Abstract
The anaphase-promoting complex or cyclosome (APC/C) is a large E3 RING-cullin ubiquitin ligase composed of between 14 and 15 individual proteins. A striking feature of the APC/C is that only four proteins are involved in directly recognizing target proteins and catalyzing the assembly of a polyubiquitin chain. All other subunits, which account for > 80% of the mass of the APC/C, provide scaffolding functions. A major proportion of these scaffolding subunits are structurally related. In metazoans, there are four canonical tetratricopeptide repeat (TPR) proteins that form homo-dimers (Apc3/Cdc27, Apc6/Cdc16, Apc7 and Apc8/Cdc23). Here, we describe the crystal structure of the N-terminal homo-dimerization domain of Schizosaccharomyces pombe Cdc23 (Cdc23Nterm). Cdc23Nterm is composed of seven contiguous TPR motifs that self-associate through a related mechanism to those of Cdc16 and Cdc27. Using the Cdc23Nterm structure, we generated a model of full-length Cdc23. The resultant “V”-shaped molecule docks into the Cdc23-assigned density of the human APC/C structure determined using negative stain electron microscopy (EM). Based on sequence conservation, we propose that Apc7 forms a homo-dimeric structure equivalent to those of Cdc16, Cdc23 and Cdc27. The model is consistent with the Apc7-assigned density of the human APC/C EM structure. The four canonical homo-dimeric TPR proteins of human APC/C stack in parallel on one side of the complex. Remarkably, the uniform relative packing of neighboring TPR proteins generates a novel left-handed suprahelical TPR assembly. This finding has implications for understanding the assembly of other TPR-containing multimeric complexes.
Abbreviations: APC/C, anaphase-promoting complex or cyclosome; TPR, tetratricopeptide repeat; EM, electron microscopy; SAD, single-wavelength anomalous dispersion; PDB, Protein Data Bank; FOM, figure of merit
Keywords: anaphase-promoting complex, tetratricopeptide repeat (TPR), cell cycle, crystallography, single-particle electron microscopy
Graphical Abstract
Highlights
-
•
The paper addresses the structure of the TPR subunits (Cdc23 and Apc7) of the APC/C.
-
•
We determine the crystal structure of the N-terminus of Cdc23 showing that Cdc23 homo-dimerizes similar to other canonical TPR subunits of the APC/C Cdc16 and Cdc27.
-
•
We generate full-length models of Cdc23 and Apc7 and suggest that the mode of dimerization of Apc7 published previously is incorrect. We propose that Apc7 will homo-dimerize similar to Cdc23, Cdc16 and Cdc27.
-
•
We dock Cdc23 and Apc7, as well as Cdc16 and Cdc27, into the negative stain EM structure of the APC/C. This reveals a novel stacking assembly of the four homo-dimeric TPR proteins that forms a left-handed suprahelix.
-
•
We show the Cdc23–Cdc16 assemble into a left-handed suprahelix, with implications for TPR assembly and TPR-containing multimeric assemblies.
Introduction
The anaphase-promoting complex or cyclosome (APC/C) is a multi-subunit E3 ubiquitin ligase whose primary function is to control cell cycle progression by mediating the destruction of proteins whose activities inhibit specific cell cycle transitions.1–4 The APC/C recognizes most of its substrates through conserved destruction motifs, the most predominant being the D (destruction) box and KEN box;5,6 however, rarer degrons such as the O box also serve as APC/C recognition signals.7 Substrate selection is dependent on coactivator subunits (either Cdc20 or Cdh1) that interact with degrons to recruit substrates to APC/C–coactivator complexes.8–12 A core APC/C subunit—Apc10—cooperates with the coactivator subunits to confer high-affinity D box binding,13–15 although coactivators alone interact with the KEN box.16
The APC/C is an unusually large E3 ubiquitin ligase comprising 13 (yeast) and 14 (metazoan) individual proteins in addition to the regulatory coactivator subunits. Since many proteins are present in two copies per complex, the total number of subunits is close to 20 with a combined molecular mass of 1.2 MDa.17 APC/C catalytic activity is conferred by the RING domain subunit Apc11 that is tightly associated with the cullin subunit Apc2. Together with the coactivator subunit and Apc10, these four subunits constitute the catalytic and substrate recognition module of the complex. Intriguingly, these subunits account for only 15–20% of the overall APC/C mass. All other subunits provide scaffolding functions to coordinate the organization of the catalytic and substrate recognition subunits and also act to mediate interactions with the mitotic checkpoint complex.18
Many scaffolding subunits are structurally related. The three canonical tetratricopeptide repeat (TPR) subunits (Cdc16/Apc6, Cdc23/Apc8 and Cdc27/Apc3) are homo-dimers based on a simple architecture of multiple contiguous copies of the 34-amino-acid TPR motif.19 Metazoan APC/C incorporates an additional TPR subunit, Apc7, a paralog of Cdc23/Apc8, whereas Apc5 is composed of 13 copies of an extended TPR motif (termed eTPR)17 recently identified in Rpn6, a subunit of the proteasomal 19S subunit.20 Multiple repeat motifs dominate other APC/C subunits such as the 11 proteosome–cyclosome repeats of Apc121 also found in Rpn1 and Rpn2 subunits of the 19S subunit,22 the three cullin repeats of Apc2 and the seven WD40 repeats of coactivators.16 Whereas the canonical TPR subunits Cdc16, Cdc23 and Cdc27 are essential APC/C components, the function of Apc7 is unclear. Deletion of Apc7 in Drosophila caused only mild chromosome defects and anaphase delay.23 Cdc23 and Cdc27 engage the coactivator subunits responsible for substrate recognition.24–27 Altering the binding of Cdc20 to Cdc23 and Cdc27 between prometaphase and metaphase modulates APC/CCdc20 specificity for its substrates.27 The conserved IR tails of coactivator and Apc10 interact with Cdc27,10,24,26,28 whereas the Cdc23-binding site on coactivator is not defined.
Early studies on the canonical Saccharomyces cerevisiae TPR subunits Cdc16, Cdc23 and Cdc27 first suggested that these proteins self-associate within a large complex.29 More recent crystallographic analyses of Schizosaccharomyces pombe Cut9 (Cdc16/Apc6) and the N-terminal region of Encephalitozoon cuniculi Cdc27 showed that both these TPR proteins homo-dimerize through a structurally conserved N-terminal dimerization domain comprising seven TPR motifs.30,31 Biochemical data also indicated that purified Cdc23 forms homo-dimers through an N-terminal dimerization domain,17 consistent with the intragenic complementation of Cdc23 heteroalleles.32 Furthermore, the localization of Cdc23 within the density maps of electron microscopy (EM)-derived structures of both S. cerevisiae and human APC/C at 10 and 20 Å resolution, respectively, showed that Cdc23 forms a V-shaped homo-dimer related in structure to those of Cdc16 and Cdc27.17,33 These results suggested that although the N-terminus of Cdc23 shares relatively little sequence similarity with either Cdc16 or Cdc27, all canonical TPR subunits incorporate related N-terminal dimerization domains.
To obtain a molecular model of Cdc23, we have determined the crystal structure of its N-terminal dimerization domain and used this to generate a full-length model of the protein. We show that the homo-dimer contact residues of Cdc23, Cdc16 and Cdc27 are structurally equivalent and we propose that Apc7 will also form a homo-dimeric structure similar to other canonical APC/C TPR proteins. We show that this model of Apc7 is consistent with its strong correspondence to the Apc7-assigned density of the EM structure of human APC/C. Finally, from analysis of the arrangement of the four TPR homo-dimers constituting the TPR lobe of human APC/C, we reveal a novel supra-TPR helical structure generated by the uniform geometric stacking of contiguous TPR homo-dimers.
Results and Discussion
Cdc23 forms a homo-dimer structurally related to Cdc16 and Cdc27
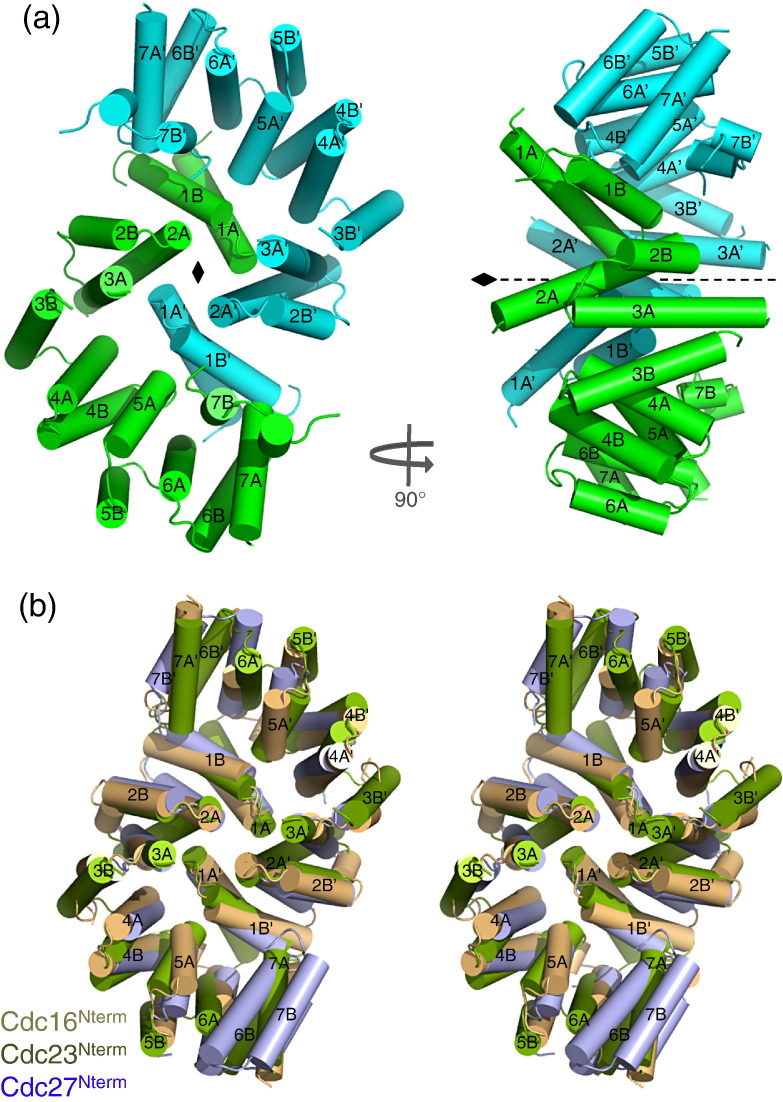
The N-terminal dimerization domain of S. pombe Cdc23 (Cdc23Nterm, residues 19–302) was crystallized and its structure was determined by means of single-wavelength anomalous dispersion (SAD) phasing using a SeMet (selenomethionine) derivative (Materials and Methods and Supplementary Table 1). Two Cdc23Nterm molecules homo-dimerize through a 2-fold symmetric homotypic interface (Fig. 1a). By several criteria, this dimer is likely to be biologically relevant. First, the dimer interface buries an extensive ~ 6000 Å2 of accessible surface area. Second, the interface is structurally related to the N-terminal dimerization interface of Cdc27 and Cdc1630,31 (Fig. 1b). Third, the interface is generated from residues conserved across all orthologs of Cdc23 (Fig. 2a). Fourth, molecular size analysis of the N-terminal dimerization domain of Cdc23 (residues 1–308) using multiangle light scattering is consistent with a Cdc23 homo-dimer in solution,17 and lastly, a model of full-length of Cdc23 docks into the protein density defined for Cdc23 within the molecular envelopes of both S. cerevisiae and human APC/C as determined by single-particle EM17,33 with a high degree of correspondence (see below).
Fig. 1.
Cdc23 self-associates through an N-terminal dimerization domain similar to Cdc16 and Cdc27. (a) Two orthogonal views of the N-terminal domain of the Cdc23Nterm homo-dimer. The two subunits are shown in green and cyan. α-Helices of the TPR motifs are labeled 1A to 7B for the A and B helices of TPR motifs 1–7. (b) Stereoview show a superimposition of Cdc23Nterm, Cdc16Nterm (PDB ID: 2XPI)30 and Cdc27Nterm (PDB ID: 3KAE)31 N-terminal dimerization domains. Figure produced using PyMOL.
Fig. 2.
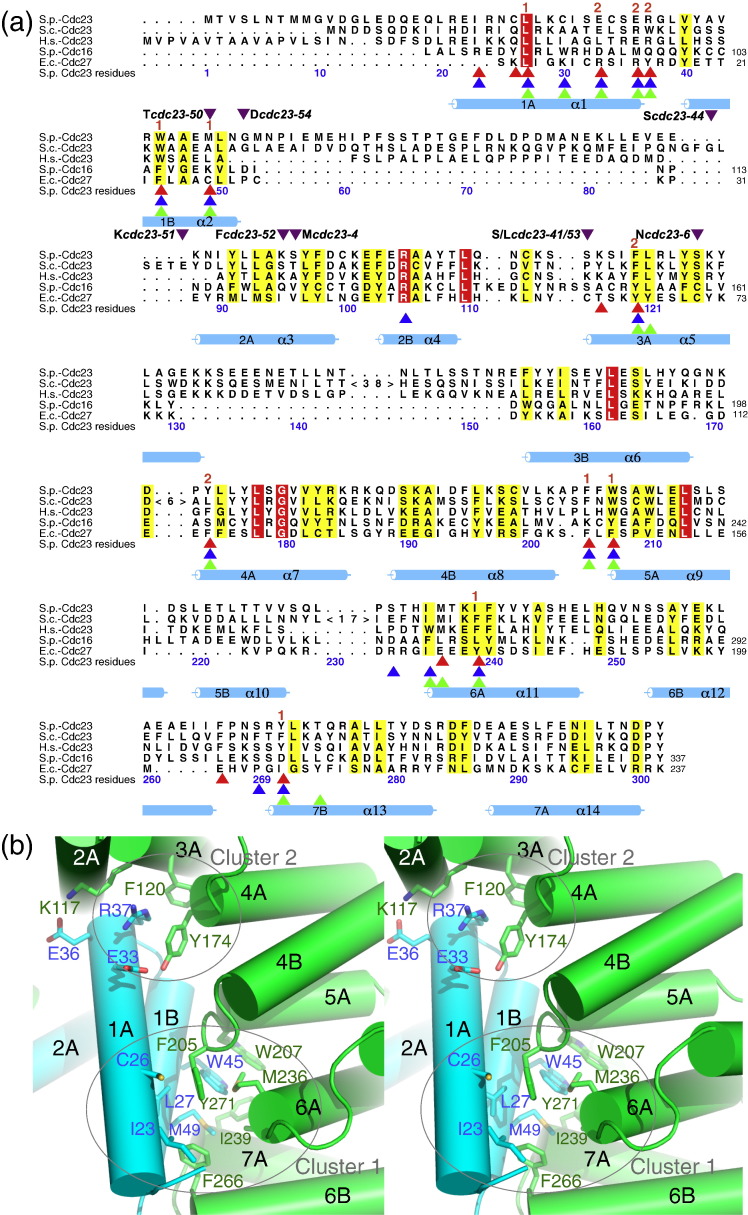
Residues of the Cdc23 homo-dimer interface of Cdc23 are conserved with Cdc16 and Cdc27. (a) Multiple sequence alignment of the dimerization domains of Cdc23, Cdc16 and Cdc27. Invariant and conserved residues are colored red and yellow, respectively. Residues at the dimer interface are indicated with green (Cdc23), blue (Cdc16) and red (Cdc27) arrows. Positions conserved in all three proteins are assigned to a specific cluster (either 1 or 2) indicated in (b). Essential S. cerevisiae Cdc23 residues that define the N-terminal mutable domain of Cdc2332 are indicated with purple arrows. The mutated residue is shown. Residues invariant in all four sequences are shown in red. Positions with either conserved polar or nonpolar residues are shown in yellow. Figure produced using ALSCRIPT.51 (b) Stereoview showing details of the Cdc23 homo-dimer interface. Contact residues are localized to two discrete clusters, labeled “1” and “2”.
S. pombe Cdc23Nterm closely resembles a TPR superhelix comprising seven consecutive TPR motifs that generate 14 α-helices organized into an antiparallel arrangement (Figs. 1a and 2a). TPR motifs 3–7 of S. cerevisiae Cdc23Nterm were correctly predicted using the TPRpred server†. A DALI search34 indicated that S. pombe Cdc23Nterm is most closely related in structure to the N-terminal dimerization domain of S. pombe Cdc16 [Cut9; Protein Data Bank (PDB) ID: 2XPI] (Z-score = 14; rmsd of 3.2 Å for 207 aligned residues)30 (Fig. 1b). Cdc23Nterm forms an intimate homotypic dimer interface (Fig. 1). In essence, the concave surface of each Cdc23Nterm domain encircles its dimer counterpart in an interlocking clasp-like arrangement. This is achieved as the N-terminal TPR motif 1 (TPR1) of each Cdc23 subunit interacts with residues lining the inner groove TPR α-helices (A-helices) and inter-TPR loops of TPR1–TPR7 of the opposite subunit through two clusters of residues centered on Arg37 (helix 1A of TPR1: cluster 2) and Trp45 (helix 1B of TPR1: cluster 1) (Fig. 2). Equivalent clusters of conserved, predominantly nonpolar residues constitute the dimer interfaces of Cdc16Nterm and Cdc27Nterm.30,31 For example, in cluster 1 of Cdc23Nterm, the aromatic side chain of Trp45 of TPR1B (helix B of TPR1) participates in hydrophobic packing interactions with Phe205 and Trp207 of TPR5A of the opposite subunit. In Cdc27Nterm, Phe23 of TPR1B (structurally equivalent to Trp45 of Cdc23) interacts with Phe145 and Phe147 (equivalent to Phe205 and Trp207 of Cdc23). In the less well conserved cluster 2, Phe120 (TPR3A) and Tyr174 (TPR4A) of Cdc23Nterm contact the intra-helix turn of TPR1 of the opposite subunit (Arg37), a role played by Tyr66 and Phe115 of Cdc27Nterm (Fig. 2a).
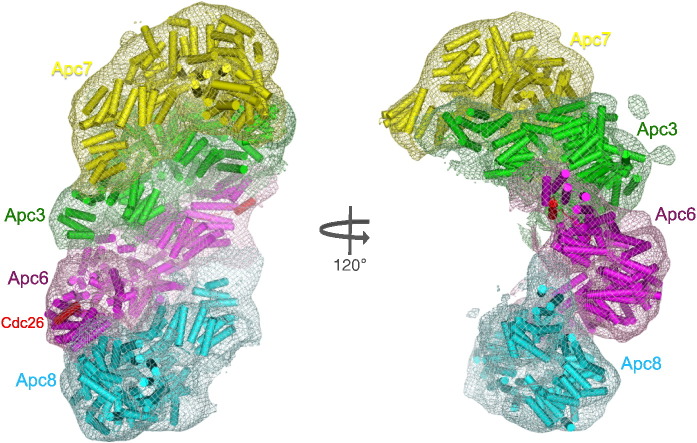
The C-terminus of Cdc23 is predicted to form seven additional TPR motifs, similar to that observed for Cdc16Cterm30 (Cdc16Cterm and Cdc23Cterm share 15% sequence identity) and that predicted for Cdc2731 and Apc7 (this work; see below) (Supplementary Fig. 1a). Thus, all canonical TPR subunits will adopt TPR superhelical structures incorporating 14 contiguous TPR motifs with a capping C-terminal α-helix. These 14 TPR motifs and C-terminal capping α-helix generate a right-handed TPR superhelix comprising an array of 29 antiparallel α-helices composed of two complete turns of helix with an overall length of 110 Å and a width of 60 Å. Their self-association into homo-dimers generates elongated V-shaped molecules with maximum dimensions of approximately 150 Å (Fig. 3). Two mutable domains of S. cerevisiae Cdc2332 map to the N-terminal dimerization domain and C-terminal TPR superhelix, respectively (Supplementary Figs. 1a and 2a). Mutations within the dimerization domain would disrupt Cdc23 function either by disrupting the Cdc23 dimer interface or by destabilizing the packing between adjacent TPR α-helices (Supplementary Fig. 2a). To test our model of the Apc8/Cdc23 homo-dimer, we assessed its structural similarity to the Apc8/Cdc23-assigned density of a human APC/CCdh1–Hsl1 ternary complex determined using negative stain EM and single-particle analysis (Fig. 4). Our docking was guided by the previous assignment of Cdc23, Cdc16 and Cdc27 into the EM envelope of S. cerevisiae APC/C.17 Figures 4b and 5 show the docking of the Apc8/Cdc23 atomic model, together with those of Apc6/Cdc16 and Apc3/Cdc27, into their corresponding assigned densities extracted from the three-dimensional structure of a human APC/CCdh1–Hsl1 ternary complex. The high correlation coefficients between docked coordinates and their respective densities in the negative stain EM map indicate a good fit between the TPR homo-dimers and the EM map: 0.79 for Apc8/Cdc23, 0.79 for Apc6/Cdc16 and 0.78 for Apc3/Cdc27.
Fig. 3.
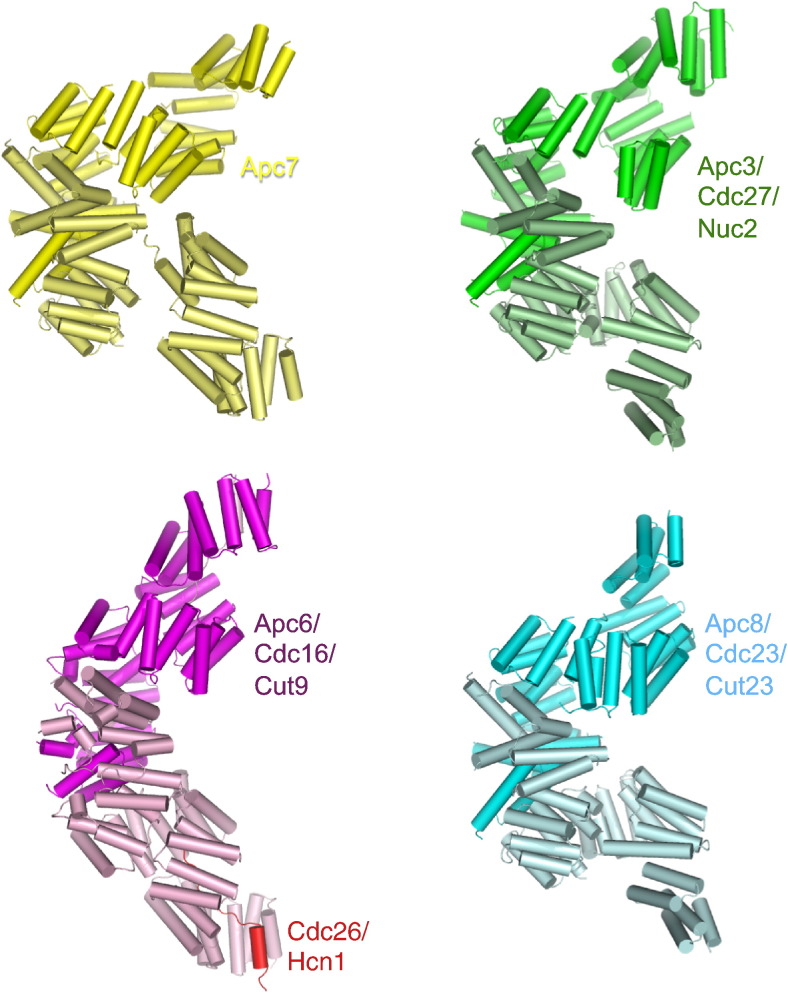
Four canonical TPR proteins form related “V”-shaped homo-dimers and assemble in parallel in a hierarchical fashion on one side of human APC/C. (a) Comparison of the four canonical TPR proteins. Cdc16 (PDB ID: 2XPI),30 Cdc27 (N-terminal domain based on PDB ID: 3KAE),31 Cdc23 and Apc7 (this study). Human, S. cerevisiae and S. pombe names are given.
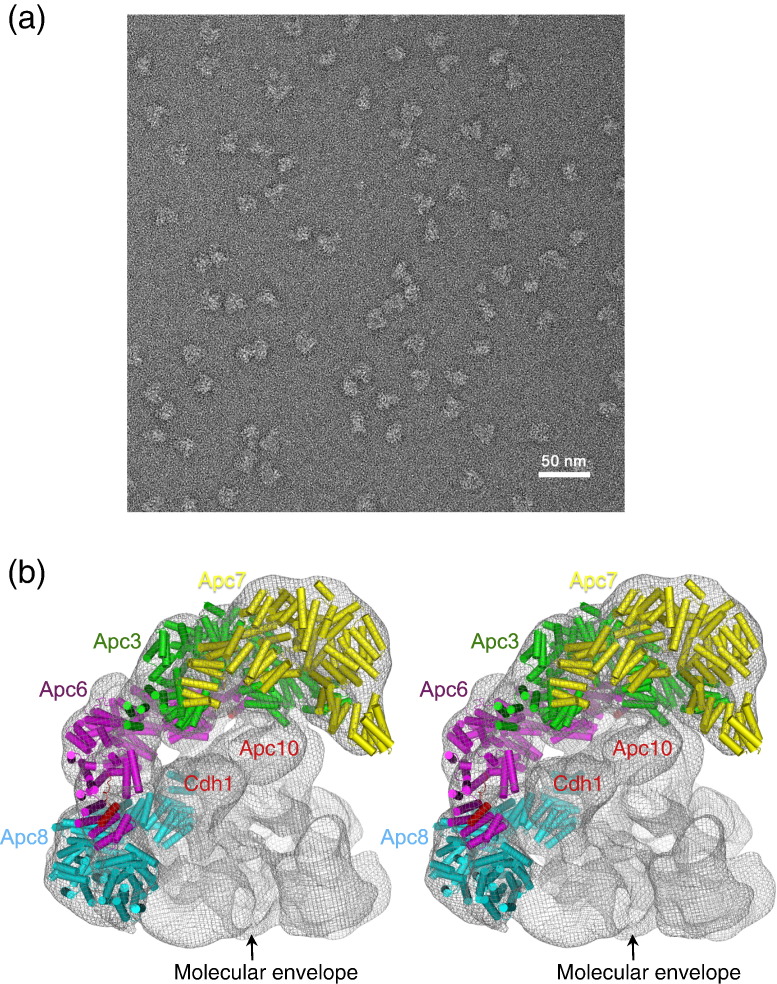
Fig. 4.
(a) Example of a negatively stained electron micrograph of recombinant human APC/CCdh1.Hsl1 ternary complex. (b) Stereoview showing the TPR subunits docked into the negative stain EM structure of the human APC/CCdh1-Hsl1 ternary complex represented in mesh. Resolution is 21 Å by the 0.5 FSC (Fourier shell correlation) criterion. The molecular envelope is shown in mesh representation. In addition to the TPR homo-dimers, Cdh1 and Apc10 have been assigned to density and their associated densities are labeled. The remaining densities correspond to the large subunits Apc1, Apc2, Apc4 and Apc5.3 Their locations have not been definitively identified.
Fig. 5.
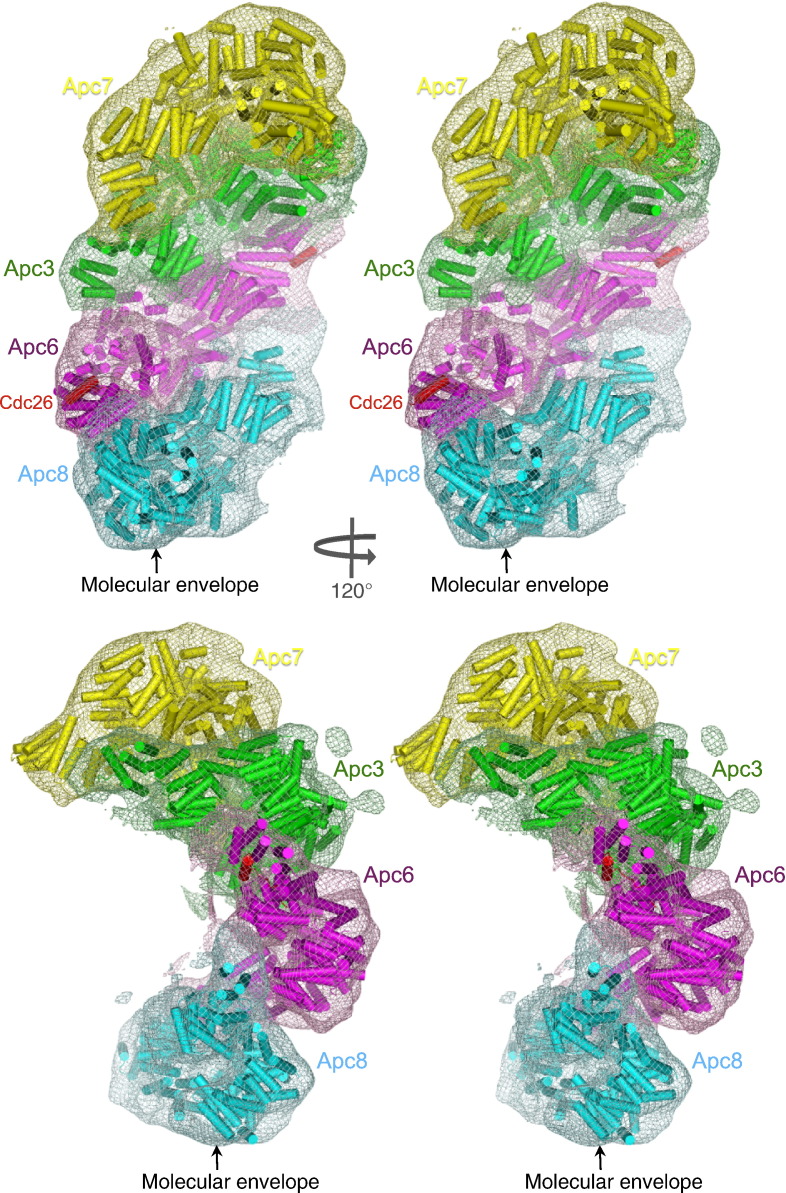
Four canonical TPR proteins stack in a suprahelical arrangement. Two stereoviews of the TPR subunits docked into their assigned density (represented in mesh) of the negative stain EM structure of the human APC/CCdh1-Hsl1 ternary complex. The complementary packing of the ridges and grooves of TPR superhelices of adjacent TPR dimers at the inter-TPR interface is indicated. Human, S. cerevisiae and S. pombe TPR subunit names are given.
Apc7 is structurally related to Cdc16, Cdc23 and Cdc27
Apc7 is a paralog of Cdc27 unique to metazoan APC/C, likely to incorporate 14 TPR motifs and adopt a homo-dimeric architecture. TPR motif 4 and motifs 6–14 equivalent to Cdc16, Cdc23 and Cdc27 are clearly identifiable through multiple sequence analysis and TPRpred predictions (Supplementary Fig. 1a). Furthermore, threading the human Apc7 sequence onto the crystal structure of S. pombe Cdc16 (PDB ID: 2XPI; 15% sequence identity) using Phyre35 is consistent with the notion that Apc7 incorporates 14 contiguous TPR motifs with a C-terminal capping α-helix (Supplementary Fig. 1a).
The crystal structure of residues 1–174 (comprising TPR motifs 1–3 and helix A of TPR4) of human Apc7 described a head-to-head homo-dimer mediated through nonpolar residues of TPR1.36 This structure differed significantly from the clasp-like dimerization mode of all other APC/C TPR subunits (Supplementary Fig. 3a). Moreover, the buried accessible surface area is 586 Å2, significantly smaller than the extensive ~ 6000 Å2 buried surface at the dimer interfaces of Cdc16, Cdc27 and Cdc23 involving TPR motifs 1–730,31 (this work). We propose that the dimer interface described for the Apc7 crystal structure does not represent the physiologically relevant mode of Apc7 dimerization. The clasp-like dimerization of Cdc16, Cdc23 and Cdc27 involves contacts between TPR1 and TPR motifs 2–7 of the opposite subunit. Residues of Apc7 (TPR1) that mediate the reported dimer interface are equivalent to residues of Cdc16, Cdc23 and Cdc27 TPR1 that mediate the clasp-like dimer interface (Supplementary Fig. 1a). Thus, because TPR motifs 4–7 are missing from the crystal structure of the Apc7 N-terminus, a clasp-like interface is precluded. However, the dimerization residues of TPR1 are exposed thereby promoting a nonphysiological dimer interface. Importantly, our model of the Apc7 homo-dimer corresponds closely to the density assigned to Apc7 at the top of the TPR lobe of the negative stain EM structure of human APC/CCdh1.Hsl1 ternary complex. A model of Apc7 is readily docked into the Apc7-assigned density (with a correlation coefficient of 0.80) (Figs. 4b and 5), density that is absent from the yeast APC/C structures that lack the Apc7 subunit.15,17,37
Assembly of the TPR homo-dimers forms a left-handed TPR suprahelix
The three conserved canonical TPR subunits (Cdc16/Apc6, Cdc23/Apc8 and Cdc27/Apc3), together with the metazoan-specific TPR subunit Apc7, are structurally related homo-dimers that self-associate through their N-terminal seven TPR motifs (Fig. 3). Docking of these proteins into the EM density map APC/CCdh1.Hsl1 indicates that the four TPR subunits stack in a contiguous parallel array on one side of the APC/C (Figs. 4b and 5). Interestingly, neighboring TPR dimers stack relative to each other through a related geometric arrangement. This uniform angular and spatial arrangement of TPR homo-dimers defines a left-handed suprahelical structure. Analysis of the inter-TPR dimer interface reveals the complementary fit of the grooves and ridges of TPR superhelices of neighboring TPR dimers, reminiscent of the packing of grooves and ridges of α-helices (Fig. 5). The 30-Å lateral displacement of neighboring TPR dimers results from the 60-Å pitch of the TPR superhelix.38 Neighboring TPR dimers rotate and tilt by ~ 35º, in order to optimize interactions. Model building multiple contiguous TPR dimers shows that this uniform geometric stacking of TPR dimers creates a TPR suprahelix that would comprise 10 TPR dimers in one complete turn, with an overall helix diameter of 300 Å and a pitch of 165 Å (Fig. 6a).
Fig. 6.
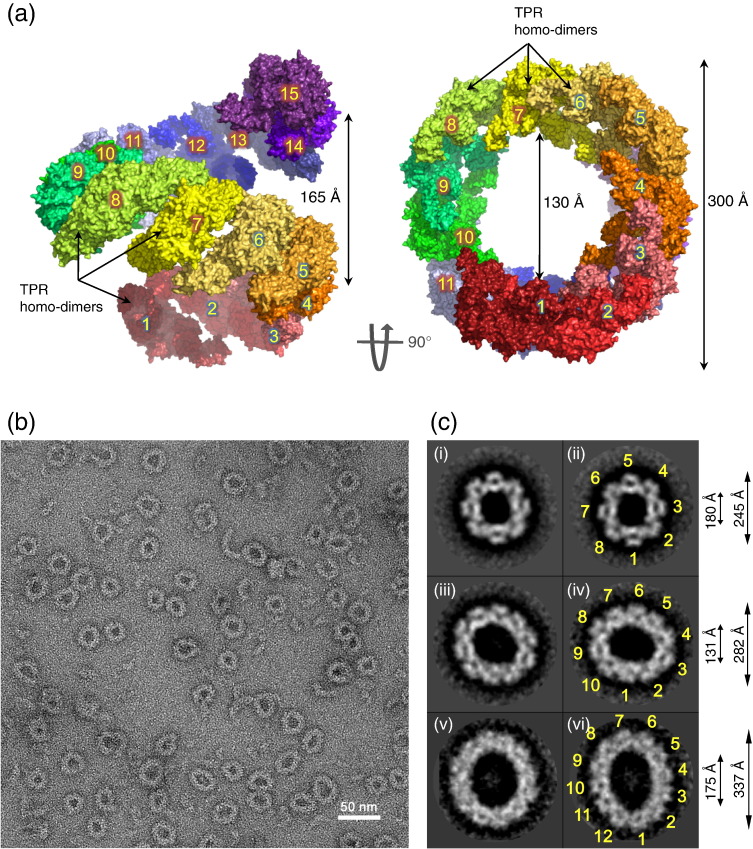
TPR homo-dimers assemble into a TPR suprahelix. (a) Two views showing a model of the TPR suprahelix based on the four TPR homo-dimers of human APC/C generated by superimposing Cdc23 and Apc7 models of contiguous TPR complexes. The model shown includes 15 TPR homo-dimers. Homo-dimers are colored-ramped from blue to red indicating the start and end of the suprahelix, respectively, and numbered. Representative examples are labeled. (b) A negative stain micrograph of human Apc6/Cdc16–Apc8/Cdc23–Cdc26–Apc13 complex. These proteins assemble into ring-shaped structures with dimensions consistent with the modeled TPR suprahelix. (c) Representative two-dimensional class averages generated from 8000 selected particles. Three class types of varying ring diameter are observed with 8-, 10- and 12-fold symmetry. Classes (i) and (ii) from 2523 and 1788 particles, respectively, show 8-fold symmetry; classes (iii) and (iv) from 1733 and 2002 particles, respectively, show 10-fold symmetry; classes (v) and (vi) from 305 and 311 particles, respectively, show 12-fold symmetry. Repeating units (i.e., TPR homo-dimers) are numbered.
The potential for TPR homo-dimers to form suprahelices explains a curious oligomeric structure of a Cdc16–Cdc23 complex, together with their accessory subunits Cdc26 and Apc13, observed using negative stain EM (Fig. 6b). A purified assembly of human Cdc16–Cdc23–Apc13–Cdc26 was visualized as large ring-like structures measuring between 240 and 340 Å in diameter. Two-dimensional classification indicated three ring sizes. Approximately 50% of the rings have an outer dimension of 240 Å with clearly defined 8-fold symmetry. Some 45% of the rings have an outer dimension of 280 Å and show 10-fold symmetry, and there is a small fraction (7%) of larger rings of diameter 340 Å with 12-fold symmetry (Fig. 6c). The ring has a thickness of 65 Å formed from clearly defined repeating protomers. The 280-Å-diameter ring incorporating 10 repeating promoters matches closely the dimensions of the TPR suprahelix modeled on the four TPR homo-dimers of the APC/C (Fig. 6a). The ring-like structures are consistent with a view projected coincident with the TPR suprahelix axis with the oligomeric assembly resulting from alternating repeats of Cdc16 and Cdc23. However, to explain the occurrence of 8-, 10- and 12-protomer rings, it is also possible that closed ring structures rather than helices are generated, dependent on variations in the relative orientation of neighboring TPR homo-dimers. Our findings that a complex of Cdc16 and Cdc23 generates a TPR suprahelix with similar geometric parameters to those that define the relative packing of adjacent TPR homo-dimers of the APC/C indicate that TPR homo-dimers have a tendency to assemble into suprahelices.
Concluding remarks
Our analysis shows that the four canonical TPR subunits of metazoan APC/C self-associate to form related V-shaped homo-dimeric structures. Each TPR subunit of the dimer comprises 14 contiguous TPR motifs, with the N-terminal seven motifs responsible for the homo-dimer interface. The geometry of the TPR helix is ideally suited in forming an inter-clasp-like arrangement. The four TPR homo-dimers stack in parallel on one side of the complex allowing the ridges and grooves of the TPR superhelices to inter-digitate. Because of the uniform geometric arrangement of the parallel stacking of adjacent TPR homo-dimers, a left-handed suprahelical structure is generated. TPR proteins represent the ninth largest protein family of the three kingdoms of life,39 and their predominant function is to mediate a diverse array of protein–protein interactions and the assembly of multi-protein complexes.40 Thus, our finding that TPR proteins have the potential to assemble into suprahelices has implications for understanding the assembly of a variety of TPR-mediated multimeric complexes.
Materials and Methods
Cloning, expression, purification and crystallization of Cdc23Nterm
To clone the putative N-terminal dimerization domain of Cdc23 (Cdc23N19-302,Y302A) (S. pombe Cut23; 565 residues), we amplified and modified cDNA by PCR from a cDNA library pTN-TH7 (a gift from T. Nakamura, National BioResource Project, Osaka City University, Japan). This choice of domain boundary was based on a screen of Cdc23 constructs. A larger construct (residues 1–308) resulted in poorly diffracting crystals, whereas a protein comprising residues 1–342 was unstable. Residues 1–18, predicted to be disordered and poorly conserved, were truncated, whereas selection of the C-terminus of Tyr302 was based on the C-terminus of TPR7. A double StrepII tag together with a TEV (tobacco etch virus) cleavage site were attached to the C-terminus of Cdc23N19-302,Y302A (hence, termed Cdc23Nterm). The protein was expressed in either the insect cell baculovirus system or Escherichia coli. For insect cell expression, Cdc23Nterm was cloned into a pFBDM − vector41 to generate baculovirus that was then amplified to a suitable titer (> 1 × 108 plaque-forming units per milliliter). High5 cells were grown in Sf-900™ II SFM medium (Invitrogen) to a cell density of 2.0 × 106 cells per milliliter and were infected at a multiplicity of infection of 2. High5 cells were harvested after 72 h and the cell pellets were frozen at − 80 °C. For E. coli expression, Cdc23Nterm was cloned into pET28 and the protein was expressed at 25 °C for 16 h in E. coli B834 pRare2 cells. The same purification procedure was used for Cdc23Nterm expressed from insect cells and E. coli. The protein was purified using a combination of Strep-Tactin (Qiagen), TEV cleavage, anion-exchange chromatography Mono Q (GE Healthcare) and Superdex 200 size-exclusion chromatography (GE Healthcare). SeMet-labeled Cdc23Nterm was produced by using SeMet Medium Base Plus Nutrient Mix and SelenoMethionine Solution (Molecular Dimensions Ltd.) and purified as for native Cdc23Nterm. Crystals were grown by vapor diffusion in a buffer containing 200 mM sodium formate and 20% (w/v) polyethylene glycol 3350 and incubated in a cryoprotection buffer comprising 200 mM sodium formate, 22% (w/v) polyethylene glycol 3350 and 25% glycerol prior to freezing in liquid nitrogen.
Structure determination of Cdc23Nterm
Data analysis and detection of twinning
Native crystals, obtained from protein expressed in insect cells, diffracted to 2.0 Å resolution. The data could be indexed, processed and scaled nearly as well in P422 as in P4, hinting at possible hemihedral twinning. When twinned data exhibit near-422-point group symmetry, the presumptive true space group is the tetragonal one with rotational 4-fold symmetry since these space groups allow true merohedral twinning. Hence, the data were processed in P4 and further analyzed for twinning with L test and self-rotation function using PHENIX42 and MOLREP,43 respectively. These examinations clearly showed the presence of near-perfect twinning with the twin fractions related by the twin law h, − k, − l (Supplementary Fig. 4a and b). The L test and H test of PHENIX42 gave an estimated twin fraction of 0.48 (Supplementary Fig. 4b). After soaking with heavy-atom compounds such as EMTS (ethylmercury thiosalicylate), crystals showed significant shrinkage in the unit cell volume along with varied twin factions, ranging from 0.28 to 0.49. However, it is interesting to observe that one of the crystals soaked with EMTS did not show any twinning but failed to provide adequate phase information (Supplementary Fig. 4c and d).
Subsequently, the protein was expressed in E. coli and, as observed with the insect-cell-expressed protein crystals, both native and SeMet-labeled crystals had twin fractions ranging from 0.30 to 0.44 (Supplementary Fig. 4e and f). Diffraction data from all crystals showed systematic absences (00 l = 4n) indicating the presence of either a 41 or a 43 crystallographic symmetry axis. Therefore, subsequent analyses were carried out with the data processed using both of these symmetries.
SeMet SAD phasing in P1
Attempts to determine the structure of Cdc23Nterm crystals by molecular replacement using the N-terminal domains of either Cdc27 or Cdc16 as search models failed, presumably due to their low sequence identities of ~ 10% with Cdc23. We therefore determined the Cdc23Nterm crystal structure by means of SeMet SAD phasing. SeMet crystals diffracted to 3.0 Å and had twin fractions ranging from 0.28 to 0.46 (Supplementary Fig. 4e and f). Similar to EMTS-soaked crystals (data not shown), twinning suppressed the anomalous signal and the anomalous data failed to provide phase information.
Often when crystals are twinned, treating data with lower symmetry aids structure solution, although we found that reducing the symmetry from P422 to P4 failed to improve phases. Hence, we tried to further reduce the symmetry and solve the structure in P1. We collected 350° of data for a SeMet crystal at the Se edge and processed the data in P1. The estimated twin fraction for this crystal was 0.28 (Supplementary Fig. 4e and f). Compared to other datasets from all other crystals, these data had a better anomalous signal with a ΔFano/σ(ΔFano) value close to 1.02. Of 48 expected Se sites corresponding to 8 molecules in the asymmetric unit, 23 were detected with HySs42 and refined with Phaser.44 Subsequently, density modification was carried out with PARROT.45 Contrary to expectations, PARROT did not find non-crystallographic symmetry for the heavy atoms. The figure of merit (FOM) after density modification was 0.21 and the maps produced were not interpretable. Similar results were obtained with the same date processed in P41. However, with P41 data, 6 out of 12 Se atoms were located and density modification produced maps with FOM of 0.28. The electron density map showed a few rod-like features with discontinuous density features (Supplementary Fig. 5a). Slightly better maps were obtained with the de-twinned P41 data (Supplementary Fig. 5b). The handedness was resolved with de-twinned P41 using density modification. However, the electron density was discontinuous and difficult to interpret. Remarkable improvement was achieved when the data processed in P1 were de-twinned by applying the twin law specific to P41 (h − k − l). With the twin-treated P1 data, HySs42 detected 38 out of 48 Se sites and FOM after density modification increased to 0.52 (Supplementary Fig. 5c). The map showed clear molecular boundaries with contiguous electron density and helical densities corresponding to the TPR motifs were well resolved. Using this map, we traced the backbone with Buccaneer46 and manually built side chains with Coot.47 The resulting coordinates of one of the Cdc23Nterm molecules were used as a search model and the final structure was solved with molecular replacement using the un-twinned data obtained from EMTS-soaked crystal. Phaser44 was employed in molecular replacement. The correct solution corresponds to the space group P43212 with one molecule per asymmetric unit. The structure was further improved with manual model building using Coot47 and refined with PHENIX.42 Data collection and refinement statistics are given in Supplementary Table 1.
Modeling of Apc7 and C-terminus of Cdc23
The N-terminus of Apc7 was modeled on Cdc23Nterm, and the C-terminal seven TPRs of Cdc23 and Apc7 were modeled on the equivalent TPR motifs of S. pombe Cut9 (Cdc16)30 using Phyre.35
Human APC/C expression and purification
Recombinant human APC/C was expressed and purified as described previously,33 except that Cdh1 and Hsl1 (residues 667–872) were co-expressed with APC/C to generate the ternary APC/CCdh1.Hsl1 complex.
Human Apc6–Apc8–Apc13–Cdc26 expression and purification
Recombinant human Apc6(Cdc16)–Apc8(Cdc23)–Apc13–Cdc26 were co-expressed in High5 cells. For affinity purification, Apc8 was fused to a C-terminal TEV-cleavable StrepII 2 × tag. The proteins form a stable complex that was purified using Strep-Tactin chromatography. The complex composition was verified by mass spectrometric analysis of SDS PAGE excised bands.
Electron microscopy
The human APC/CCdh1.Hsl1 complex at ~ 20 μg/ml was loaded on glow-discharged Quantifoil 2/2 EM grids coated with a second layer of thin carbon. After 30 s, the grids were negatively stained with 2% (w/v) uranyl acetate. Data were collected at room temperature in a FEI Tecnai TF20 electron microscope at an accelerating voltage of 200 kV under low-dose conditions with an exposure of ~ 30 e−/Å− 2, a nominal magnification of 80,000 × and an underfocus of ~ 1.2 μm generating a first minimum in the contrast transfer function at ~ 17 Å. Images were recorded using a Tietz F415 CCD camera and adjacent boxes of 2 pixels × 2 pixels were averaged, resulting in a calibrated sampling of 2.36 Å/pixel.
Particles were automatically selected using the EMAN48 boxer software. A total of 12,757 particles were selected. The human APC/CCdh1.Hsl1 structure was determined using the human apo APC/C reconstruction33 as the initial reference for refinement. Multiple rounds of multi-reference alignment were performed using EMAN48 software, and angular assignment was performed by projection matching in IMAGIC.49 Atomic coordinates were docked into the human APC/C maps using Chimera.50 Grids for the Cdc16–Cdc23–Cdc26–Apc13 complex were prepared and data were collected using a similar protocol. Two-dimensional class average was determined using Refine2D in EMAN.48
Accession numbers
Coordinates and structure factors have been deposited in the PDB with accession numbers 3zn3 and r3zn3sf3zn3r3zn3sf, respectively.
Acknowledgements
The work was funded by a Cancer Research UK program grant to D.B. We thank staff at Diamond Light Source I04 and European Synchrotron Radiation Facility beamlines ID29 and ID14-4 for help with data collection. The S. pombe cDNA library pTN-TH7 was a gift from T. Nakamura, National BioResource Project, Osaka City University, Japan.
Edited by J. H. Naismith
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2013.04.004
Contributor Information
Kiran Kulkarni, Email: ka.kulkarni@ncl.res.in.
David Barford, Email: David.Barford@icr.ac.uk.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Peters J.M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev., Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan M., Morgan D.O. Finishing mitosis, one step at a time. Nat. Rev., Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 3.Barford D. Structural insights into anaphase-promoting complex function and mechanism. Philos. Trans. R. Soc. Lond., B Biol. Sci. 2011;366:3605–3624. doi: 10.1098/rstb.2011.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev., Mol. Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 5.Glotzer M., Murray A.W., Kirschner M.W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 6.Pfleger C.M., Kirschner M.W. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 7.Araki M., Yu H., Asano M. A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev. 2005;19:2458–2465. doi: 10.1101/gad.1361905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton J.L., Solomon M.J. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilioti Z., Chung Y.S., Mochizuki Y., Hardy C.F., Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- 10.Kraft C., Vodermaier H.C., Maurer-Stroh S., Eisenhaber F., Peters J.M. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 11.Pfleger C.M., Lee E., Kirschner M.W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwab M., Neutzner M., Mocker D., Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passmore L.A., McCormack E.A., Au S.W., Paul A., Willison K.R., Harper J.W., Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll C.W., Enquist-Newman M., Morgan D.O. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr. Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 15.da Fonseca P.C., Kong E.H., Zhang Z., Schreiber A., Williams M.A., Morris E.P., Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 17.Schreiber A., Stengel F., Zhang Z., Enchev R.I., Kong E.H., Morris E.P. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 18.Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sikorski R.S., Boguski M.S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 20.Pathare G.R., Nagy I., Bohn S., Unverdorben P., Hubert A., Korner R. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc. Natl Acad. Sci. USA. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J., Kulkarni K., da Fonseca P.C., Krutauz D., Glickman M.H., Barford D., Morris E.P. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric α-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Lupas A., Baumeister W., Hofmann K. A repetitive sequence in subunits of the 26S proteasome and 20S cyclosome (anaphase-promoting complex) Trends Biochem. Sci. 1997;22:195–196. doi: 10.1016/s0968-0004(97)01058-x. [DOI] [PubMed] [Google Scholar]
- 23.Pal M., Nagy O., Menesi D., Udvardy A., Deak P. Structurally related TPR subunits contribute differently to the function of the anaphase-promoting complex in Drosophila melanogaster. J. Cell Sci. 2007;120:3238–3248. doi: 10.1242/jcs.004762. [DOI] [PubMed] [Google Scholar]
- 24.Vodermaier H.C., Gieffers C., Maurer-Stroh S., Eisenhaber F., Peters J.M. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 25.Matyskiela M.E., Morgan D.O. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol. Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornton B.R., Ng T.M., Matyskiela M.E., Carroll C.W., Morgan D.O., Toczyski D.P. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izawa D., Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat. Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendt K.S., Vodermaier H.C., Jacob U., Gieffers C., Gmachl M., Peters J.M. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat. Struct. Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 29.Lamb J.R., Michaud W.A., Sikorski R.S., Hieter P.A. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Kulkarni K., Hanrahan S.J., Thompson A.J., Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Roe S.M., Diogon M., Kong E., El Alaoui H., Barford D. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. J. Mol. Biol. 2010;397:1316–1328. doi: 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 32.Sikorski R.S., Michaud W.A., Hieter P. p62cdc23 of Saccharomyces cerevisiae: a nuclear tetratricopeptide repeat protein with two mutable domains. Mol. Cell. Biol. 1993;13:1212–1221. doi: 10.1128/mcb.13.2.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Yang J., Kong E.H., Chao W.C., Morris E.P., da Fonseca P.C., Barford D. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) Biochem. J. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 34.Holm L., Kaariainen S., Rosenstrom P., Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 36.Han D., Kim K., Kim Y., Kang Y., Lee J.Y., Kim Y. Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7. J. Biol. Chem. 2009;284:15137–15146. doi: 10.1074/jbc.M804887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohi M.D., Feoktistova A., Ren L., Yip C., Cheng Y., Chen J.S. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol. Cell. 2007;28:871–885. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das A.K., Cohen P.W., Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian J., Stenger B., Wilson C.A., Lin J., Jansen R., Teichmann S.A. PartsList: a web-based system for dynamically ranking protein folds based on disparate attributes, including whole-genome expression and interaction information. Nucleic Acids Res. 2001;29:1750–1764. doi: 10.1093/nar/29.8.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeytuni N., Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Berger I., Fitzgerald D.J., Richmond T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 42.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 44.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowtan K. Recent developments in classical density modification. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 49.van Heel M., Gowen B., Matadeen R., Orlova E.V., Finn R., Pape T. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C.C., Pettersen E.F. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barton G.J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material