ~7.5 ms. A naive approach might simply move the piezo and wait for vibration to damp out, but we found this takes many tens of ms, and would thus limit the speed of our Z-stacks. An improved approach might feed the piezo stage a smoothed signal to reduce vibration, but we found this still leads to unacceptable vibration over many resonant periods. The best approach we found applies three voltage steps per Z piezo motion. Supplementary Fig. 6 shows a zoomed view of our piezo control voltage.
The first voltage step overshoots the desired position, causing faster motion of the stage but also substantial vibration. The second voltage jump overshoots even further, and causes further vibration. However, these two motions are separated by half of the resonant vibration period, so the vibrations largely cancel. The last voltage jump brings us back to the desired voltage, separated by another half-resonant period. We experimented with various jump amplitudes, and found the fastest motion with least vibration resulted when the first jump is twice as large as the desired motion, the second jump takes the stage to 2.4x the desired motion, and the final jump returns the stage to the desired position. The combination of voltage overshoot and vibration cancellation allows us to move the stage rapidly and precisely in just over one resonant vibrational period.
All volumes presented in this paper contained 12 imaging planes, with 0.25 μm spacing between planes.
Deconvolution
Although the raw images produced by instant SIM already possess improved resolution relative to conventional widefield fluorescence images, achieving the full 2x resolution gain in SIM requires deconvolution.
For all 3D deconvolution (Fig. 2, Supplementary Videos 1–9, we used the Richardson-Lucy algorithm32,33, assuming a symmetrical PSF:
For i = 1, 2, .... N
End
where Blur() indicates convolution with the PSF. We assumed theoretical Gaussian PSFs with lateral and axial FWHMs derived from measurements on 100 nm yellow green beads (Table 1 lists the parameters used for instant SIM, line-scanning confocal and spinning disk confocal images), and ran each deconvolution for 40 iterations. For 2D deconvolution, we used the Huygens deconvolution software (Scientific Volume Imaging) which seemed to give better results than our deconvolution algorithm. For deconvolving ER datasets (Fig. 3), we used the following parameters: ‘confocal’ PSF type, 55.5 nm pixel dimensions, 1.45 NA objective, 1.51 immersion oil index of refraction, 1.33 index of refraction of the imaging media, and 40 iterations. For deconvolving fish embryo datasets (Fig. 4), we used the following parameters: ‘confocal’ PSF type, 55.5 nm pixel dimensions, 1.3 NA objective, 1.4 immersion oil index of refraction, 1.33 index of refraction of the imaging media, and 40 iterations. It is unclear what the Huygens processing algorithm is, but we suspect it is Richardson-Lucy deconvolution with presmoothing and a regularization step.
Image Display
All movies were bleach-corrected using an ImageJ plugin that normalized the intensity values in each image to the average intensity present in that image. The plugin is available online here: http://www.embl.de/eamnet/html/bleach_correction.html
For Figure 3, images were displayed using the ‘HotRed’ look up table for ImageJ. All other images are presented in grayscale.
Molecular biology and cloning procedures
In order to label peroxisomes we replaced the mTurqoise2 fragment with the GFP or Cherry fragment as NheI/BsrGI in frame in the peroxisome targeting vector (ref.34, Addgene).
Endoplasmic reticulum was labeled with two constructs marking either the whole ER entity or the transitional ER exit sites for dual labeling with peroxisomes. For whole ER we used the translocon subunit SEC61A1. Human SEC61A cDNA was obtained from ASUS repository (Arizona State University) (clone HsCD00350497) and subcloned by PCR into the EGFP C1 vector as SalI/KpnI using the following set of primers: SEC61A Forw ACAGTCGACATGGCAATCAAATTTCTG; Sec61A Rev ATAGGTACCTTAGAAGAGCAGGGCCCCC. For the labeling of transitional ER exit sites we used the Sec16B-GFP (pmGFP-Sec16S, ref.35) chimera (obtained from Addgene).
Labeling of mitochondrial subunits with fluorescent proteins was performed by tagging the precursors at the c-terminus. The outer mitochondrial membrane was labeled with the human Tom20 subunit of the translocator with mCherry and for mitochondrial nucleoids we used the mouse mitochondrial Transcription Factor A (TFAM). Untagged cDNAs were obtained from Origene and the coding sequences were PCR amplified digested with BglII/Sac-1 and subcloned in frame into pmCherry-N1 or EGFP-N1 vectors (Clontech). For PCR amplification we used the following sets of primers: Tom20 Forw CACAGATCTATGGTGGGTCGGAACAGCGC; Tom20 Rev ATAGAGCTCTTCCACATCATCTTCAGCC; TFAM Forw CACAGATCTATGGCGCTGTTCCGGGGAAT; TFAM Rev ATAGAGCTCATGCTCAGAGATGTCTCCGG.
Rab8A tagged with Cherry was a kind gift of Dr. Lymarie Maldonado-Baez (NHLBI, NIH). H-Ras was obtained from a cDNA library created from MRC-5 human lung fibroblasts by RT-PCR procedures using the Superscript kit (Life Technologies) and the following primers: H-Ras Forw TTCGAATTCAGAGGAGCGATGACGGAATATAAGCTGGT; H-Ras Rev GGTGGATCCCGGGGTGACTGGGCTCCAGCAG. The cDNA was cloned in frame as EcoRI/BamH1 in the EGFP C1 vector tagging the molecule at the N-terminus.
Bead and fluorescent slide samples
Bead samples were prepared as previously described13, except that we mounted the beads in the immersion oil used for each microscope. When conducting optical sectioning experiments (Supplementary Fig. 13) we acquired z stacks of a yellow-green, thick fluorescent plastic slide (Chroma Technology, 92001).
Cell culture and cellular sample preparation
Routine cell culture procedures were followed, keeping cells in DMEM supplemented with 10% FBS (Life technologies) in a CO2 supplied incubator (5%) at 37°C. Experiments were performed in a human transformed cell line designated as MRL-TR. This line was derived from primary lung MRC-5 fibroblasts (ATCC). For live imaging, cells were plated in either 4-well chambers (Labtek II) with coverglass bottoms, or in Bioptechs dishes (#1.5 glass thickness in either case). DMEM supplemented with 10% FBS (without phenol red) was used as the imaging medium in all live cell experiments. Data collection for Fig. 2, Fig. 4, and Supplementary Videos 1–9, 11 was performed at room temperature. For ER experiments (Fig. 3, Supplementary Video 10), we used a Bioptechs Delta T system to perform experiments at 37°C.
Comparative confocal microscopy
We performed line-scanning confocal imaging on a Live 5 duoscan microscope (Zeiss), equipped with galvanometric scanning mirrors and 488 nm (for GFP excitation) and 561 nm (for Cherry excitation) diode lasers. A Planapo 100X oil immersion lens with 1.46 NA (Zeiss) was used for line-scanning confocal imaging. The confocal aperture was set to 1 AU for both colors, Kalman averaging was set to 2, and scanning speed ranged from 20 to 40 frames per second (fps) depending on the intensity of the signal. In order to obtain imaging speeds equivalent to instant SIM, scanning was performed at a field of view of 512×512 pixels, yielding a pixel size of 130 nm. Acquisition was simultaneous with appropriate pre-acquisition single channel corrections for cross excitation, bleed through and saturation. A 565 nm RTNT1 NFT dichroic mirror was used to split emitted light and direct it into appropriate acquisition channels using a BP 495–555 nm (bandpass) filter for GFP emission and a LP 580 nm (longpass) filter to detect signal from mCherry. For z stacks, a piezo focus lens positioner (Physik Instrumente, P-725.2CD and E-665) was used to obtain 250 nm axial steps, thus obtaining 12 slices (for 3 μm depth, identical to instant SIM imaging).
Spinning disk confocal imaging was performed on a TE2000 microscope stand (Nikon) equipped with CCD (Photometrics CoolSnap HQ2) and CSU- X10 spinning disc confocal head. Images were acquired using a 60X, 1.49 TIRF objective (Nikon), with exposure times ranging from 20–50 ms per frame. Imaging volumes were acquired using a piezoelectric stage (Mad City Labs). All hardware was controlled with MetaMorph (Molecular Devices).
When performing comparative spinning disk, line-scanning confocal experiments, and instant SIM experiments (Fig. 2, Supplementary Fig. 11, Supplementary Videos 1–3), we fixed the imaging volume thickness (3 μm) and number of timepoints (20) while adjusting laser power to produce the brightest signals we could without overly bleaching the sample.
Zebrafish embryo sample preparation
For live zebrafish imaging, Tg(XlEef1a1:dclk2-GFP)io008 embryos13 were collected by natural spawning and maintained at 28°C. Prior to imaging, embryos at 72–78 hpf were anesthetized in Tricaine (Sigma, E10521) at a final concentration of 600 mM in embryo media (60 mg RedSea Coral Pro Salt (Drs Foster and Smith Pet Supplies) per liter ddH2O). Anesthetized embryos were immobilized in 0.75% low-melt agarose (Cambrex, 50080) on 25 mm #1.5 round coverslips (Warner Instruments, 64-0715), placed in a round coverslip holder (ASI, I-3033-25D), and imaged at room temperature.
Supplementary Material
Acknowledgments
We thank G. Patterson for encouragement and the use of his cell culture facilities, L. Maldonado-Baez (US National Heart, Lung, and Blood Institute) for the Cherry tagged Rab8A plasmid, C. Combs for lending us his objective lenses, S. Parekh for useful discussions and for help in sample preparation, E. Tyler and A. Hoofring for help with illustrations, and Y. Wu for help with Huygens deconvolution software. This work was supported by the Intramural Research Programs of the US National Institutes of Health National Institute of Biomedical Imaging and Bioengineering, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Heart, Lung and Blood Institute, and the National Institute of Child Health and Human Development.
Footnotes
Author contributions
A.G.Y. and H.S. conceived idea and designed optical system. A.G.Y. built optical system, designed and implemented data acquisition software, and performed simulations. A.G.Y., P.C., D.D.N., J.H., R.S.F., and H.S. acquired data. P.C., D.D.N, R.S.F., and A.C. provided guidance on biological experiments. P.C., D.D.N., J.H., P.W., and R.S.F. prepared samples. P.C., D.D.N., J.H., R.S.F., and A.C. provided biological reagents. All authors analyzed data. A.G.Y., P.C., and H.S. wrote the paper with input from all authors. H.S. supervised research.
‘Competing Financial Interests’
We have no competing financial interests.
References and Notes
- 1.Hell SW. Far-Field Optical Nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 2.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–505. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westphal V, et al. Video-Rate Far-Field Optical Nanoscopy Dissects Synaptic Vesicle Movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 5.Huang F, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nature Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heintzmann R, Cremer C. Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating. Proc SPIE. 1999;3568:185–196. [Google Scholar]
- 7.Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 8.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao L, Kner P, Rego EH, Gustafsson MGL. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- 10.Sheppard CJR. Super-resolution in Confocal Imaging. Optik. 1988;80:53–54. [Google Scholar]
- 11.Muller CB, Enderlein J. Image Scanning Microscopy. Physical Review Letters. 2010;104:198101. doi: 10.1103/PhysRevLett.104.198101. [DOI] [PubMed] [Google Scholar]
- 12.Heintzmann R, Benedetti PA. High-resolution image reconstruction in fluorescence microscopy with patterned excitation. Applied Optics. 2006;45:5037–5045. doi: 10.1364/ao.45.005037. [DOI] [PubMed] [Google Scholar]
- 13.York AG, et al. Resolution Doubling in Live, Multicellular Organisms via Multifocal Structured Illumination Microscopy. Nature Methods. 2012;9:749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MGL. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc Natl Acad Sci USA. 2012;109:5311–5315. doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown TA, et al. Superresolution Fluorescence Imaging of Mitochondrial Nucleoids Reveals Their Spatial Range, Limits, and Membrane Interaction. Mol Cell Biol. 2011;31:4994–5010. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurm CA, et al. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc Natl Acad Sci U S A. 2011;108:13546–13551. doi: 10.1073/pnas.1107553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt R, et al. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 20.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neuspiel M, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 22.Yonekawa S, et al. Sec16B is involved in the endoplasmic reticulum export of the peroxisomal membrane biogenesis factor peroxin 16 (Pex16) in mammalian cells. Proc Natl Acad Sci U S A. 2011;108:12746–12751. doi: 10.1073/pnas.1103283108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wozniak MJ, et al. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grotjohann T, et al. rsEGFP2 enables fast RESOLFT nanoscopy of living cells. eLIFE. 2012;1:e00248. doi: 10.7554/eLife.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundd P, et al. ‘Slings’ enable neutrophil rolling at high shear. Nature. 2012;488:399–403. doi: 10.1038/nature11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. Aberration-free optical refocusing in high numerical aperture microscopy. Optics Letters. 2007;32:2007–2009. doi: 10.1364/ol.32.002007. [DOI] [PubMed] [Google Scholar]
- 27.Brakenhoff GJ, Visscher K. Confocal imaging with bilateral scanning and array detectors. Journal of Microscopy. 1992;165:139–146. [Google Scholar]
- 28.Rego EH, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc Natl Acad Sci USA. 2011;109:E135–E143. doi: 10.1073/pnas.1107547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grotjohann T, et al. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature. 2011;478:204–208. doi: 10.1038/nature10497. [DOI] [PubMed] [Google Scholar]
- 31.Oliphant T. Python for Scientific Computing. Comput Sci Eng. 2007;9:10–20. [Google Scholar]
- 32.Richardson WH. Bayesian-Based Iterative Method of Image Restoration. JOSA. 1972;62:55–59. [Google Scholar]
- 33.Lucy LB. An iterative technique for the rectification of observed distributions. Astronomical Journal. 1974;79:745–754. [Google Scholar]
- 34.Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.