Abstract
Background
Despite the strong evidence of HPV infection as the etiological agent in a subset of oral cancer, oral α-HPV detection is rare in healthy individuals, and little is known of the existing of novel HPV types in oral cavity.
Objective
We determined whether novel HPV types can be isolated from oral rinse samples collected from healthy individuals.
Study design
We performed rolling circle amplification (RCA) coupled with degenerated PCR assay on 48 oral rinse samples to amplify novel HPV types. Full length HPV DNA was cloned using long range PCR. Quantitative type specific Taqman assays were used to determine the prevalence of novel HPV types in 158 archived oral tissue samples.
Results
We were able to isolate four novel human papillomavirus types. Full length HPV DNA was cloned for three of the four novel HPV types. All four HPV types belong to the genus Gammapapillomavirus (γ-PV), where HPV 171 is most closely related to HPV 169, showing 88% similarity; HPV 172 is most closely related to HPV 156, showing 70% similarity; HPV 173 is most closely related to HPV 4, showing 73% similarity; oral sample lavage (OSL) 37 is most closely related to HPV 144, showing 69% similarity. Finally, we showed that HPV 173 was rarely present in oral tissues (2/158), HPV 172 was only detected in normal oral tissues (25/76), and HPV 171 was more prevalent in malignant oral tissues (17/82 vs 10/76, p=0.21).
Conclusions
Novel γ-HPV types are present in oral cavity of healthy individuals.
Keywords: HPV, oral lavage, oral squamous cell carcinoma
Background
Human papillomavirus (HPV) infection is the etiological agent for cervical cancer. Recent studies suggest that it is also the causal agent for a subset of head and neck cancer 1–3, the prevalence of HPV infection in oral squamous cell carcinoma (OSCC) ranges from 10–20% in oral cavity cancer to 6–60% in oropharyngeal cancer 4–11, with HPV 16 being the most prevalent type. Detection of HPV 16 infection in oral exfoliated cells increases the odds of oropharyngeal cancer more than 14 fold 3, and HPV integration and expression of E6 and E7 oncogenes have been detected in OSCC tissues 12–15. In summary, HPV is a significant etiological factor for OSCC, with potential to influence the prevention, diagnosis and treatment of OSCC.
On the other hand, oral α-HPV detection so far is rare in healthy individuals. A recent systemic review of literature reported the prevalence of any oral HPV detected below 5%, and HPV 16 accounted for 28% of all HPV detected in the oral region 16, 17. We recently determined the incidence and prevalence of oral HPV infection in a cohort of male university students 18. The prevalence at enrollment was 7.5% and 12-month cumulative incidence was 12.3%. However, our published study, like most others, was limited to testing for α-HPV types. As current evidence indicates that types from other genera infect oral epithelium19, the true prevalence of HPV infection in oral cavity is likely underestimated.
Of over 160 HPV types identified so far, most are categorized into one of three genera based on sequence differences: Alphapapillomavirus (α-PV), Betapapillomavirus (β-PV), and Gammapapillomavirus (γ-PV). Thus far, only a few β-PVs 20–22 but no γ-PVs have been shown to participate in tumorigenesis. Recent studies suggest the presence of novel beta and gamma genra HPV types present in oral cavity. For example, Bottalico et al identified 12 novel gamma and 8 beta HPV types in an HIV+ population, and a novel HPV type 120 was identified in the oral rinse sample that has showed a wide range of tropism19.
To determine whether novel HPV types are present in oral rinse samples, we first used rolling circle amplification to enrich for full length circular HPV genomes then used degenerate PCR to identify novel HPV types present in oral rinse samples collected from healthy young men.
Objectives
We plan to isolate novel HPV types from oral cavity of healthy individuals and determine their prevalence in normal and malignant oral tissues.
Study design
Clinical samples
A total of 48 archived oral rinse samples from 41 subjects were selected for the isolation of novel HPV types. These samples were selected from a longitudinal study investigating the natural history of HPV infection in male population18. The demographics of the 41 subjects whose oral rinse samples were used in the current study was similar to the entire study population published before 18. Briefly, oral specimens were collected via gargle/rinse and swabbing of the oropharynx. The median age of subjects was 20, and over 80% of them were Caucasians.
In addition, a total of 158 oral tissue blocks were selected from University of Washington Department of Pathology, including 76 normal oral tissue blocks (56 from oral cavity and 20 from oropharynx), 82 malignant oral tissue blocks (66 from patients with oral squamous cell carcinoma (OSCC), 16 from patients with oropharyngeal squamous cell carcinoma (OPSCC)). Cancer patients on average were significantly older than normal patients (58.5 vs 45.1, p<.0001). The majority of the population was male (63.5%) and Caucasian (65.7%).
Genomic DNA isolation
Genomic DNA was extracted from oral rinse samples by the QIAamp DNA mini kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). Tissue sections were prepared by the Department of Pathology of University of Washington. Special care was taken to minimize cross-contamination between tissue blocks: microtome was cleaned and blade was replaced after processing each block/ Total DNA was extracted from 80 µm oral tissue block sections using the RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE Tissues according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA).
Multiply primed Rolling circle amplification (MP-RCA)
MP-RCA was performed on each oral rinse DNA sample with the TempliPhi 100 amplification kit (GE healthcare, Piscataway, NJ) with modifications optimized for papillomavirus amplification 23. Specifically, 1 µl purified sample DNA (∼100–300 ng) was denatured at 95ºC for 3 minutes in 5 µl Sample Buffer, then cooled to 4ºC. Subsequently, TempliPhi premix containing 5 µl Reaction Buffer, 0.047 µl of 50 mM dNTPs, and 0.2 µl enzyme mix was added to each denatured sample. Reaction was performed at 30ºC for 16 hrs, then heat inactivated at 65ºC for 10 minutes. For the first 33 oral rinse samples, every three RCA reactions were pooled for subsequent consensus PCR reaction, while consensus PCR was performed for individual RCA samples for oral rinse samples 34–48. Therefore, a total 26 RCA samples went through consensus PCR reactions.
HPV detection by consensus PCR amplification
Four consensus PCR assays were performed on MP-RCA amplified samples, following published protocols: nested PGMY/GP5+/6+ PCR assay 24, FAP 25, CP 26, and a newly described broad spectrum (BS) PCR assay 27. PCR product was cloned using TA cloning kit (Invitrogen, Life technologies, Grand Island, NY) and at least two clones from each PCR product were sequenced. The presence of HPV sequence was determined through BLAST search (http://blast.ncbi.nlm.nih.gov/).
Full length HPV cloning by long range PCR and sequencing
The cloned HPV fragment sequence was used to design primers for long range PCR reaction using Stratagene kit (Stratagene, La Jolla, CA). The presence of full length HPV PCR product (∼6–7kb) was subsequently cloned and sequenced. The primers used for long range PCR are listed in Table 1.
Table 1. Primers and probes used in this study.
Both primers used to clone new HPV types as well as primers and probe for Taqman assays were listed.
| Primers and probe sequences | |
|---|---|
| HPV171 | |
| Long range PCR | AAGCAGGATTATGCCCTCCT GGACCACCTCTGTCAATTTCA |
| Real time PCR | F: TGTGTGTGTAGTTGCTTCAGAAGGA Probe: 6FAM-CAATACACCTGCTGCAGC-MGB R: GGAGAGATCGGTCGACAAAAGT |
| HPV172 | |
| Long range PCR | TGGCCTGTTGCTCCTATACC CACACCACCAACAGGACAGT |
| Real time PCR | F: AACCAGCACCGGTTACTATAAAAGA Probe: 6FAM-TGTTCTGCAAAGCTC-MGB R: CCAGCCATCGAAAAGAGAGAA |
| HPV173 | |
| Long range PCR | CCTAAGGGTCCTCCTCTGCT CCAGTTATTGGAGAGCATTGG |
| Real time PCR | F: AGGAGGTGTTTTCTATTTAGTTCGAAGT Probe: 6FAM-AGTGGAAAGGGTGTTGCA-MGB R: GTCCCTCTCATTGTTCAATCATACA |
Phylogenetic analysis of novel HPV types
Maximum likelihood tree was estimated by PhyML v3 30. ModelTest v3.7 29 reported GTR+I+G substitution model as the best fit model. HPV16, an α-HPV type was used as the out group for the tree analysis, and the tree was based on L1 sequences of 50 gamma HPV reference clones (www.hpvcenter.se).
HPV detection by real time PCR in oral tissue samples
Type-specific Taqman assays were designed for HPV 171, 172 and 173 based on the HPV E6/E7 region. Primers and probe sequences were listed in Table 1. These Taqman assays were extensively validated using plasmids containing individual HPV type E7 gene. Each assay can specifically detect down to 1 copy HPV but did not detect nonspecific HPV types present at 10,000 copies. Normalization of input DNA was performed using the quantitative Taqman assay on Alu sequence. Absolute quantification was performed on archived oral tissue samples for each HPV and Alu 31. For each run, both negative (K562 genomic DNA) and positive (plasmid containing specific HPV type) controls were included. Serial dilutions of human genomic DNA as well as full length HPV plasmids of known concentration were used as standard curves.
Statistical analysis
Pearson’s Chi-Square Tests were used to compare the prevalence of HPV detection. Fisher’s Exact Tests were used when one or more cells had expected counts less than five. HPV viral load was log10 transformed in order to normalize the distribution for all statistical analyses. T-tests were used to compare the level of HPV detection among samples with detectable HPV viral loads. A two-sided 0.05 test level determined statistical significance for all analyses. All analyses were conducted using SAS version 9.2 (SAS Institute Inc., Cary, NC).
Results
MP-RCA preferentially amplifies circular HPV DNA
In order to improve the sensitivity to detect low copy number novel HPV types present in oral rinse samples, we determined whether MP-RCA can preferentially amplify circular HPV over human genomic DNA in oral rinse samples. We first performed RCA on Siha DNA and Siha DNA mixed with HPV 16 plasmid. Both β-actin (ACTB) and HPV 16 was quantitated by real time Taqman assays before and after MP-RCA. The enrichment of viral DNA relative to human genomic DNA after MP-RCA amplification was calculated using the 2-ΔΔCt relative quantification method (ABI, Foster City, CA). We showed that both β-actin (ACTB) and linear HPV 16 present in Siha DNA were either not amplified or modestly amplified (4 folds) respectively, while HPV 16 plasmid DNA was amplified 40 fold (Table 2). Subsequently, we determined the efficiency of MP-RCA to amplify circular HPV DNA in 9 clinical samples, including 3 anal swabs, 3 cervical swabs, and 3 oral rinse samples. Regardless of the source of samples, MP-RCA preferentially amplified HPV DNA 100-107 fold. Of three oral rinse samples, HPV 16 was amplified 105 and 106 fold in CS7 and CS8 respectively, and HPV 59 was amplified 7000 fold in CS9. We concluded that multiply primed RCA could be used to efficiently amplify HPV viral DNA in clinical samples.
Table 2. Multiply primed RCA preferentially amplifies HPV DNA from a variety of clinical samples.
Eleven samples went through MP-RCA. Both HPV and β-actin were quantified before and after RCA reaction, relative enrichment for HPV is calculated.
| Sample | Diagnosis | Virus present | HPV | ACTB | Enrichment* | |||
|---|---|---|---|---|---|---|---|---|
| ID | Type | Before RCA (Ct) |
After RCA (Ct) |
Original (Ct) |
After RCA (Ct) |
|||
| C1 | Siha | HPV 16 | 25.81 | 23.22 | 25.90 | 25.34 | 4.1 | |
| C2 | Siha+HPV16p | HPV 16 | 26.20 | 19.37 | 26.98 | 25.48 | 40.2 | |
| CS1 | Anal swab | Neg | HPV 16 | 22.50 | 15.34 | 31.19 | 35.66 | 3184.0 |
| CS2 | Anal swab | Neg | HPV 16 | 31.29 | 27.95 | 30.35 | 34.20 | 146.3 |
| CS3 | Anal swab | Neg | HPV 16 | 21.02 | 12.44 | 29.89 | 33.30 | 4093.9 |
| CS4 | Cervical swab | Neg | HPV 16 | 29.99 | 27.52 | 25.62 | 33.93 | 1756.4 |
| CS5 | Cervical swab | Neg | HPV 16 | 37.18 | 24.88 | 26.19 | 30.54 | 103418.3 |
| CS6 | Cervical swab | Neg | HPV 16 | 32.71 | 14.20 | 25.82 | 30.79 | 11725814.7 |
| CS7 | Oral lavage | Neg | HPV16 | 33.48 | 14.07 | 25.71 | 25.39 | 558036.5 |
| CS8 | Oral lavage | Neg | HPV16 | 36.70 | 18.34 | 25.30 | 26.59 | 822696.1 |
| CS9 | Oral lavage | Neg | HPV 59 | 29.71 | 21.64 | 28.57 | 33.28 | 7033.4 |
Enrichment was calculated using the 2−ΔΔCt method, where ΔΔCt= [CtHPV(after RCA)- CtHPV(before RCA)]- [CtACTB(after RCA)-CtACTB(before RCA)]
Identification and cloning of three novel gamma HPV types in oral rinse samples
Because different consensus PCR primers are optimized to detect different HPV types, we used four different PCR systems to detect HPV present in oral rinse samples after MP-RCA amplification. Of 26 MP-RCA samples, 13 were HPV positive and 17 different HPV types were detected. PGMY/GP5+/6+ PCR detected HPV in 2 samples, FAP PCR system detected HPV in 12 samples, CP PCR detected HPV in 7 samples, and BS PCR detected HPV only in one sample (Table 3). Of all the HPV types detected by the four PCR systems, 4 HPV types detected by the FAP PCR system (from specimens 19–21, 28–30, 37, and 42) represented potential new HPV types. Based on FAP PCR product sequence (∼500 bp), these HPV types belong to the gamma genus, showing 70% homology to existing HPV types in the database.
Table 3. Degenerate HPV PCR on MP-RCA samples.
Forty-eight clinical samples went through MP-RCA reaction. These samples were HPV genotyped by liquid bead microarray assay (LBMA) assay 37. Either pooled RCA reactions (every three samples, samples 1–33) or individual RCA reactions were used for subsequent degenerate PCR reactions: PGMY/GP5+/6+, FAP, CP and BS systems. HPV types detected by each PCR system were listed.
| Sample Number | PCR by LBMA | MP-RCA | PGMY/GP5+/6+ | FAP | CP | BS |
|---|---|---|---|---|---|---|
| 1 | 31, 39 | 1 | - | - | - | - |
| 2 | 16, 31, 33, 35, 39 | 2 | ||||
| 3 | 18 | 3 | ||||
| 4 | 51, 61, 84 | 4 | 16 | 120 | 120 | 120 |
| 5 | 16 | 5 | ||||
| 6 | 35, 39 | 6 | ||||
| 7 | 51 | 7 | 16 | 16 | - | - |
| 8 | 16 | 8 | ||||
| 9 | 16 | 9 | ||||
| 10 | 6 | 10 | - | - | - | - |
| 11 | 33, 39, 81 | 11 | ||||
| 12 | 31 | 12 | ||||
| 13 | 16 | 13 | - | 51 | 55 | - |
| 14 | 18 | 14 | ||||
| 15 | 51, 55, 59 | 15 | ||||
| 16 | - | 16 | - | - | - | - |
| 17 | - | 17 | ||||
| 18 | - | 18 | ||||
| 19 | - | 19 | - | 171 | - | - |
| 20 | - | 20 | ||||
| 21 | - | 21 | ||||
| 22 | - | 22 | - | - | - | - |
| 23 | - | 23 | ||||
| 24 | - | 24 | ||||
| 25 | - | 25 | - | - | - | - |
| 26 | - | 26 | ||||
| 27 | - | 27 | ||||
| 28 | - | 28 | - | 172 | 17 | - |
| 29 | - | 29 | ||||
| 30 | - | 30 | ||||
| 31 | - | 31 | - | - | - | - |
| 32 | - | 32 | ||||
| 33 | - | 33 | ||||
| 34 | - | 34 | - | FA88 | - | - |
| 35 | - | 35 | - | 57b | 57b | - |
| 36 | - | 36 | - | - | 27 | - |
| 37 | - | 37 | - | OSL37 | - | - |
| 38 | - | 38 | - | - | - | - |
| 39 | - | 39 | - | - | - | - |
| 40 | - | 40 | - | - | - | - |
| 41 | - | 41 | - | - | - | - |
| 42 | - | 42 | - | 173 | - | - |
| 43 | - | 43 | - | - | - | - |
| 44 | 16 | 44 | - | - | - | - |
| 45 | 18 | 45 | - | - | - | - |
| 46 | 55, 59 | 46 | - | 59 | 59 | - |
| 47 | 18 | 47 | - | 50 | 105 | - |
| 48 | 39, 53 | 48 | - | 31, 39 | - | - |
Subsequently, we were able to clone full length HPV genomes from three new HPV types (HPV 171, 172, 173 from specimens 19–21, 28–30, and 42, respectively) using long range PCR. Consistent with being a member of gamma HPV genera, these HPV genomes contain only 7 ORFs, including L1, L2, E1, E2, E4, E6, E7, but lacking E5 ORF. The E6 proteins from the three HPV types contain five conserved zinc-binding domains (Cys-X-X-Cys) separated by 25, 0, 36, 29 amino acids respectively. The E7 proteins of these three HPV types contain two zinc-binding domains, separated by 29–30 amino acids. The E1 proteins contain the conserved ATP-binding site (HPV 171: GVPDSGKS; HPV 172: GPSDTGKS; HPV 173: GPPDTGKS). The putative binding sites for E2 and polyA signals are identified in the LCR regions as well as binding sites for transcription factors NFI, AP1, YY1, Oct-1 and Tef-1 (Table 5).
Table 5. Similarity between novel HPV type ORFs and other representative PVs.
Putative ORFs (E6, E7, E1, E2, E4, L1, and L2) of novel HPV types (HPV 171, 172, and 173) are compared to each other, as well as representative γ-HPV types (HPV 4, 60, 103, 156, 163, 169), β-HPV (HPV 5) and α-HPV (HPV 16). The most similar HPV type for each novel HPV type is highlighted in bold.
| (A) HPV 171 | Genome length | E6 | E7 | E1 | E2 | E4 | L2 | L1 |
|---|---|---|---|---|---|---|---|---|
| Number of nucleotides (nt)* | 7261 | 432 | 300 | 1815 | 1173 | 519 | 1491 | 1578 |
| Position of ORF in genome | - | 19–450 | 453–752 | 736–2550 | 2486–3658 | 2913–3431 | 3660–5152 | 5159–6736 |
| Number of amino acids (aa) | - | 143 | 99 | 604 | 390 | 172 | 496 | 525 |
| Sequence similarity percentage nt (percentage aa) |
γ1-HPV 4 | 52.7 (43.6) | 48.6 (40.0) | 64.6 (58.8) | 53.8 (44.5) | 51.6 (30.3) | 50.5 (43.9) | 63.2 (58.2) |
| γ4-HPV 60 | 57.3 (49.6) | 54.0 (44.8) | 64.3 (53.9) | 57.6 (48.0) | 53.3 (32.7) | 51.5 (44.6) | 62.5 (58.5) | |
| γ6-HPV 103 | - | 45.6 (36.1) | 59.9 (48.0) | 54.2 (42.8) | 49.9 (24.3) | 46.1 (37.4) | 57.8 (52.4) | |
| γ23-HPV 156 | 56.3 (42.9) | 57.6 (54.7) | 62.6 (52.9) | 59.5 (49.2) | 56.7 (35.9) | 49.5 (38.8) | 62.0 (57.7) | |
| γ22-HPV 163 | 54.1 (46.0) | 49.5 (42.7) | 62.6 (54.4) | 53.7 (45.0) | 49.9 (24.1) | 52.2 (44.1) | 61.5 (59.0) | |
| γ11-HPV 169 | 92.4 (94.4) | 96.0 (98.0) | 92.4 (97.4) | 93.2 (94.9) | 95.6 (93.6) | 89.8 (92.3) | 88.3 (97.0) | |
| γ23-HPV 172 | 54.9 (46.2) | 51.9 (47.9) | 60.4 (47.7) | 56.7 (44.1) | 54.3 (35.2) | 50.1 (36.5) | 60.1 (54.2) | |
| γ1-HPV 173 | 50.1 (45.0) | 47.9 (40.4) | 63.4 (52.9) | 53.9 (43.5) | 49.8 (27.5) | 53.7 (43.2) | 62.0 (56.2) | |
| β-HPV 5 | 43.6 (30.3) | 44.1 (35.1) | 56.8 (46.4) | 46.6 (42.1) | 46.0 (23.5) | 45.1 (34.8) | 59.1 (54.7) | |
| α-HPV 16 | 41.7 (23.1) | 40.5 (28.4) | 54.8 (42.4) | 48.5 (30.7) | 42.2 (13.5) | 45.1 (29.7) | 40.5 (48.1) | |
| (B) HPV 172 | Genome length | E6 | E7 | E1 | E2 | E4 | L2 | L1 |
| Number of nucleotides (nt)* |
7203 | 552 | 297 | 1803 | 1188 | 420 | 1554 | 1551 |
| Position of ORF in genome | - | 106–657 | 638–934 | 921–2723 | 2659–3846 | 3191–3610 | 3846–5152 | 5410–6960 |
| Number of amino acids (aa) | - | 183 | 98 | 600 | 395 | 139 | 517 | 516 |
| Sequence similarity percentage nt (percentage aa) |
γl-HPV 4 | 53.9 (42.9) | 53.1 (46.4) | 60.9 (50.2) | 56.3 (44.0) | 53.8 (27.1) | 50.1 (39.9) | 60.3 (52.6) |
| γ4-HPV 60 | 52.4 (43.7) | 54.0 (46.9) | 63.6 (53.2) | 60.6 (49.4) | 51.7 (34.4) | 53.0 (42.0) | 63.2 (55.0) | |
| y6-HPV 103 | - | 50.2 (48.0) | 55.9 (45.0) | 52.2 (40.5) | 47.0 (23.4) | 44.0 (37.3) | 54.9 (47.0) | |
| γ23-HPV 156 | 62.4 (52.9) | 65.3 (58.8) | 68.2 (61.3) | 67.6 (60.2) | 66.7 (48.1) | 58.2 (48.7) | 68.2 (66.2) | |
| γ22-HPV 163 | 60.1 (47.1) | 56.9 (49.0) | 61.7 (48.6) | 54.3 (41.4) | 47.9 (21.1) | 52.0 (41.0) | 59.5 (53.0) | |
| γll-HPV 169 | 53.9 (45.5) | 51.6 (49.0) | 60.1 (47.8) | 56.9 (43.9) | 55.3 (35.2) | 50.1 (36.0) | 59.8 (55.0) | |
| γ11-HPV 171 | 54.9 (46.2) | 51.9 (47.9) | 60.4 (47.7) | 56.7 (44.1) | 54.3 (35.2) | 50.1 (36.5) | 60.1 (54.2) | |
| γ1-HPV 173 | 59.3 (48.6) | 55.6 (45.8) | 59.6 (48.8) | 56.1 (41.3) | 48.9 (22.6) | 49.7 (40.3) | 60.1 (51.9) | |
| β-HPV 5 | 46.2 (30.1) | 44.9 (36.1) | 55.2 (43.4) | 47.1 (40.8) | 43.7 (28.7) | 42.6 (33.7) | 57.4 (48.9) | |
| γ-HPV 16 | 45.9 (25.8) | 44.3 (32.2) | 53.9 (41.3) | 47.8 (30.2) | 36.3 (18.0) | 43.3 (31.8) | 37.2 (44.2) | |
| (C) HPV 173 | Genome length | E6 | E7 | E1 | E2 | E4 | L2 | L1 |
| Number of nucleotides (nt)* | 7297 | 423 | 294 | 1812 | 1176 | 357 | 1584 | 1542 |
| Position of ORF in genome | - | 31–453 | 450–743 | 730–2541 | 2483–3658 | 3066–3422 | 3689–5272 | 5287–6828 |
| Number of amino acids (aa) | - | 140 | 97 | 603 | 391 | 118 | 527 | 513 |
| Sequence similarity percentage nt (percentage aa) |
γ1-HPV 4 | 61.0 (52.9) | 67.7 (57.3) | 71.6 (66.7) | 70.2 (67.8) | 66.1 (44.6) | 64.4 (60.4) | 71.0 (73.6) |
| γ4-HPV 60 | 54.6 (46.8) | 54.4 (41.5) | 63.6 (52.7) | 57.5 (46.6) | 51.5 (32.7) | 55.0 (46.8) | 63.6 (59.9) | |
| γ6-HPV 103 | - | 53.2 (45.8) | 59.9 (47.2) | 53.2 (40.0) | 50.7 (27.9) | 48.2 (47.3) | 61.0 (58.6) | |
| γ23-HPV 156 | 64.8 (52.1) | 58.4 (53.7) | 61.0 (49.6) | 57.3 (42.6) | 50.9 (25.2) | 53.6 (43.1) | 62.3 (55.2) | |
| γ22-HPV 163 | 61.0 (48.9) | 64.0 (57.7) | 68.3 (58.0) | 67.3 (61.1) | 65.8 (51.7) | 64.2 (58.8) | 67.6 (69.8) | |
| γ11-HPV 169 | 49.9 (44.3) | 47.9 (39.4) | 63.6 (53.0) | 54.4 (43.2) | 48.3 (26.5) | 52.2 (42.1) | 60.8 (55.8) | |
| γ11-HPV 171 | 50.1 (45.0) | 47.9 (40.4) | 63.4 (52.9) | 53.9 (43.5) | 49.8 (27.5) | 53.7 (43.2) | 62.0 (56.2) | |
| γ23-HPV 172 | 59.3 (48.6) | 55.6 (45.8) | 59.6 (48.8) | 56.1 (41.3) | 48.9 (22.6) | 49.7 (40.3) | 60.1 (51.9) | |
| β-HPV 5 | 47.3 (34.3) | 44.8 (32.6) | 56.7 (44.8) | 46.5 (35.9) | 40.0 (19.5) | 45.0 (41.3) | 60.8 (57.7) | |
| α-HPV 16 | 46.3 (27.9) | 41.6 (35.2) | 54.5 (42.0) | 48.8 (32.6) | 38.6 (15.0) | 45.8 (34.6) | 37.4 (49.9) |
Including STOP codon
For all ORFs, HPV 171 has about 90% similarity to HPV 169, about 60% similarity to other γ-HPV types and β-HPV (HPV 5), only 40% similarity to α-HPV (HPV 16). For HPV 172 and HPV 173, all ORFs have 60–70% similarity to other HPV types except only 40% similarity to α-HPV (HPV 16) (Table 4).
Table 4. Presence of novel HPV types in oral tissue samples.
Formalin-fixed paraffin-embedded oral tissue blocks from both normal and cancer patients were included. Quantitative Taqman assays were performed on each sample. Any samples amplified were considered positive for the HPV type tested and samples not amplified were negative for that HPV type.
| Normal | Cancer | p-value Cancer vs. Normal |
|||
|---|---|---|---|---|---|
| Oral Cavity n=56 |
Oropharynx n=20 |
Oral Cavity n=66 |
Oropharynx n=16 |
||
| HPV 16 | 9 (16%) | 1 (5%) | 6 (9%) | 3 (19%) | 0.67 |
| HPV 18 | 0 (0%) | 0 (0%) | 1 (2%) | 0 (0%) | 1.00 |
| HPV171 | 8 (14%) | 2 (10%) | 16 (24%) | 1 (6%) | 0.21 |
| HPV172 | 22 (39%) | 3 (15%) | 0 (0%) | 0 (0%) | <.0001 |
| HPV173 | 2 (4%) | 0 (0%) | 0 (0%) | 0 (0%) | 0.23 |
Phylogenetic analysis of novel HPV types
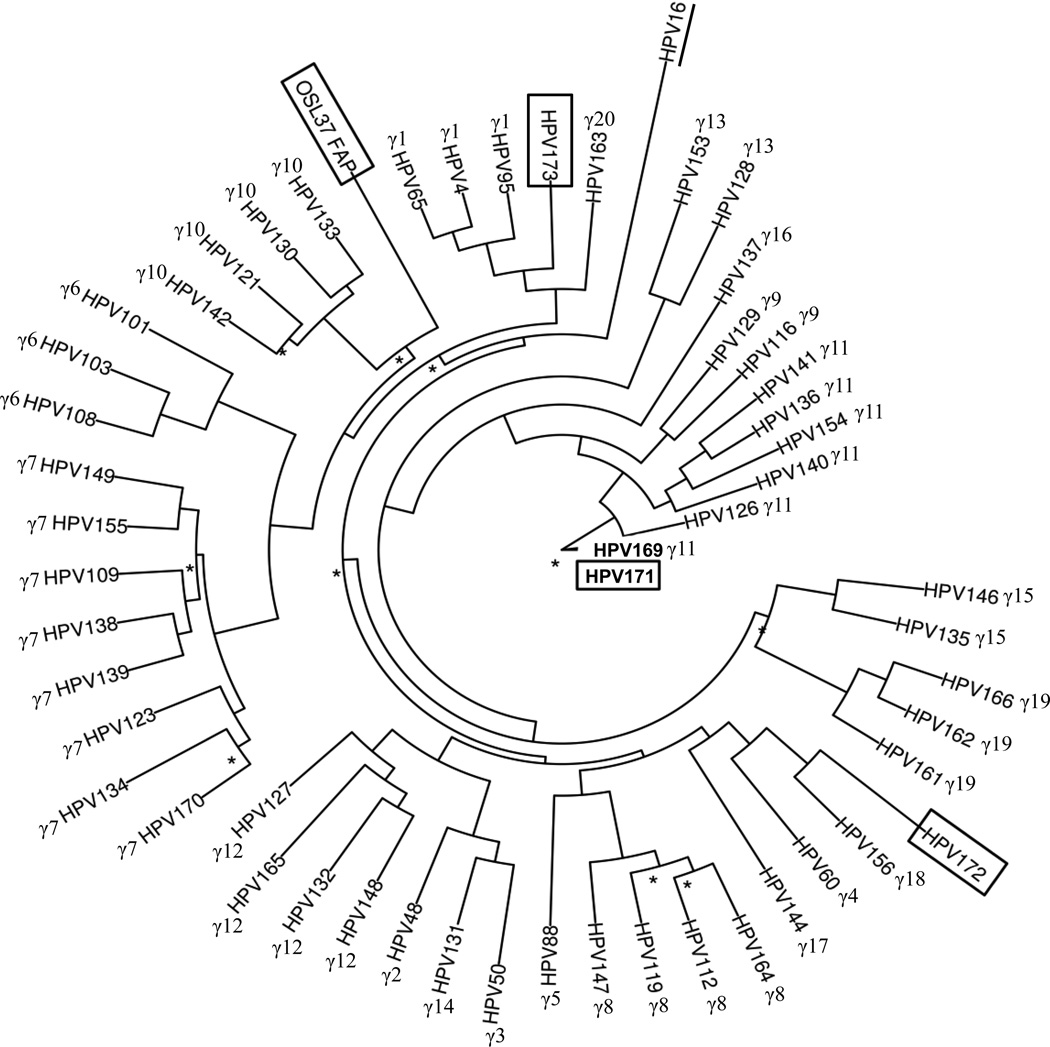
We performed a phylogenetic analysis of the four new novel HPV types with the 50 γ-HPV types of reference clones accessed from International Human Papillomavirus Reference Centre (www.hpvcenter.se), using HPV 16 as the out group. The phylogenetic tree is shown in Figure 1. The four HPV types belong to different branches in the gamma genus. HPV 171, HPV 172 and HPV 173 clustered closely to γ-PV species 11, γ-PV species 18, and to γ-PV species 1, respectively. OSL 37 belonged to γ-PV species 10.
Figure 1. Phylogenetic tree of four novel HPV types among Gammapapillomaviruses.
Maximum likelihood tree was estimated by PhyML v3 30. ModelTest v3.7 29 reported GTR+I+G substitution model as the best fit model. HPV16, an α-HPV type (underlined) was used as the out group for the tree analysis, and the tree was based on L1 sequences of 50 gamma HPV reference clones (www.hpvcenter.se). Nodes having bootstrap values less than 0.7 were labeled with an asterisk, and new HPV types identified in this study were marked by boxes.
Prevalence of novel HPV types in oral tissues
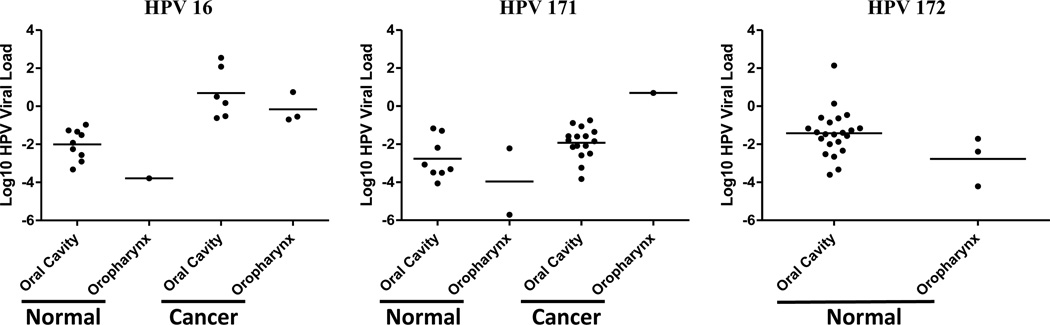
Finally, we determined the prevalence and viral loads of these three novel HPV types in oral tissue samples using type specific Taqman PCR assays targeting the E6/E7 region. As a positive control, we also determined the prevalence and viral load for HPV 16 and HPV 18. As shown in Table 4, HPV 16, HPV 171 and HPV 172 were detected in 12–17% of oral tissue samples. The prevalence of HPV 16 was similar for malignant and normal tissue samples (11% vs 13%, p=0.67), HPV 171 was detected marginally more often in malignant tissue samples compared to normal tissue samples (21% vs 13%, p=0.21), and HPV 172 was only detected in normal tissue samples (33% vs 0%, p<.0001). On the other hand, HPV 18 and HPV 173 were rarely detected in oral tissues: HPV 18 was detected in one malignant tissue sample, and HPV 173 was present in 2 normal tissue samples. Although there was a similar percentage of samples positive for HPV 16, among HPV 16 positive samples, the average viral load was significantly higher in malignant tissue samples compared to normal oral tissues (p<.0001), even after adjusting for age (p=0.0005) (Figure 2). Similarly, among HPV 171 positive samples, the viral load was also significantly higher in malignant oral tissue samples compared to normal oral tissues (p=0.01), but not after adjusting for age (p=0.12).
Figure 2. HPV viral load in oral tissue samples.
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were selected to include both normal (oral cavity and oropharynx) and cancerous (oral cavity and oropharynx) tissues. Viral loads for three HPV types (HPV 16, HPV 171 and HPV 172) by tissue types were presented by scatter plot. The horizontal lines represent the mean viral load in each type of tissue group.
Discussion
We report the isolation of four and characterization of three novel HPV types in oral rinse samples from health male university students. These four HPV types showed 70% homology to existing HPV types, and all belong to the gamma HPV genus. The whole genome encodes 7 ORFs, lacking E5 as a signature of gamma HPV genera. Phylogenetic tree analysis suggests that these four HPV types belong to different branches in the gamma genus, though OSL 37 and HPV 173 are more closely related. Finally, we determined the prevalence and viral load of three HPV types in oral tissue samples. HPV 173 was rarely present in oral tissues, HPV 171 was present more frequently in malignant tissues than in normal tissues, while HPV 172 was only present in normal oral tissues.
Our data suggest that MP-RCA is a valid method to use for preferentially amplifying circular HPV DNA present in oral rinse samples. The lower viral load of HPV 171, 172 and 173 in oral tissue samples compared to HPV 16 confirmed the ability of MP-RCA to increase the sensitivity of consensus PCR to detect novel HPV infections present at low copy numbers. Comparison of four different consensus PCR systems suggests that the FAP-PCR system has the highest sensitivity to detect novel gamma HPV types.
A majority of studies of oral HPV infection has been focused on detecting existing known oncogenic mucosal HPV types, and HPV 16 is the most frequent HPV type detected in oral rinse and oral tissue samples. Our data are consistent with recent reports on the detection of abundant beta and gamma HPV types in oral cavity 19, 32. Bottalico et al reported the presence of a wide spectrum of beta and gamma HPV types in oral cavity specimens 19, and cloned a novel beta HPV 120 from the oral cavity, which subsequently showed that could be detected in multiple anatomical sites 32. Although beta and gamma HPV types are commonly considered cutaneous types, the presence of gamma HPVs in oral cavity specimens suggest that they might have a broader spectrum of tropism.
Unlike α-HPV types, there are no conclusive data demonstrating that beta and gamma HPV types are oncogenic. In conjunction with UVR, beta HPVs may participate in skin carcinogenesis 33. Vairisio D et al reported that HPV 38 E6 and E7 act as promoter and progression factors in multi-stage skin carcinogenesis initiated by DNA damage agent DMBA 34. Wallace et al indicated that HPV 5 and 8 E6 can increase the persistence of UVB induced damage through abrogation of ATR activity 22. Cornet et al reported the ability of HPV 49 E6 and E7 to immortalize primary human keratinocytes and disrupt both p53 and pRb pathways 35. Very little is known about gamma HPV in tumorigenesis. Smith et al reported a complex etiology involving HPV, tobacco and alcohol in the risk and survival from oral cancer 36. The low viral load of gamma HPV present in oral cavity specimens raises the question as to whether these oral HPVs could participate in tumorigenesis. Alternatively, it has been hypothesized that oral HPV might induce tumorigenesis via a mechanism that is different than that of α-HPVs: they might initiate oral tumorigenesis but not tumor progression, thus they might not be detected more frequently in tumor tissues than in normal tissues. Although HPV 171 was more prevalent in malignant than in normal oral tissues and the viral load was significantly higher in malignant than in normal oral tissues, it was not significant after adjusting for age. Interestingly, HPV 172 was only detected in normal but not malignant oral tissues. Whether these two novel HPVs play a role in oral carcinogenesis requires further investigation. It is possible that oncogenic gamma HPV types are rarely present in healthy individuals, thus can only be identified by surveying oral rinse samples collected from oral cancer patients.
Our study has several limitations: we identified new HPV types using oral rinse samples collected from young health male subjects, thus might not extend to other populations; we only analyzed several γ-HPV types in oral tissue blocks, which could not be generalized to other γ-HPV types; the lack of association of these novel HPV types with oral cancer suggest that new oncogenic HPV types could only be identified from oral rinse samples collected from oral cancer patients.
Our study further confirms previously published observations that the oral cavity harbors unknown HPV types, mainly in the genera of beta and gamma. Further investigation of the natural history of these novel HPV types will no doubt enrich our understanding of HPV tropism, as well as identification of potential novel oncogenic gamma HPV types.
Nucleotide sequences
Nucleotide sequences of full length new HPV types have been submitted to Genbank with following accession number: HPV 171: KF006398; HPV 172: KF006399; HPV 173: KF006400.
Table 6. Frequency of conserved motifs in novel HPV types.
Various conserved motifs in HPV LCR, E6 and E7 regions are identified. The codes for degenerate nucleotides are as follows: K=G or T; W= A or T; M=A or C; R=G or A; Y=T or C; B=G, T, C; D=G, A, T. N represents any nucleotide, and X represents any amino acid.
| Region | Motif | HPV 171 | HPV 172 | HPV 173 |
|---|---|---|---|---|
| LCR | PolyA signal [AATAAA] | 2 | 2 | 1 |
| E2 [ACC(N)6GGT] | 6 | 3 | 1 | |
| NFI [TTGGC] | 3 | 1 | 1 | |
| AP1 [TKWNTMA] | 3 | 3 | 3 | |
| YY1 [MCATNKT] | 0 | 1 | 1 | |
| Oct-1 [AANWGYAB] | 0 | 0 | 1 | |
| Tef-1 [YRCATDBYDB] | 1 | 1 | 1 | |
| E6 | Zinc-binding [CXXC] | 5 | 5 | 5 |
| PDZ domain [X(T/S)X(L/V)] | 4 | 2 | 3 | |
| E7 | Zinc-binding [CXXC] | 2 | 2 | 2 |
| PDZ domain [X(T/S)X(L/V)] | 2 | 3 | 3 |
Acknowledgement
We thank Ms. Donna Kenney and Dr. Viorica Popov for support.
Funding
The study is supported by the FHCRC/UW Cancer Consortium Cancer Center Support Grant of the National Institutes of Health under Award Number P30 CA015704, R01CA105181 (L.K.), and T32-MH19139 (ZE) from National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Declarations
Competing interests:
The authors have no commercial or other associations that might pose a conflict of interest.
Ethical approval:
Informed consent was obtained according to procedures approved by the Human Subjects Committee of University of Washington.
References
- 1.Hobbs CG, Sterne JA, Bailey M, Heyderman RS, Birchall MA, Thomas SJ. Human papillomavirus and head and neck cancer: a systematic review and meta-analysis. Clin Otolaryngol. 2006;31:259–266. doi: 10.1111/j.1749-4486.2006.01246.x. [DOI] [PubMed] [Google Scholar]
- 2.Herrero R, Castellsague X, Pawlita M, Lissowska J, Kee F, Balaram P, et al. Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. J Natl Cancer Inst. 2003;95:1772–1783. doi: 10.1093/jnci/djg107. [DOI] [PubMed] [Google Scholar]
- 3.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 4.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 5.Hennessey PT, Westra WH, Califano JA. Human papillomavirus and head and neck squamous cell carcinoma: recent evidence and clinical implications. J Dent Res. 2009;88:300–306. doi: 10.1177/0022034509333371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans M, Newcombe R, Fiander A, Powell J, Rolles M, Thavaraj S, et al. Human Papillomavirus-associated oropharyngeal cancer: an observational study of diagnosis, prevalence and prognosis in a UK population. BMC Cancer. 2013;13:220. doi: 10.1186/1471-2407-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahl A, Kumar P, Dar L, Mohanti BK, Sharma A, Thakar A, et al. Prevalence and trends of human papillomavirus in oropharyngeal cancer in a predominantly north Indian population. Head Neck. 2013 doi: 10.1002/hed.23317. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigo JP, Heideman DA, Garcia-Pedrero JM, Fresno MF, Brakenhoff RH, Molina JP, et al. Time trends in the prevalence of HPV in oropharyngeal squamous cell carcinomas in northern Spain (1990–2009) Int J Cancer. 2013 doi: 10.1002/ijc.28355. [DOI] [PubMed] [Google Scholar]
- 9.Kreimer AR, Johansson M, Waterboer T, Kaaks R, Chang-Claude J, Drogen D, et al. Evaluation of Human Papillomavirus Antibodies and Risk of Subsequent Head and Neck Cancer. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.47.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boscolo-Rizzo P, Del Mistro A, Bussu F, Lupato V, Baboci L, Almadori G, et al. New insights into human papillomavirus-associated head and neck squamous cell carcinoma. Acta Otorhinolaryngol Ital. 2013;33:77–87. [PMC free article] [PubMed] [Google Scholar]
- 11.De Stefani A, Boffano P, Averono G, Ramella A, Pia F, Bongioannini G. Prevalence and characteristics of HPV infection in oropharyngeal cancer. The Journal of craniofacial surgery. 2013;24:e40–e43. doi: 10.1097/SCS.0b013e31826cfffa. [DOI] [PubMed] [Google Scholar]
- 12.Ragin CC, Reshmi SC, Gollin SM. Mapping and analysis of HPV16 integration sites in a head and neck cancer cell line. Int J Cancer. 2004;110:701–709. doi: 10.1002/ijc.20193. [DOI] [PubMed] [Google Scholar]
- 13.Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 14.Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene. 2002;21:1510–1517. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]
- 15.van Houten VM, Snijders PJ, van den Brekel MW, Kummer JA, Meijer CJ, van Leeuwen B, et al. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer. 2001;93:232–235. doi: 10.1002/ijc.1313. [DOI] [PubMed] [Google Scholar]
- 16.Kreimer AR, Bhatia RK, Messeguer AL, Gonzalez P, Herrero R, Giuliano AR. Oral human papillomavirus in healthy individuals: a systematic review of the literature. Sex Transm Dis. 2010;37:386–391. doi: 10.1097/OLQ.0b013e3181c94a3b. [DOI] [PubMed] [Google Scholar]
- 17.Syrjanen S, Lodi G, von Bultzingslowen I, Aliko A, Arduino P, Campisi G, et al. Human papillomaviruses in oral carcinoma and oral potentially malignant disorders: a systematic review. Oral diseases. 2011;17(Suppl 1):58–72. doi: 10.1111/j.1601-0825.2011.01792.x. [DOI] [PubMed] [Google Scholar]
- 18.Edelstein ZR, Schwartz SM, Hawes S, Hughes JP, Feng Q, Stern ME, et al. Rates and determinants of oral human papillomavirus infection in young men. Sex Transm Dis. 2012;39:860–867. doi: 10.1097/OLQ.0b013e318269d098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bottalico D, Chen Z, Dunne A, Ostoloza J, McKinney S, Sun C, et al. The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J Infect Dis. 2011;204:787–792. doi: 10.1093/infdis/jir383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akgul B, Cooke JC, Storey A. HPV-associated skin disease. The Journal of pathology. 2006;208:165–175. doi: 10.1002/path.1893. [DOI] [PubMed] [Google Scholar]
- 21.Bouwes Bavinck JN, Feltkamp M, Struijk L, ter Schegget J. Human papillomavirus infection and skin cancer risk in organ transplant recipients. The journal of investigative dermatology Symposium proceedings / the Society for Investigative Dermatology, Inc [and] European Society for Dermatological Research. 2001;6:207–211. doi: 10.1046/j.0022-202x.2001.00048.x. [DOI] [PubMed] [Google Scholar]
- 22.Wallace NA, Robinson K, Howie HL, Galloway DA. HPV 5 and 8 E6 abrogate ATR activity resulting in increased persistence of UVB induced DNA damage. PLoS pathogens. 2012;8:e1002807. doi: 10.1371/journal.ppat.1002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rector A, Tachezy R, Van Ranst M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J Virol. 2004;78:4993–4998. doi: 10.1128/JVI.78.10.4993-4998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Leon DC, Montiel DP, Nemcova J, Mykyskova I, Turcios E, Villavicencio V, et al. Human papillomavirus (HPV) in breast tumors: prevalence in a group of Mexican patients. BMC Cancer. 2009;9:26. doi: 10.1186/1471-2407-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80(Pt 9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 26.Berkhout RJ, Tieben LM, Smits HL, Bavinck JN, Vermeer BJ, ter Schegget J. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. Journal of clinical microbiology. 1995;33:690–695. doi: 10.1128/jcm.33.3.690-695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Koning M, Quint W, Struijk L, Kleter B, Wanningen P, van Doorn LJ, et al. Evaluation of a novel highly sensitive, broad-spectrum PCR-reverse hybridization assay for detection and identification of beta-papillomavirus DNA. Journal of clinical microbiology. 2006;44:1792–1800. doi: 10.1128/JCM.44.5.1792-1800.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 31.Walker JA, Kilroy GE, Xing J, Shewale J, Sinha SK, Batzer MA. Human DNA quantitation using Alu element-based polymerase chain reaction. Anal Biochem. 2003;315:122–128. doi: 10.1016/s0003-2697(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 32.Bottalico D, Chen Z, Kocjan BJ, Seme K, Poljak M, Burk RD. Characterization of human papillomavirus type 120: a novel betapapillomavirus with tropism for multiple anatomical niches. J Gen Virol. 2012;93:1774–1779. doi: 10.1099/vir.0.041897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiviat NB. Papillomaviruses in non-melanoma skin cancer: epidemiological aspects. Semin Cancer Biol. 1999;9:397–403. doi: 10.1006/scbi.1999.0143. [DOI] [PubMed] [Google Scholar]
- 34.Viarisio D, Muller Decker K, Aengeneyndt B, Flechtenmacher C, Gissmann L, Tommasino M. Human papillomavirus type 38 E6 and E7 act as tumour promoters during chemically induced skin carcinogenesis. J Gen Virol. 2013 doi: 10.1099/vir.0.048991-0. [DOI] [PubMed] [Google Scholar]
- 35.Cornet I, Bouvard V, Campo MS, Thomas M, Banks L, Gissmann L, et al. Comparative analysis of transforming properties of E6 and E7 from different beta human papillomavirus types. J Virol. 2012;86:2366–2370. doi: 10.1128/JVI.06579-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith EM, Rubenstein LM, Haugen TH, Pawlita M, Turek LP. Complex etiology underlies risk and survival in head and neck cancer human papillomavirus, tobacco, and alcohol: a case for multifactor disease. Journal of oncology. 2012:571862. doi: 10.1155/2012/571862. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Q, Cherne S, Winer RL, Balasubramanian A, Lee SK, Hawes SE, et al. Development and evaluation of a liquid bead microarray assay for genotyping genital human papillomaviruses. Journal of clinical microbiology. 2009;47:547–553. doi: 10.1128/JCM.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]