Abstract
The Plasmodium falciparum var multigene family encodes P. falciparum erythrocyte membrane protein 1, which is responsible for the pathogenic traits of antigenic variation and adhesion of infected erythrocytes to host receptors during malaria infection. Clonal antigenic variation of P. falciparum erythrocyte membrane protein 1 is controlled by the switching between exclusively transcribed var genes. The tremendous diversity of the var gene repertoire both within and between parasite strains is critical for the parasite's strategy of immune evasion. We show that ectopic recombination between var genes occurs during mitosis, providing P. falciparum with opportunities to diversify its var repertoire, even during the course of a single infection. We show that the regulation of the recombined var gene has been disrupted, resulting in its persistent activation although the regulation of most other var genes is unaffected. The var promoter and intron of the recombined var gene are not responsible for its atypically persistent activity, and we conclude that altered subtelomeric cis sequence is the most likely cause of the persistent activity of the recombined var gene.
Abbreviations: PfEMP1, P. falciparum erythrocyte membrane protein 1; PFGE, pulsed-field gel electrophoresis; TARE, telomere-associated repetitive element; Q-RT-PCR, quantitative reverse transcription–polymerase chain reaction; ICAM-1, intercellular adhesion molecule 1; CI, confidence interval
Keywords: malaria, recombination, var, switch, transcription
Introduction
Each Plasmodium falciparum parasite possesses approximately 60 members of the var multigene family, which encode variants of P. falciparum erythrocyte membrane protein 1 (PfEMP1), the immunodominant variant antigen of P. falciparum. PfEMP1 is expressed on the surface of the infected erythrocyte and is the parasite ligand that binds numerous host receptors in vascular tissues, thus mediating the pathogenic traits of infected erythrocyte sequestration in these tissues and rosetting. Var genes show tremendous diversity among parasites,1 and it has been proposed that much of this diversity arises from ectopic recombination between regions of homology within var genes leading to a vast repertoire of chimaeric var gene sequences.2 This has been demonstrated for meiosis and has been proposed to occur during mitosis.3
A single parasite abundantly transcribes a single var gene at one time and P. falciparum evades the host immune response through antigenic variation by switching between the dominant transcribed var genes. Partially defined epigenetic processes control the transcription of a single var gene. Histone deacetylation has been shown to silence some var genes,4 and specific histone methylation and acetylation modifications are associated with var gene silencing and activation.5,6
Var genes are present as clusters or single copies at chromosome internal or subtelomeric sites with upstream sequences (ups) defined by homology as being upsA, B, C, or E.2,7 The different ups types have a restricted genomic distribution: upsC are present in chromosome internal var genes; upsB are present in the most subtelomeric var genes, which are transcribed towards the centromere and in some chromosome internal var genes; upsA are present in var genes that are transcribed towards the telomere and that are also at subtelomeric sites but are centromeric to upsB var genes; upsE, which are always upstream of the conserved var2csa gene, have the same chromosomal location and orientation as the upsA var genes.2 The different orientations of the different ups type var genes have been proposed to restrict their ability to recombine to other var genes of the same type, thus forming compartments of var genes within the genomic repertoire.2 The rate at which var genes switch on and off is a fixed, gene-specific characteristic.8 The on and off rates are different so that genes that switch on rapidly and off slowly should predominate in a parasite in the absence of any selective pressure on PfEMP1. A recent study revealed that different ups types have different on- and off-switch rates with upsC var genes switching off much more slowly than upsA, upsB, or upsE.9 Whether the switch rate was dependent on the ups type, on the chromosomal location of the var gene, or on some other cis sequence element associated with the var gene locus could not be determined. Some cis sequence elements have been associated with var gene silencing including upstream elements10 and the var gene intron that possesses both bidirectional promoter activity11 and the ability to repress the activity of var gene promoters.12
We report the first evidence of a parasite that has undergone ectopic recombination within a var gene during mitosis to generate progeny carrying a chimaeric var gene. This recombination has occurred within the var gene intron and has generated a functional chimaeric var gene that has altered activity in that it is more rapidly turned on and persistently transcribed. The ability to switch the expression of var genes on and off at other loci appears unaffected, suggesting that the effect on var gene switching is due to the recombination. The chimaeric var gene remains in its original locus and its promoter is unaffected. We showed that subtelomeric repeat sequence could decrease var promoter activity and conclude that var gene regulation is not determined solely by upstream sequence or genomic location.
Results
Ectopic recombination within a var gene during mitosis
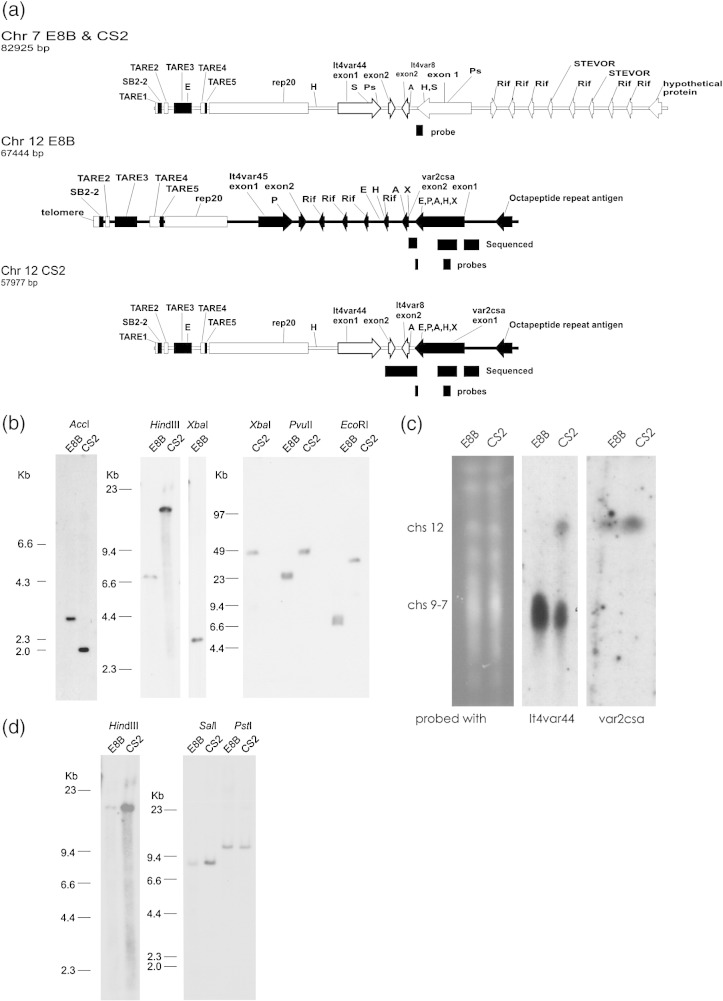
Ectopic recombination during mitotic division of the CS2 clonal parasite has replaced the var2csa intron, the second exon, and sequence downstream of var2csa with sequences originating within the var gene It4_var8 (Fig. 1a). The CS2 parasite is a mitotic progeny of the parental E8B clone and was derived from E8B by selection for adhesion to chondroitin sulfate A. The E8B parasite was itself cloned from ItG parasites. The CS2 parasites described in this study were cloned again prior to analysis. PCR using a degenerate second exon primer was used to obtain CS2 var2csa intron and second exon sequence. Universal Fast Walking PCR13 was used to obtain 2645 nucleotides of unknown sequence downstream of the var2csa second exon (Fig. 1a). Recombination within the var2csa intron was confirmed by Southern blotting AccI-digested DNA (Fig. 1b) and sequencing cloned PCR products that were amplified using specific primers on gDNA. Pulsed-field gel electrophoresis (PFGE) was used to confirm retention of var2csa on chromosome 12 in CS2 parasites and to identify the original location of the sequence integrated into the var2csa locus as either chromosome 7 or chromosome 8 (Fig. 1c). PFGE revealed that the genetic rearrangement was nonreciprocal, with the original sequence also remaining in the original locus on chromosome 7or 8 in CS2.
Fig. 1.
(a) Diagrams of the left-hand telomeric fragment of chromosome 7 and the var2csa locus on the left-hand telomeric fragment of chromosome 12 in wild-type E8B clonal parasites and in their clonal progeny CS2 following ectopic recombination between chromosome 7 and chromosome 12. The filled bar indicates wild-type chromosome 12, and the unfilled bar indicates the sequence from chromosome 7 introduced following recombination. Positions are indicated for TARE 1–5, TARE 6 (rep20), the second telomere-associated repeat sequence from subtelomeric block 2 (SB2-2),7 several members of the rif and var multigene families, and the restriction sites AccI (A), EcoRI (E), HindIII (H), PvuII (P), XbaI (X), SalI (S), and PstI (Ps). The positions of probes used for Southern and Northern blots and regions that were directly sequenced are indicated beneath the diagrams. (b) Southern blots of CS2 and E8B gDNA digested with the enzymes indicated above the figure and separated by conventional gels (AccI, HindIII, and XbaI) and PFGE (XbaI, PvuII, and EcoRI) were probed with sequence from the 3′ end of var2csa exon 1. (c) Southern blots of PFGE-separated chromosomes probed with sequence from the 3′ ends of var2csa and It4_var44. (d) Southern blots of CS2 and E8B gDNA digested with the enzymes indicated above the figure were probed with sequence from the 3′ end of It4_var8 exon 1.
We used ItG sequence data produced by the P. falciparum Sequencing Group at the Sanger Institute that can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/falciparum/IT_strain/ to identify sequences contiguous with those we had cloned. The ItG Supercontig_0002004 starts at telomere-associated repetitive element (TARE) 1, which is usually adjacent to the approximately 1.5-kb-long P. falciparum telomere sequence,14 and continues through TARE 1–6,15 It4_var44, and It4_var8. There is 41 kb between the start of Supercontig_0002004 and the recombination site within the It4_var8 intron. The Supercontig_0002007 available at ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/falciparum/IT_strain/ contains sequence including the 50 kb between the left-hand end of chromosome 12 and the recombination site in the var2csa intron. From this resource, a single copy conserved gene contiguous with It4_var8 was identified on the Sanger supercontig_0002004 that is also present as PF07_0004 at a subtelomeric site on chromosome 7 in 3D7 parasites. We concluded that the sequence integrated into var2csa came from chromosome 7.
The Southern blot hybridisation patterns of CS2 and E8B gDNA digested with EcoRI, HindIII, PvuII, and XbaI and probed with sequence from the 3′ end of var2csa exon 1 all confirmed that a duplicative transposition had occurred (Fig. 1b). An 18-kb HindIII fragment and a 36.8-kb EcoRI fragment from CS2 parasites hybridised to a probe from the 3′ end of var2csa exon 1 and confirmed that the left-hand end of chromosome 12 from the var2csa intron to at least the subtelomeric repeats had been replaced with chromosome 7 donor sequence that extended from near the start of the It4_var8 intron through the HindIII site between the It4var44 exon 1 and rep20 repeat sequences and to at least the EcoRI site in TARE 3. Replacement of the wild-type ItG chromosome 12 sequence with chromosome 7 resulted in loss of the PvuII and XbaI sites that generated the predicted fragments of 22 and 3.1 kb, respectively, from chromosome 12 in wild-type E8B parasites. The observed PvuII and XbaI fragments from chromosome 12 in CS2 parasites that hybridised to var2csa exon 1 were consistent with the predicted 43.6 XbaI and 42.5 PvuII fragments that would span from var2csa to the telomere if recombination with Supercontig_0002004 chromosome 7 sequence had resulted in replacement of the entire left-hand subtelomeric sequence of chromosome 12 with that of chromosome 7. PCR confirmed the loss from CS2 of the rif genes located between It4var_45 and var2csa on chromosome 12 of E8B (Supplementary Fig. 1)
Sequence from the first exon of It4_var8 hybridised on Southern blots to 11.35-kb PstI, 8.9-kb SalI, and 17.1-kb HindIII fragments of both CS2 and E8B gDNA, proving that the left-hand telomeric chromosome 7 sequence remained intact in CS2 from the PstI site halfway through the first exon of It4_var8 to at least the HindIII site between the It4var44 exon 1 and rep20 repeat sequences (Fig. 1d).
The recombined var2csa intron had a similar overall structure to other var introns (Supplementary Fig. 2). The three regions previously defined by strand asymmetry were present as were the repeats in the G-rich region 1 that were complementary to repeats present in the C-rich region 3.11 The number of these repeats was the same between CS2 var2csa, E8B var2csa, and It4_var8, although the spacing between the repeats was different. The intron region 2 that possesses bidirectional promoter activity was 526 nucleotides long in E8B var2csa and 449 nucleotides long in CS2 var2csa.
The chimaeric var2csa gene is persistently transcribed
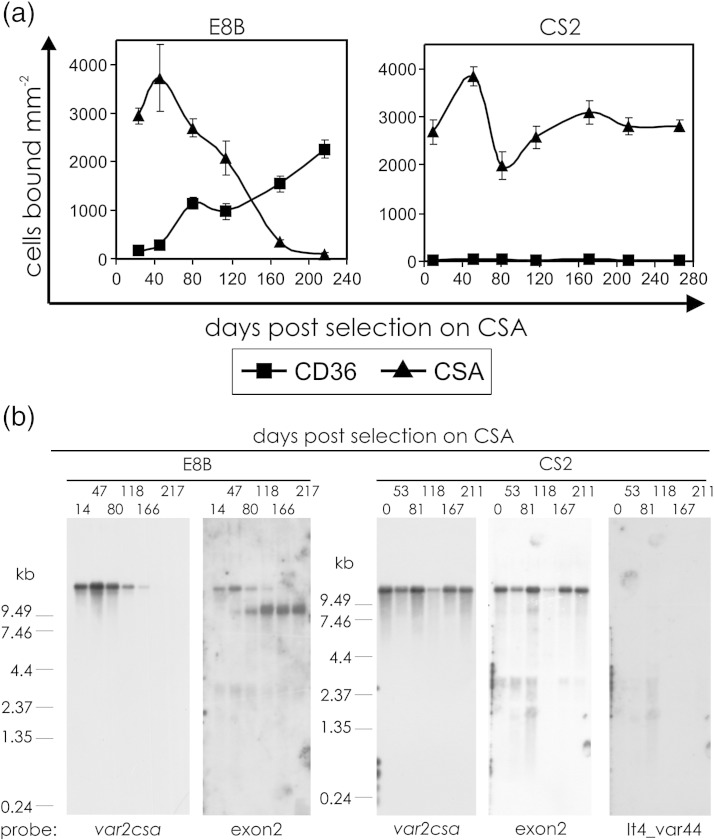
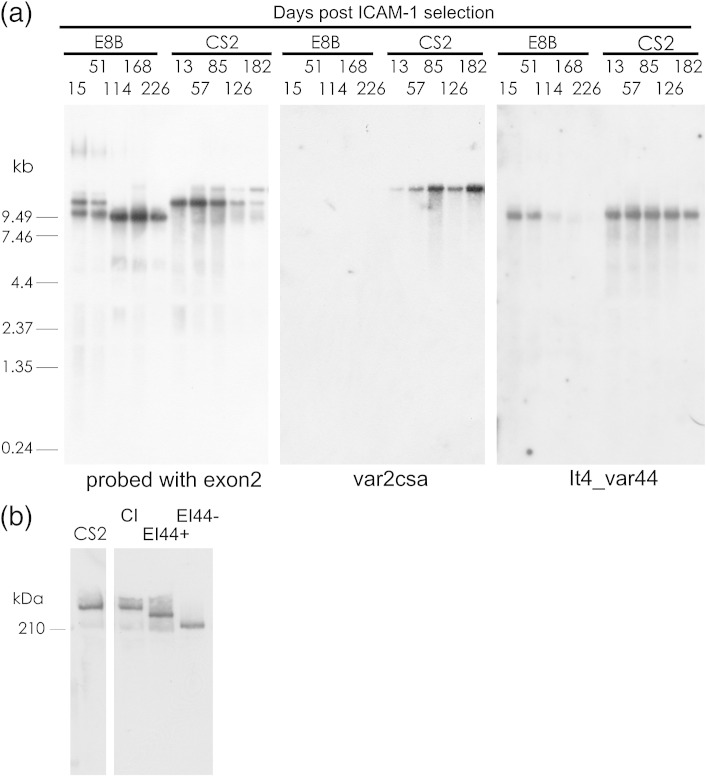
CS2 and E8B clonal parasites were selected for adhesion to CSA and grown for 266 and 217 days, respectively, in the absence of selection. Their adhesion phenotype and var gene transcriptional profile were tested at regular intervals over this time. CS2 parasites did not lose the ability to adhere to CSA over this period; however, the E8B parasites gradually lost the ability to adhere to CSA and reverted to the default CD36 adhesion phenotype (Fig. 2a). This reversion to CD36 adhesion by E8B parasites was accompanied by a decrease in transcription of var2csa and an increase in transcription of other var genes whilst CS2 parasites maintained their transcription of var2csa over the entire period (Fig. 2b).
Fig. 2.
(a) The numbers of CS2 and E8B parasites that bound per square millimeter to immobilised CSA and CD36 at different time points following selection on CSA. Means of three replicates for each of two assays conducted on separate days; error bars are standard error of the mean. (b) Northern blots of RNA from E8B and CS2 parasites collected at different time points following selection on CSA and probed with sequence from the centre of the first exon of var2csa (Fig. 1a), from the 3′ end of IT4_var44, and with a conserved var exon 2 probe.
Persistent activity of var2csa is not associated with promoter mutation
To determine whether mutation or recombination of the upsE sequence upstream of the var2csa in CS2 may have caused the persistent var2csa activity, we amplified, cloned, and sequenced 2381 bp of upstream sequence in both CS2 and E8B parasites. The sequences were amplified on two separate occasions using different methods, and 5 clones for each upsE were fully sequenced. Over the 2381 bp, there was an average of 2.5 mismatches and one insertion or deletion per clone relative to the consensus derived from all 10 clones. In all but two instances, the mutations were present in only a single clone; in the two exceptions, there were single and dinucleotide insertions in homopolymeric tracts that were present in only 2 of the 5 clones available for that upsE sequence. Thus, we concluded that the E8B and CS2 upsE sequences were identical over the 2381 bp and that persistent var2csa transcription in CS2 was not due to differences in the upsE sequence.
Persistent transcription of var2csa in CS2 parasites is not due to a general deficit in the var gene switching machinery
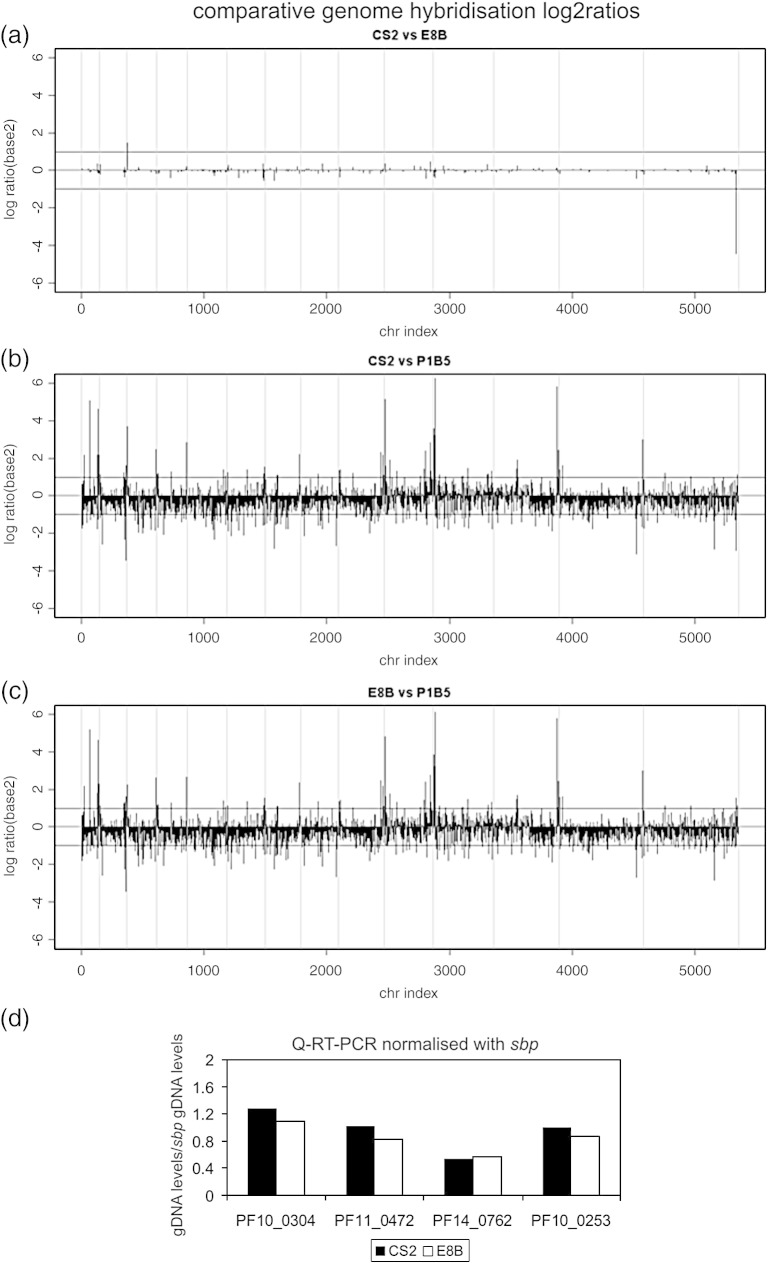
To determine whether the persistent var2csa transcription was caused by a loss of genes controlling epigenetic regulation, we compared genomic DNA from CS2 and E8B parasites by microarray hybridisation. The 3D7 array used would not detect loss of genes specific to ItG lineage parasites, but we assumed that genes controlling the var gene switching mechanism would be conserved among P. falciparum isolates. There was very little genetic variation between E8B and CS2, confirming their mitotic, clonal lineage (Fig. 3). This finding contrasted with the polymorphism previously observed between Ituxi (IT) subclones including the sequenced clone P1B5. Comparative analysis with P1B5 revealed much more polymorphism between it and either CS2 or E8B than between CS2 and E8B, suggesting that the previously observed polymorphisms between IT subclones related to the source from which they were derived.16
Fig. 3.
Comparative genome hybridisation log2ratios of (a) CS2 compared to E8B, (b) CS2 compared to P1B5, and (c) E8B compared to P1B5. The genes (vertical black bars) are plotted as physical order on the x-axis whilst the log ratios are on the y-axis. The zero line shows the distribution of the genes if no variation was to be found between the two strains compared. The horizontal lines at + 1 and − 1 are limits indicating amplification (log2ratio > 1) and deletion (log2ratio < − 1), respectively. The grey vertical bars delimit the chromosomes. (d) The skeleton binding protein gene sbp and genes identified as possibly deleted or duplicated by microarray were quantitated in both CS2 and E8B gDNA using standard curves of serially diluted gDNA. The levels of sbp were used to normalise the levels of the other genes so that the resulting relative values compare the level of each gene in equivalent amounts of CS2 and E8B gDNA.
Five genes had log2 CS2/E8B ratios ≥ 1 or ≤ − 1, indicating duplication or deletion (Supplementary Table 1). PFC0015c log2ratio CS2/E8B = 1.452014 is a subtelomeric VARC pseudogene, that is, var exon 2 sequence, which is unlikely to be conserved between 3D7 and ItG. The probeset for PFC0015c was limited to two oligonucleotides (PF03.4611c_st and PF03.4611w_st) that were, in fact, complementary; thus, the probeset only covered a single 25-nucleotide sequence that was 80% identical with exon 2 of It4_var8, 84% identical with exon 2 of It4_var44, and 88% identical with exon 2 of It4_var45 but had no identity to the wild-type E8B var2csa exon 2. Therefore, the recombination at the var2csa locus in CS2 led to a net gain of one sequence that could hybridise to these probes and PFC0015c was not investigated further.
The other genes that varied between CS2 and E8B were PF14_0762, a predicted exported protein of unknown function with a log2ratio(CS2/E8B) = − 4.45878, and three predicted proteins of unknown function, PF10_0304, PF11_0472, and PF10_0253, which had log2ratio(CS2/E8B) values of 1.032, 1, and 0.984, respectively. Quantitative reverse transcription–polymerase chain reaction (Q-RT-PCR) primers that amplified a region covered by the microarray probes were designed and used to quantitate the levels of these genes in CS2 and E8B gDNA (Fig. 3d). All of the genes were shown to be present at equivalent levels in CS2 and E8B gDNA; hence, we concluded that microarray analysis had not detected any amplifications or deletions in CS2 versus E8B apart from the net gain of one var exon 2 sequence with homology to the 3D7 VARC PFC0015c. Although this analysis was a useful screen for candidate regulators of switching, it could not detect recombinations or point mutations within genes encoding regulators of var gene switching.
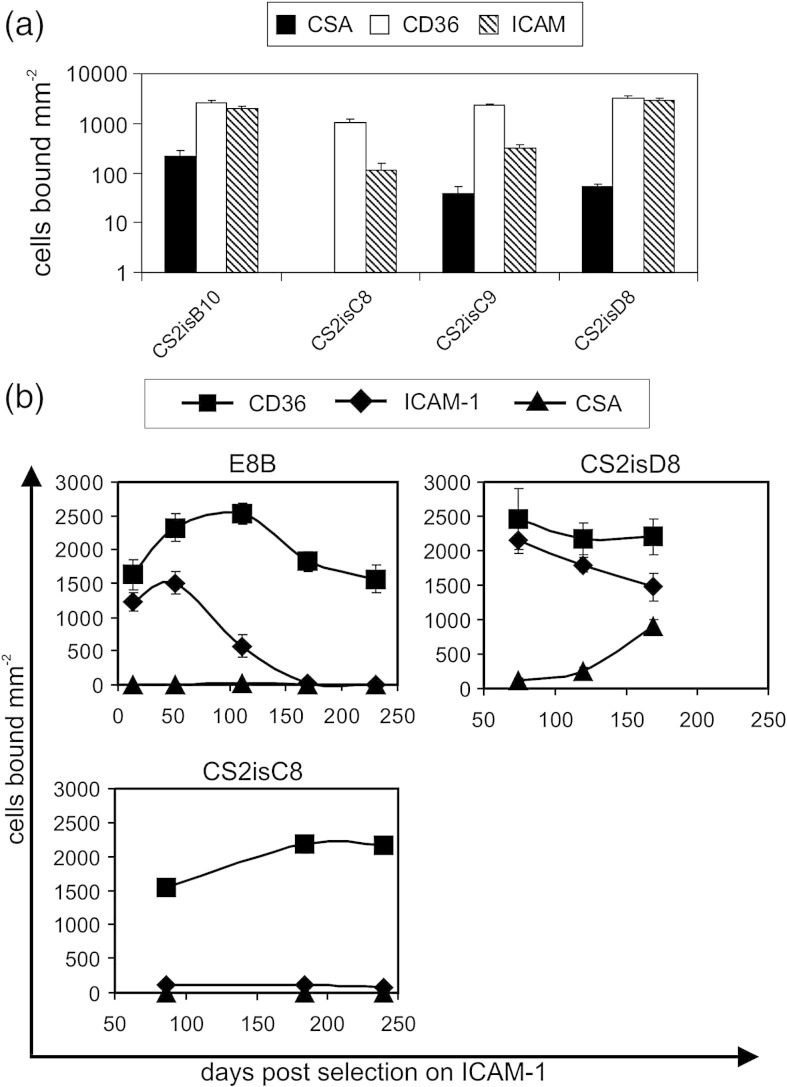
To confirm that loss of trans factors involved in general epigenetic regulation was not responsible for the persistent transcription of var2csa in CS2, we analysed the ability of CS2 and E8B parasites to switch from expression of var genes encoding an intercellular adhesion molecule 1 (ICAM-1) adhesion phenotype. CS2 and E8B parasites were selected for adhesion to ICAM-1 and then the CS2 parasites were cloned prior to further analysis. This excluded the possibility that a minor population of CSA binding CS2 parasites persisted through the selection for ICAM-1 and then outgrew the ICAM-1 binding CS2 parasites. The four CS2 clones analysed bound predominantly to ICAM-1 and CD36, which can be encoded by the same var gene.17 However, three clones (CS2isB10, CS2isC9, and CS2isD8) also bound at low levels to CSA (Fig. 4a) and had therefore switched on var2csa expression during the 65 days between ICAM-1 selection and recovery of sufficient culture of the CS2 clones to perform adhesion assays. The ICAM-1-selected E8B and two of the ICAM-1-selected CS2 clones were then grown continuously in vitro. Both the ICAM-1-selected E8B and the CS2 clone CS2isD8 progressively lost adhesion to ICAM-1; the CS2 clone CS2isC8 had presumably already lost ICAM-1 adhesion during the period of cloning and subsequent growth (Fig. 4b). Both E8B and CS2 switched to transcription of other var genes (Figs. 5a and 6), proving that the persistent var2csa transcription was not due to a general defect in the ability to switch between transcription of different var genes due to the loss of factors regulating var gene switching. The ICAM-1-selected CS2 parasites spontaneously switched to transcription of var2csa when grown in the absence of phenotype selection but the parental E8B parasites selected on ICAM-1 did not (Figs. 5a and 6), demonstrating that var2csa had a higher on switch in CS2 than in the wild-type E8B.
Fig. 4.
(a) CS2 parasites were selected on ICAM-1 and then cloned, and the number of infected erythrocytes of each clone that bound per square millimeter to immobilised CSA, CD36, and ICAM-1 was determined. (b) The numbers of erythrocytes infected with CS2 clones and E8B parasites that bound per square millimeter to immobilised CSA, CD36, and ICAM-1 at different time points following selection on ICAM-1. Means of three replicates for each of two assays conducted on separate days; error bars are standard error of the mean.
Fig. 5.
(a) Northern blot of RNA collected from E8B and CS2 parasites at different times after selection for adhesion to ICAM-1 and sequentially probed with sequences from var2csa, the 3′ end of exon 1 of It4_var44, and the conserved var exon 2. (b) Western blot probed with rabbit antiserum to the conserved exon 2 of PfEMP1; samples were CS2, CS2 selected on ICAM-1 and cultured until both var2csa and It4_var44 were transcribed (CI), E8B soon after selection on ICAM-1 when It4_var44 was transcribed (EI44+), and E8B selected on ICAM-1 and grown until transcription of It4_var44 had ceased (EI44-).
Fig. 6.
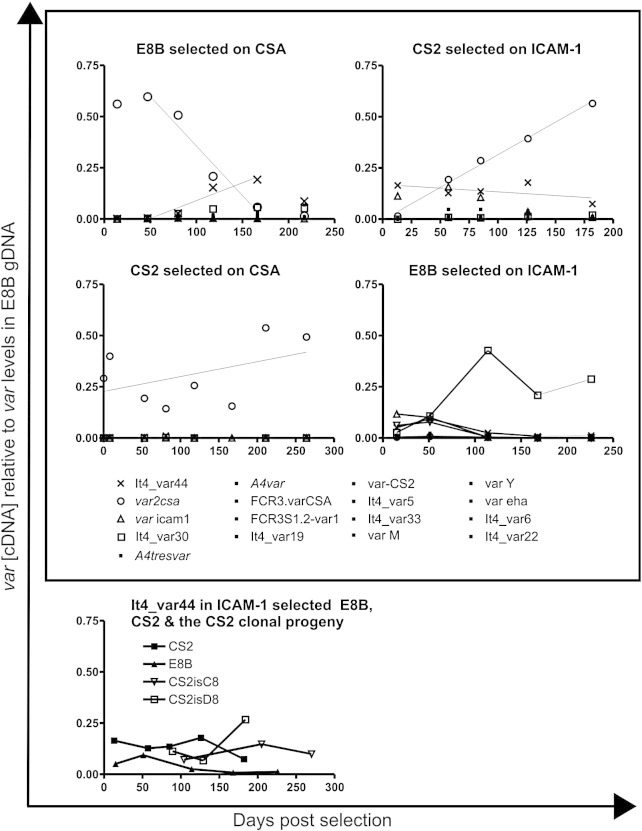
Q-RT-PCR was used to determine the levels of 17 var genes in the cDNAs of E8B and CS2 parasites at different time points during continuous growth following selection for adhesion to CSA and ICAM-1. Quantitation was performed using efficiency correction for each PCR and expressed relative to the levels of each gene in E8B gDNA following normalisation with the skeleton binding protein gene sbp. Linear regression curves are indicated on the plots of E8B selected on CSA and CS2 selected on CSA and ICAM-1.
The var2csa on-switch rate is different between CS2 and E8B parasites
The off-switch rates for var genes in 3D7 were determined previously as approximately 1% for var2csa and between 0% and 0.3% for the upsC var genes.9 We estimated switch rates for var2csa using Q-RT-PCR. The cDNA levels of 17 ItG var genes were determined at different time points over several months of continuous culture following selection of CS2 and E8B parasites for adhesion to CSA or ICAM-1. The ratio of the quantity of cDNA of each var gene relative to the quantity of that gene present in a constant amount of E8B gDNA was determined by normalising with the skeleton binding protein gene.18 We assumed that each gene was present as a single copy in the E8B genome, and therefore, fold difference relative to gDNA allowed us to compare expression between genes as well as between samples. Where possible, we fitted linear regression curves to the data and used the equation of the resulting curves to estimate the on-switch rate of var2csa in ICAM-1-selected CS2 as 1.02% (0.84%, 1.35%) per generation [95% confidence interval (CI)] and the var2csa off-switch rate in CSA-selected E8B as 1.56% (1.35%, 1.76%) per generation (95% CI) (Fig. 6). The calculation of all gene switch rates is detailed in Methods. The var2csa on-switch rate in E8B was much slower than that in CS2 parasites, preventing detection of spontaneous on-switching in the absence of selection on CSA, and therefore, the E8B var2csa on-switch rate could not be calculated. The levels of var2csa cDNA in the serial samples of CSA-selected CS2 parasites fluctuated too greatly to allow accurate fitting of a linear regression curve and calculation of a switch rate.
The mutant var intron is not less effective at repressing upsE
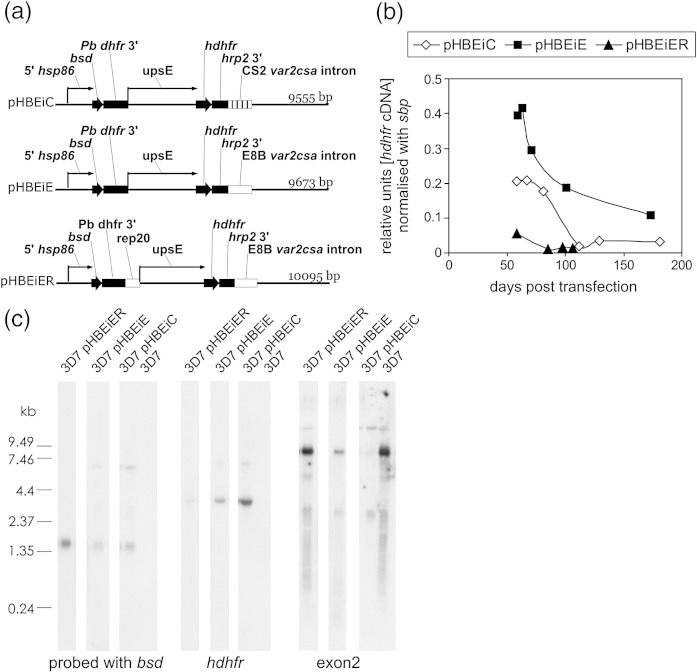
It is known that the var intron can repress var genes,12 and thus, recombination within the CS2 var2csa intron was a potential cause of persistent var2csa activity. To determine whether the replacement of the var2csa intron with the chimaeric intron contributed to the persistent var2csa activity in CS2, we transfected 3D7 parasites with plasmids that carried either intron. These plasmids contained the upsE sequence from CS2 parasites driving transcription of hdhfr, followed by either the mutant var2csa intron from CS2 (pHBEiC) or the wild-type var2csa intron (pHBEiE). A third plasmid (pHBEiER) differed from pHBEiE in that it carried the TARE 6 (rep20) preceding the upsE (Fig. 7a). The plasmids also contained the blasticidin deaminase gene driven by the P. falciparum hsp86 promoter and were stably maintained by selection for blasticidin resistance.
Fig. 7.
(a) Plasmids transfected into 3D7-infected erythrocytes carried the P. falciparum heat shock protein 86 promoter (5′ hsp86) driving transcription of the blasticidin deaminase gene (bsd) followed by the Plasmodium berghei dihydrofolate reductase transcriptional terminator (Pb dhfr 3′) followed by the ItG upsE driving transcription of human dihydrofolate reductase (hdhfr) followed by the P. falciparum histidine-rich protein 2 transcriptional terminator (hrp2 3′) followed by either the E8B or CS2 var2csa intron in plasmids pHBEiE and pHBEiC, respectively. The plasmid pHBEiER is the same as pHBEiE except for the inclusion of the TARE 6 (rep20) upstream of the upsE. (b) Plasmids were maintained in 3D7 parasites by growth in the presence of blasticidin-S, and the levels of hdhfr transcription were determined at different time points following transfection by Q-RT-PCR. Absolute quantitation was performed using standard curves of serially diluted, cloned hdhfr DNA of known concentration. The results were normalised using skeleton binding protein 1 cDNA levels to allow comparison of equivalent quantities of parasite material and then expressed as relative units of hdhfr cDNA per plasmid by dividing by the number of plasmids per genome. Plasmid number was determined by using Q-RT-PCR of transfected parasite gDNA to determine hdhfr normalised with sbp. (c) Northern blots of RNA from transfected parasites and nontransfected 3D7 parasites serially probed with sequence from bsd, hdhfr, and a conserved var exon 2.
The level of hdhfr transcripts per plasmid clearly decreased over time in parasites carrying pHBEiC, pHBEiE, and pHBEiER (Fig. 7b). Linear regression curves fitted to the first four time points of the pHBEiE and pHBEiC hdhfr cDNA levels (Fig. 7b) gave rates of decay of 1.49% per generation for pHBEiE and 1.65% per generation for pHBEiC, both of which were similar to the observed rate of 1.56% per generation for var2csa in E8B. The level of hdhfr transcribed per plasmid by parasites carrying the mutant var intron on pHBEiC was actually lower than the level transcribed by the wild-type intron pHBEiE parasites. Therefore, the mutant intron did not fail to silence the upsE in these constructs and we concluded that it was not responsible for the persistent var2csa activity in CS2 parasites.
The level of hdhfr transcripts in parasites carrying the rep20 construct pHBEiER was much lower than that in the other transfected parasites (Fig. 7b and c), and the hdhfr transcripts declined more rapidly in pHBEiER parasites than in the other transfectants to reach a low steady state (Fig. 7b). This indicated that the upsE was more effectively silenced when associated with rep20. However, Southern blots and the Sanger supercontig_0002004 sequence predicted an increase in the length of rep20 in the left-hand subtelomeric region of CS2 chromosome 12. Therefore, loss of rep20 was not responsible for decreased silencing of var2csa in CS2, but the observed silencing effect of rep20 on the episomal upsE indicates that subtelomeric cis sequence elements can repress var genes.
Attempts to select transfected parasites for expression of hdhfr by treating with the drug WR99210 did not succeed despite detectable hdhfr transcripts (Fig. 7c) that, at their peak in pHBEiC and pHBEiE, were present at 24% and 23%, respectively, of the maximum level of upsE transcripts in CS2 (as determined by absolute Q-RT-PCR of var2csa DBL3x in CS2 using standard curves of purified plasmid containing the DBL3x sequence). Repeated independent transfections with new plasmids also generated blasticidin-resistant transfectants that transcribed hdhfr at similar levels but were not WR99210 resistant (data not shown). We concluded that the recently described posttranscriptional silencing exerted by an open reading frame within the upsE probably prevented translation of functional protein from the hdhfr transcripts.19 This prevented examination of parasites exclusively expressing the episomal upsE, but the observed levels of upsE activity were sufficient to down-regulate endogenous var gene transcription in pHBEiE and pHBEiC parasites (Figs. 7c and 8), which confirmed that the upsE activity we observed in the transfected parasites was in the biologically relevant range.
Fig. 8.
Fold differences in the cDNA levels of the transcribed var repertoire of transfected parasites relative to 3D7 nontransfected parasites calculated from 2− ΔΔCt values normalised using arginyl tRNA synthetase.
The var gene introduced downstream of recombined var2csa is also frequently activated
The greatly diminished transcription of hdhfr by parasites carrying plasmids with rep20 (pHBEiER) indicated that altered subtelomeric sequences could modify gene regulation and may contribute to persistent var2csa expression in CS2 parasites. If differences in the subtelomeric sequences cis to var2csa were responsible for its persistent activity in CS2, then persistent activation might also be observed for the It4var44 gene that was duplicated from chromosome 7 to chromosome 12 downstream of var2csa in CS2. Both ICAM-1- and CSA-selected E8B parasites express It4_var44, and linear regression of It4_var44 expression over four time points in CSA-selected E8B revealed a moderate on-switch rate of approximately 0.6% per generation (Fig. 6). In both CSA- and ICAM-1-selected E8B, It4_var44 was rapidly switched off. In contrast, the ICAM-1-selected CS2 parasites maintained similar, intermediate levels of It4var44 transcripts for the 182 days examined [0.14 ± 0.01 (mean ± standard error of the mean) = 23% of the maximal levels of var2csa transcripts] (Figs. 5 and 6) and the CS2isC8 and CS2isD8 subclones continued to express It4_var44 at similar levels for the 270 and 184 days, respectively, they were monitored (Fig. 6). Therefore, It4_var44 is switched on rapidly in both E8B and CS2 and switched off rapidly in E8B but its activity is maintained in CS2.
Discussion
We have provided the first evidence of ectopic recombination between var genes during mitosis. This mechanism and meiotic recombination of var genes3 together generate the mosaic var genes that contribute to the tremendous diversity of the var gene repertoire.1–3 Ectopic recombination during mitosis could increase the rate at which P. falciparum diversifies its repertoire of subtelomeric contingency genes and also provides parasites with the capability to generate novel variant antigens during the course of a single infection. The var2csa gene had recombined with an upsA gene that shared its subtelomeric location and its orientation towards the centromere, consistent with previous observations that var gene recombination is restricted by genomic location.2 The recombination initiated within the intron of var2csa and replaced the second exon; hence, the recombination of var2csa did not alter either the VAR2CSA adhesion phenotype or the VAR2CSA epitopes important for immunity.
The presence of factors required for homologous recombination and the absence of factors required for nonhomologous end joining7 suggests that mitotic recombination within var2csa in CS2 arose from double-strand break repair by non-allelic homologous recombination.20 Consistent with this model, there are 206 nucleotides at the start of the recombination that share 97% homology between CS2 and ItG wild-type var2csa and which immediately precede the divergence between CS2 and ItG var2csa sequences (Supplementary Fig. 1). This region of homology contains four polymorphic nucleotides between wild-type and CS2 var2csa that are conserved between CS2 var2csa and It4_var8, indicating that it corresponds to the It4_var8 sequence that was invaded by the 3′ protruding strand of the 5′ resected, broken var2csa duplex.21
The var gene switch rates are an intrinsic characteristic of each gene,8 but we have shown that intact, identical upsE promoters in the same locus in two isogenic parasites differ in their switch rate. Thus, the most probable determinants of var gene switch rate previously reported, the var gene promoter type9 and chromosomal location,8,9 do not alone account for a parasite's predisposition to express a particular var gene in the absence of any selective pressure. Var2csa is translated in CS2 parasites (Fig. 5b); thus, the increased activation of var2csa in CS2 is not caused by the recently described repression of PfEMP1 translation that is dependent on continual transcription of untranslated var2csa mRNA.22 Neither is it caused by loss of a gene encoding a generic var gene regulator because CS2 parasites can switch on and off transcription of var genes encoding adhesion to ICAM-1. The reversion to expression of var2csa by cloned ICAM-1-selected CS2 parasites showed that the increased var2csa on-switch rate persisted through silencing of var2csa and switch to expression of other var genes. This indicates that the altered switch rate of var2csa in CS2 is due to a genetic change because epigenetic memory of a var gene's activity is erased after silencing.23 Whether the var2csa off-switch rate is decreased in CS2 is unclear. The protracted expression of var2csa in CS2 could result from a normal off-switch rate paired with an on-switch rate higher than that for other var genes. However, there is no evidence that the continual switching predicted from such an arrangement occurred during 280 days of continuous culture of CS2 parasites as CD36 adhesion did not increase at any stage; neither were transcripts of other var genes detected on Northern blots probed with a conserved exon 2 probe, nor were increased levels of transcripts of any of the 17 var genes assayed by Q-RT-PCR detected. There are two probable explanations for the increased activation of var2csa in CS2. Firstly, cis sequence elements that contribute to regulation of the var2csa locus have been altered. Second, the parasites may have lost a trans factor that, when present, represses only var2csa.
The only cis sequence elements shown to regulate var gene transcription to date are the ups upstream sequence and the intron. The var intron can repress var promoters.12,24 The var intron also has bidirectional promoter activity11,25 and there are precedents in other organisms for transcription of non-coding RNAs repressing transcription of heterologous genes.26 The CS2 and E8B upsE were identical but recombination at the var2csa locus commenced within the var2csa intron, which led us to suspect that the chimaeric intron contributed to persistent activation of var2csa. We analysed the intron by transfection studies but found that the episomal CS2 var2csa intron was not less effective at silencing the upsE than the episomal E8B var2csa intron. These data strongly suggested that the mutant CS2 var2csa intron did not confer increased upsE activity although it is possible that the introns behave differently when integrated in the var gene locus.
The plasmids we used contained the hsp86 promoter that was recently shown to repress a var promoter.27 However, episomal var introns still repress their cognate var promoters by approximately 50% despite the presence of the hsp86 promoter.28,29 Therefore, despite the presence of the hsp86 promoter, we expected to observe greater silencing of the episomal upsE by the E8B var2csa intron than by the CS2 var2csa intron if the CS2 var2csa intron was responsible for the increased var2csa activity. In fact, we observed the reverse as the episomal upsE cis to the CS2 var2csa intron had less activity than the episomal upsE cis to the E8B wild-type var2csa intron (Fig. 7b). We concluded that any repression of the upsE exerted by the hsp86 promoter had not masked failure by the CS2 var2csa intron to decrease activity of the upsE. The var promoters that may have been silenced by hsp86 in previous studies were all silenced by default.27–29 In contrast, the episomal upsE was not silenced by default but retained 24% of wild-type upsE activity 60 days after transfection, indicating that the upsE was largely unaffected by the hsp86 activity. The episomal upsE was active at biologically relevant levels because it exerted considerable repression on endogenous var gene transcription, possibly through promoter competition for a limited pool of trans acting factors.23
Differences in the sequence between the telomere and var2csa in E8B and CS2 may be responsible for the increased activation of var2csa in CS2. This is suggested by the frequent activation in both E8B and CS2 of the It4_var44 gene that was duplicated downstream of var2csa in CS2. Transposition of the readily activated It4_var44 locus downstream of the var2csa locus may have conferred on CS2 an ability to readily activate var2csa similar to It4_var44, resulting in the higher on switch of var2csa in CS2 than in wild-type E8B. Specific sequences cis to It4_var44 and/or general structural features, such as the shorter distance from var2csa to the telomere in CS2, may have affected var2csa regulation in CS2. It4_var44 activity is maintained for far longer in CS2 than in E8B, which may be due to a faster on switch or a slower off switch in CS2 than in E8B. However, the It4_var44 locus is the same on chromosomes 7 and 12 in CS2 and chromosome 7 in E8B; thus, It4var44 probably has the same moderately high on-switch rate in both CS2 and E8B, leading to frequent activation in both parasites. Because there are two copies of It4_var44 in CS2, their cumulative activity could maintain a more constant, intermediate level of It4_var44 transcript abundance in CS2 than in E8B.
The greatly reduced activity of the upsE on the plasmid carrying the rep20 proved that subtelomeric, cis sequence elements can affect upsE activity, although lack of rep20 itself was probably not responsible for the diminished silencing of var2csa in CS2. In contrast, rep20 does not affect upsB and upsC activity.28,29 Rep20 presumably constrained the upsE plasmid mobility by strong, random association with primarily silent telomeric clusters at the nuclear periphery.30 The upsB and upsC sequences must themselves form associations with silent telomeric clusters that are sufficiently strong to render the association mediated by rep20 redundant. Interestingly, the CS2 var2csa gene does not leave telomeric clusters to be transcribed,31 whereas the var2csa genes in 3D7 parasites and FCR3 parasites (which are isogenic with CS2) do leave telomeric clusters when transcribed.22,32 Subtelomeric sequences determine telomere localisation to telomeric clusters.14 Hence, differences between CS2 parasites and FCR3 and 3D7 parasites in the subtelomeric sequences contiguous with var2csa may contribute to the different mobilities of the var2csa locus in these parasites.
Subtelomeric sequences cis to the var2csa locus could determine the rate of var2csa activation by binding trans factors required for epigenetic activation or silencing. Both trans factors Sir2 and Orc1 bind subtelomeric sequences, and Sir2 binding spreads upstream and silences var2csa.4,33,34 Doubtless, other sequences cis to var2csa are also required for typical regulation of var2csa, including an unknown cis sequence that recruits the trans factors responsible for trimethylation of lysine 9 of histone 3 in silent var2csa.6 Sequences cis to var2csa could also bind trans factors required for formation of chromatin loops that are tethered in transcriptionally active or repressed subnuclear domains.35
Effective var gene switching is obviously important for evasion of the immune response but the biological significance of the different var gene switch rates is unclear. PfEMP1s expressed by parasites causing severe malaria appear to be more commonly encountered than PfEMP1s expressed by parasites causing non-severe malaria, but this frequency decreases with increasing host age and presumably with acquisition of effective immunity.36,37 Conversely, expression of the upsA and upsB var genes is associated with more severe disease, but the upsC var genes are expressed more frequently in the absence of selective pressure.9,38,39 It has been proposed that their adhesion phenotype confers a selective advantage on the more pathogenic and less frequently expressed upsA and upsB var genes, but development of host immunity removes this advantage, allowing the more frequently expressed and less pathogenic upsC var genes to predominate.9 Thus, the difference in switch rate between different var genes may be unimportant for the prevalence of severe disease in children but will contribute to the observed pattern of infection in older individuals and may protect the parasites antigenic repertoire.
We have shown that the ups type, the var intron, and chromosomal location are insufficient to determine the switch rate. We have also shown that the cis sequence element rep20 can affect var transcription and our findings indicate that other cis sequence elements lying outside subtelomeric var genes contribute to their intrinsic switch rate. Further investigation of the subtelomeric sequences of CS2 parasites will provide valuable insights into the mechanisms P. falciparum employs to initiate var gene switching, a critical component of the process of antigenic variation.
Methods
Plasmodium culture, selection for adhesion phenotype, assay of adhesion phenotype, and transfection
The Brazilian P. falciparum isolate IT was cloned twice by Dr. L. H. Miller (National Institutes of Health) to generate the clone ItG2F6. In our laboratory, ItG2F6 was cloned again to generate the clone FAF6.40 A line derived in our laboratory from FAF6 for adhesion to endothelial cells was named interchangeably FAF-EA841 or E8B.42 E8B was selected for adhesion to CSA to generate CS2.43 We cloned CS2 prior to this study. Therefore, E8B was derived from the IT isolate by three serial clonings by limiting dilution and CS2 was derived from E8B by one limiting dilution cloning. The IT parasites are isogenic with parasites frequently described as FCR3, presumably due to an early contamination.44,45
P. falciparum parasites were maintained in culture as previously described.46 The parasite culture medium was supplemented with either 0.5% albumax II (Gibco) for nontransfected parasites or 5% heat-inactivated human serum 0.25% albumax for transfected parasites. CS2 and E8B parasite lines were selected for high-level adhesion to receptors immobilized on plastic Petri dishes by repeated panning.46 The receptors bound to the dishes were 50 μg/ml bovine trachea CSA (BT-CSA) (Sigma), 8 μg/ml recombinant ICAM-1 (Bender MedSystems), and 15 μg/ml recombinant CD36 (R&D Systems). 3D7 parasites were transfected with 80 μg of purified plasmid (Qiagen)47 and selected for resistance to 2 μg ml− 1 blasticidin-S 4–6 h after transfection.
Cloning
The wild-type It4_var4 (ItG var2csa) sequence from within the first exon to within the second exon was amplified from E8B gDNA and cloned and sequenced. The mutant CS2 var2csa sequence from the end of the first exon to the end of the second exon was amplified using the forward primer GGTATTGCGTTGGCGTTAGG and the degenerate second exon reverse primer ATATCCAHTTCTTCATAYTCACTTTC. The second exon sequence was used to prime amplification of 2645 nucleotides of unknown sequence downstream of CS2 var2csa by Universal Fast Walking PCR13 using the oligonucleotides primer 1, ATACATCCCCAAACCTACG; primer 2, TATCCCTACACGTCACCTAANNNNNNNNNN; primer 3, AGTGATAGTGGACACTACTACGAA; and primer 4, CAAGTGGAAATAGTTCAACAAATACC. The var2csa upstream sequence (upsE) was amplified from both CS2 and E8B parasites as 2256 nucleotides of sequence 5′ of the var2csa coding sequence. PCR products were cloned and sequenced by standard methods.
The plasmid pHBCAMR 28 was digested with EcoRI and ligated to either CS2 or E8B var2csa intron sequences that were engineered with complementary MfeI sites at both ends. These plasmids were digested with PstI and BamHI and then ligated with the 2256-bp CS2 var2csa upstream sequence (upsE) that had been amplified with complementary PstI and BamHI sites. The resulting plasmid carrying the E8B wild-type var2csa intron was named pHBEiE, and the plasmid carrying the CS2 mutant var2csa intron was named pHBEiC. The plasmid pHBEiER was made by ligating PstI-digested pHBEiE with a 540-nucleotide rep20 sequence with complementary PstI sites at each end.
Southern blots
Separation of P. falciparum chromosomes by PFGE was performed as previously described.48 PFGE of digested gDNA was performed on the BioRad CHEF-DR II apparatus using a modification of the manufacturer's method; briefly, electrophoresis through 1% agarose in 0.5 × TBE was performed for 14 h at 6 V/cm using a ramped switch time from 1 to 6 s. Southern blots of conventional gels and PFGE were probed with [α-32P]dATP random prime-labelled DNA. Southern blots of digested gDNA were probed with sequence from nucleotides 7354 to 7607 at the 3′ end of the CS2 var2csa first exon. Southern blots of PFGE-separated chromosomes were probed with sequences from the 3′ end of It4_var44 (nucleotides 5557–5993) and the middle of ItG var2csa (It4_var4) (nucleotides 2275–3406). Southern blots were washed with 0.2 × SSC 0.1% sodium dodecyl sulfate (SDS) at 65 °C.
RNA extraction and Northern blots
Trizol reagent (Sigma)49 was used to extract RNA from early ring stage parasite cultures at approximately 8 h post-invasion. Northern blots were performed using 0.9% agarose gels and probed with [α-32P]dATP random prime-labelled DNA as previously described.50 The ItG var gene second exon probe was described previously.51 The var2csa and It4_var44 exon 1 probes were the same as those used to probe PFGE-separated chromosomes. Northern blots probed with exon 2 sequences were washed with 2 × SSC and 0.1% SDS at 55 °C. Northern blots probed with specific exon 1 sequences were washed with 0.5 × SSC and 0.1% SDS at 60 °C.
Quantitative RT-PCR
All Q-RT-PCR was performed using SYBR PCR master mix (Applied Biosystems) on a PE7900HT (Applied Biosystems).
Genes identified as possibly deleted or duplicated by microarray were quantitated in both CS2 and E8B gDNA using specific primers (Supplementary Table 2). A single gDNA serially diluted 5 times over 5 logs of concentration was used to create standard curves for each set of primers from which the levels of each gene in the CS2 and E8B gDNA samples were interpolated from their Ct values. These levels were normalised by dividing by the levels of the skeleton binding protein gene sbp determined by the same approach in each sample. The resulting relative values compare the level of each gene in equivalent amounts of CS2 and E8B gDNA.
Q-RT-PCR of 17 ItG var genes was performed to determine switch rates. Each primer pair was used to amplify a 5-point, 5-log dilution series of CS2 gDNA, and the gradient of the curve of threshold cycle versus concentration was used in the equation 10[− 1/gradient] to calculate the efficiency (E) of each PCR reaction.52 This value for each gene was then used to calculate the ratio of its expression in each cDNA relative to the number of copies of the gene present in a constant amount of E8B gDNA using the skeleton binding protein 1 gene (SBP) (PFE0065w) to normalise in the equation ratio = ((Evar gene)ΔCPvar gene(E8B gDNA–cDNA sample))/((ESBP)ΔCPSBP(E8B gDNA–cDNA sample)),52 where crossing point (CP) was equivalent to the threshold cycle (Ct) determined using SDS software (Applied Biosystems). The primer sequences and PCR efficiencies are provided in Supplementary Table 2.
Absolute quantitation of hdhfr and bsd in transfectant cDNA and gDNA by Q-RT-PCR was performed using the primers listed in Supplementary Table 2. Standard curves were made using purified plasmids of known concentration containing the target gene sequences as previously described.50 The relative quantities in transfectant cDNA and gDNA of the skeleton binding protein gene (sbp) were determined by Q-RT-PCR using the primers listed in Supplementary Table 2 to amplify standard curves of diluted 3D7 isolate gDNA.18 Relative sbp levels were used to normalise the bsd and hdhfr levels, and the normalised cDNA data were divided by the normalised gDNA data to calculate relative levels of hdhfr and bsd cDNA per plasmid.
Relative quantitation of the 3D7 var repertoire by 2− ΔΔCt analysis was performed using a previously described primer set53 supplemented with previously described additional primers.9,54,55 Analysis was performed using arginyl tRNA synthetase to normalise54 and wild-type 3D7 cDNA as a calibrator. The following genes were amplified by the same set of primers and are indicated elsewhere by the first gene accession only: PFC1120c/PFC0005w, PFA0015c/PFI1820w/MAL6P1.314, PFD1235w/MAL7P1.1, PFL1955w/PFL1970w, PFD0630c/PFD0635c, and PFD0995c/PFD1000c.
Gene switch rates
Switch rates of var2csa were determined by fitting linear regression curves to var2csa cDNA levels. Maximum var2csa transcript levels were determined as 0.62 relative to gDNA from the average of the maximum values for the CSA-selected CS2 and E8B time courses (Fig. 6). Inspection of the ICAM-selected CS2 parasites var2csa regression curve revealed a linear increase throughout the entire time period with 0 var2csa cDNA predicted for day 1 by the regression equation y = (0.003185 ± 0.0001581)x + (−0.004249 ± 0.1727) (± 95% CI) (r2 = 0.9927). Therefore, the on-switch rate/generation was determined by dividing 100 by the solution of the equation for x (number of days) divided by 2 when the maximum level of var2csa cDNA was substituted for the y value. The levels of var2csa in the CSA-selected E8B parasites did not decline in a uniformly linear fashion but was preceded by a plateau of maximal transcript levels followed by a rapid decline and then a much slower decline from low levels to silent. Linear regression was fitted to the four time points in the middle of the curve corresponding to the maximal rate of decline (Fig. 6) to generate the equation y = (− 0.004829 ± 0.0006827)x + (0.8391 ± 0.07639) (± 95% CI) (r2 = 0.9616). The off-switch rate was determined by dividing 100 by the difference of the solutions of the equation for x (number of days) at y = 0 and y = maximum var2csa [cDNA] and then dividing the result by 2. This approach assumed that no net switching away from var2csa had occurred for the first 47 days of culture during which var2csa cDNA levels remained maximal.
E8B grown after selection on CSA also transiently expressed It4_var44 at sufficient levels to allow linear regression over four time points; the slope of the resulting equation, y = (0.001736 ± 0.000353)x + (− 0.08402 ± 0.0395) (± 95% CI) (r2 = 0.9236) was 0.55 times that of var2csa in CS2 selected on ICAM-1, indicating an on-switch rate for It4_var44 of approximately 0.55 × 1.02 = 0.6% per generation in E8B.
Linear regression curves were fitted to the first four time points of the pHBEiE and pHBEiC hdhfr cDNA levels presented in Fig. 7b. The resulting equations for pHBEiE, y = (− 0.005285 ± 0.001191)x + (0.7115 ± 0.08967) (± 95% CI) (r2 = 0.9079), and for pHBEiC, y = (− 0.003643 ± 0.0007271)x + (0.4415 ± 0.05969) (± 95% CI) (r2 = 0.9262), were used to calculate the percentage of episomal upsE silencing per generation as (100/(x intercept/2)) to give values of 1.49% per generation for pHBEiE and 1.65% per generation for pHBEiC.
Western blot
Triton-X-100-insoluble, SDS-soluble fractions of parasite cultures were extracted; separated by SDS-PAGE; transferred to nitrocellulose membrane; and probed with polyclonal rabbit antiserum raised to the conserved acidic terminal sequence of PfEMP1 as previously described.56
DNA microarray hybridisation
gDNA (12 μg) from CS2, E8B, and P1B5 (IT sequence strain†) parasites were fragmented by a frequent cutter restriction enzyme ApoI for 2 h at 50 °C and then heat inactivated for 20 min at 80 °C. After phenol-chloroform extraction and isopropanol precipitation, the fragmented DNAs were resuspended in water and end labelled with 25 nmol of Biotin-N11-ddATP (NEN) using a terminal transferase (Roche) following the manufacturer's instructions. Biotin-labelled gDNA were then added to the Affymetrix hybridisation cocktail (standard eukaryotic Affymetrix protocol) and hybridised to the PFSANGER arrays overnight at 65 °C in a rotating oven (60 rpm).
The high-density 8-μm PFSANGER tiling-like array has been described elsewhere.57 Following hybridisations, the chips were washed, stained, and scanned according to the manufacturer's recommendations. P1B5 hybridisation was not performed the same day as E8B and CS2, but the three raw files were normalised together for analysis. The hybridisation intensity for each 25-mer feature was computed using Affymetrix GCOS v1.3 software, and the CEL files were transferred into the R/Bioconductor environment.58 For DNA–DNA analysis, the chips were RMA preprocessed59 using the affy package60 and a modified chip definition file generated in-house (using the altcdfenv package) containing 5677 probesets = 5358 genes (P. falciparum genome genedb-release 2005‡). Log2 ratio of CS2/E8B, CS2/P1B5, and E8B/P1B5 were calculated for each gene and compared to assess gene amplification and/or deletion [over log2 (1) or under log2 (− 1), respectively].
Acknowledgements
The authors thank Dr. Till Voss for providing plasmids and Associate Professor Stephen Rogerson for helpful suggestions on preparation of the manuscript.
Edited by J. Karn
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.04.032
Appendix A.
Supplementary Fig. 1.
(a) PCR of the four rif genes present on chromosome 12 of E8B (E) and lost following recombination in CS2 (C); a PCR of It4_var8 exon 1 was included as a positive control. (b) PCR across the var2csa intron using primers specific for the E8B wild-type sequence or the CS2 mutant sequence: CS2 (C), E8B (E), CS2 ICAM-1-selected subclones CS2isB10 (B10), CS2isC9 (C9), CS2isD8 (D8), negative control (-). Subclone C8 could not be recovered from cryopreserves for this analysis.
Alignment of intron sequences from the recombined CS2 var2csa, the donor It4_var8, and the recipient wild-type E8B var2csa (It4_var4). “Start” above the alignment indicates the start of the homologous sequence predicted to precede the var2csa breakpoint. Asterisks below the alignment indicate nucleotide polymorphisms conserved between CS2 var2csa and It4_var8 that lie within the region of homology predicted to correspond to the protruding 3′ invading strand of var2csa. The underlined sequence is var intron region 2. Blocks in black background are the conserved repeat sequences that are inverted in var intron region 3 relative to region 1.
References
- 1.Barry A.E., Leliwa-Sytek A., Tavul L., Imrie H., Migot-Nabias F., Brown S. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog. 2007;3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer S.M., Kyes S.A., Aggarwal G., Springer A.L., Nelson S.O., Christodoulou Z. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of Plasmodium falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 4.Duraisingh M.T., Voss T.S., Marty A.J., Duffy M.F., Good R.T., Thompson J.K. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A.Z., Hartl D.L., Deitsch K.W. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Rubio J.J., Gontijo A.M., Nunes M.C., Issar N., Hernandez Rivas R., Scherf A. 5' flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horrocks P., Pinches R., Christodoulou Z., Kyes S.A., Newbold C.I. Variable var transition rates underlie antigenic variation in malaria. Proc. Natl. Acad. Sci. USA. 2004;101:11129–11134. doi: 10.1073/pnas.0402347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank M., Dzikowski R., Amulic B., Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 2007;64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voss T.S., Kaestli M., Vogel D., Bopp S., Beck H.P. Identification of nuclear proteins that interact differentially with Plasmodium falciparum var gene promoters. Mol. Microbiol. 2003;48:1593–1607. doi: 10.1046/j.1365-2958.2003.03528.x. [DOI] [PubMed] [Google Scholar]
- 11.Calderwood M.S., Gannoun-Zaki L., Wellems T.E., Deitsch K.W. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 12.Deitsch K.W., Calderwood M.S., Wellems T.E. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- 13.Myrick K.V., Gelbart W.M. Universal Fast Walking for direct and versatile determination of flanking sequence. Gene. 2002;284:125–131. doi: 10.1016/s0378-1119(02)00384-0. [DOI] [PubMed] [Google Scholar]
- 14.Figueiredo L.M., Freitas-Junior L.H., Bottius E., Olivo-Marin J.C., Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueiredo L.M., Pirrit L.A., Scherf A. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 2000;106:169–174. doi: 10.1016/s0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- 16.Carret C.K., Horrocks P., Konfortov B., Winzeler E., Qureshi M., Newbold C., Ivens A. Microarray-based comparative genomic analyses of the human malaria parasite Plasmodium falciparum using Affymetrix arrays. Mol. Biochem. Parasitol. 2005;144:177–186. doi: 10.1016/j.molbiopara.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Smith J.D., Craig A.G., Kriek N., Hudson-Taylor D., Kyes S., Fagen T. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: A parasite adhesion trait implicated in cerebral malaria. Proc. Natl. Acad. Sci. USA. 2000;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy M.F., Caragounis A., Noviyanti R., Kyriacou H.M., Choong E.K., Boysen K. Transcribed var genes associated with placental malaria in Malawian women. Infect. Immun. 2006;74:4875–4883. doi: 10.1128/IAI.01978-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amulic B., Salanti A., Lavstsen T., Nielsen M.A., Deitsch K.W. An upstream open reading frame controls translation of var2csa, a gene implicated in placental malaria. PLoS Pathog. 2009;5:e1000256. doi: 10.1371/journal.ppat.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr-Weaver T.L., Szostak J.W. Fungal recombination. Microbiol. Rev. 1985;49:33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haber J.E. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair. 2006;5:998–1009. doi: 10.1016/j.dnarep.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 22.Mok B.W., Ribacke U., Rasti N., Kironde F., Chen Q., Nilsson P., Wahlgren M. Default Pathway of var2csa switching and translational repression in Plasmodium falciparum. PLoS ONE. 2008;3:e1982. doi: 10.1371/journal.pone.0001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dzikowski R., Deitsch K.W. Active Transcription is Required for Maintenance of Epigenetic Memory in the Malaria Parasite Plasmodium falciparum. J. Mol. Biol. 2008;382:288–297. doi: 10.1016/j.jmb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deitsch K.W., del Pinal A., Wellems T.E. Intra-cluster recombination and var transcription switches in the antigenic variation of Plasmodium falciparum. Mol. Biochem. Parasitol. 1999;101:107–116. doi: 10.1016/s0166-6851(99)00062-6. [DOI] [PubMed] [Google Scholar]
- 25.Gannoun-Zaki L., Jost A., Mu J., Deitsch K.W., Wellems T.E. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryotic Cell. 2005;4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang P.K., Kuroda M.I. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 27.Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voss T.S., Healer J., Marty A.J., Duffy M.F., Thompson J.K., Beeson J.G. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1009. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 29.Voss T.S., Tonkin C.J., Marty A.J., Thompson J.K., Healer J., Crabb B.S., Cowman A.F. Alterations in local chromatin environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol. Microbiol. 2007;66:139–150. doi: 10.1111/j.1365-2958.2007.05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Donnell R.A., Freitas-Junior L.H., Preiser P.R., Williamson D.H., Duraisingh M., McElwain T.F. A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. EMBO J. 2002;21:1231–1239. doi: 10.1093/emboj/21.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marty A.J., Thompson J.K., Duffy M.F., Voss T.S., Cowman A.F., Crabb B.S. Evidence that Plasmodium falciparum chromosome end clusters are cross-linked by protein and are the sites of both virulence gene silencing and activation. Mol. Microbiol. 2006;62:72–83. doi: 10.1111/j.1365-2958.2006.05364.x. [DOI] [PubMed] [Google Scholar]
- 32.Ralph S.A., Scheidig-Benatar C., Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. USA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mancio-Silva L., Rojas-Meza A.P., Vargas M., Scherf A., Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- 34.Freitas-Junior L.H., Hernandez-Rivas R., Ralph S.A., Montiel-Condado D., Ruvalcaba-Salazar O.K., Rojas-Meza A.P. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 35.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Bull P.C., Kortok M., Kai O., Ndungu F., Ross A., Lowe B.S. Plasmodium falciparum-infected erythrocytes: agglutination by diverse Kenyan plasma is associated with severe disease and young host age. J. Infect. Dis. 2000;182:252–259. doi: 10.1086/315652. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen M.A., Staalsoe T., Kurtzhals J.A., Goka B.Q., Dodoo D., Alifrangis M. Plasmodium falciparum variant surface antigen expression varies between isolates causing severe and nonsevere malaria and is modified by acquired immunity. J. Immunol. 2002;168:3444–3450. doi: 10.4049/jimmunol.168.7.3444. [DOI] [PubMed] [Google Scholar]
- 38.Jensen A.T., Magistrado P., Sharp S., Joergensen L., Lavstsen T., Chiucchiuini A. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J. Exp. Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaestli M., Cockburn I.A., Cortes A., Baea K., Rowe J.A., Beck H.P. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J. Infect. Dis. 2006;193:1567–1574. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biggs B.A., Goozé L., Wycherley K., Wollish W., Southwell B., Leech J.H., Brown G.V. Antigenic variation in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1991;88:9171–9174. doi: 10.1073/pnas.88.20.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaiyaroj S.C., Coppel R.L., Novakovic S., Brown G.V. Multiple ligands for cytoadherence can be present simultaneously on the surface of Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. USA. 1994;91:10805–10808. doi: 10.1073/pnas.91.23.10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott S.R., Duffy M.F., Byrne T.J., Beeson J.G., Mann E.J., Wilson D.W. Cross-reactive surface epitopes on chondroitin sulfate A-adherent Plasmodium falciparum infected erythrocytes are associated with transcription of var2csa. Infect. Immun. 2005;73:2848–2856. doi: 10.1128/IAI.73.5.2848-2856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooke B.M., Rogerson S.J., Brown G.V., Coppel R.L. Adhesion of malaria-infected red blood cells to chondroitin sulfate A under flow conditions. Blood. 1996;88:4040–4044. [PubMed] [Google Scholar]
- 44.Robson K.J., Hall J.R., Davies L.C., Crisanti A., Hill A.V., Wellems T.E. Polymorphism of the TRAP gene of Plasmodium falciparum. Proc. R. Soc. London, Ser. Biol. Sci. 1990;242:205–216. doi: 10.1098/rspb.1990.0126. [DOI] [PubMed] [Google Scholar]
- 45.Triglia T., Foote S.J., Kemp D.J., Cowman A.F. Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol. Cell. Biol. 1991;11:5244–5250. doi: 10.1128/mcb.11.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noviyanti R., Brown G.V., Wickham M.E., Duffy M.F., Cowman A.F., Reeder J.C. Multiple var gene transcripts are expressed in Plasmodium falciparum infected erythrocytes selected for adhesion. Mol. Biochem. Parasitol. 2001;114:227–237. doi: 10.1016/s0166-6851(01)00266-3. [DOI] [PubMed] [Google Scholar]
- 47.Maier A.G., Braks J.A., Waters A.P., Cowman A.F. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol. Biochem. Parasitol. 2006;150:118–121. doi: 10.1016/j.molbiopara.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Rubio J.P., Thompson J.K., Cowman A.F. The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J. 1996;15:4069–4077. [PMC free article] [PubMed] [Google Scholar]
- 49.Smith J.D., Kyes S., Craig A.G., Fagan T., Hudson-Taylor D., Miller L.H. Analysis of adhesive domains from the A4VAR Plasmodium falciparum erythrocyte membrane protein-1 identifies a CD36 binding domain. Mol. Biochem. Parasitol. 1998;97:133–148. doi: 10.1016/s0166-6851(98)00145-5. [DOI] [PubMed] [Google Scholar]
- 50.Duffy M.F., Byrne T.J., Elliott S.R., Wilson D., Rogerson S.J., Beeson J.G. Broad analysis reveals a consistent pattern of var gene transcription in Plasmodium falciparum repeatedly selected for a defined adhesion phenotype. Mol. Microbiol. 2005;56:774–788. doi: 10.1111/j.1365-2958.2005.04577.x. [DOI] [PubMed] [Google Scholar]
- 51.Duffy M.F., Maier A.G., Byrne T.J., Elliott S.R., Marty A.J., O'Neill M.T. VAR2CSA is the principal ligand for chondroitin sulfate A in two allogeneic isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 2006;148:117–124. doi: 10.1016/j.molbiopara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 52.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salanti A., Staalsoe T., Lavstsen T., Jensen A.T., Sowa M.P., Arnot D.E. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 54.Dzikowski R., Frank M., Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavstsen T., Magistrado P., Hermsen C.C., Salanti A., Jensen A.T., Sauerwein R. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar. J. 2005;4:21. doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duffy M.F., Brown G.V., Basuki W., Krejany E.O., Noviyanti R., Cowman A.F., Reeder J.C. Transcription of multiple var genes by individual, trophozoite-stage Plasmodium falciparum cells expressing a chondroitin sulphate A binding phenotype. Mol. Microbiol. 2002;43:1285–1293. doi: 10.1046/j.1365-2958.2002.02822.x. [DOI] [PubMed] [Google Scholar]
- 57.Cortes A., Carret C., Kaneko O., Yim Lim B.Y., Ivens A., Holder A.A. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 2007;3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 60.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of intron sequences from the recombined CS2 var2csa, the donor It4_var8, and the recipient wild-type E8B var2csa (It4_var4). “Start” above the alignment indicates the start of the homologous sequence predicted to precede the var2csa breakpoint. Asterisks below the alignment indicate nucleotide polymorphisms conserved between CS2 var2csa and It4_var8 that lie within the region of homology predicted to correspond to the protruding 3′ invading strand of var2csa. The underlined sequence is var intron region 2. Blocks in black background are the conserved repeat sequences that are inverted in var intron region 3 relative to region 1.