Abstract
Human COQ6 encodes a monooxygenase which is responsible for the C5-hydroxylation of the quinone ring of coenzyme Q (CoQ). Mutations in COQ6 cause primary CoQ deficiency, a condition responsive to oral CoQ10 supplementation. Treatment is however still problematic given the poor bioavailability of CoQ10. We employed S. cerevisiae lacking the orthologous gene to characterize the two different human COQ6 isoforms and the mutations found in patients. COQ6 isoform a can partially complement the defective yeast, while isoform b, which lacks part of the FAD-binding domain, is inactive but partially stable, and could have a regulatory/inhibitory function in CoQ10 biosynthesis. Most mutations identified in patients, including the frameshift Q461fs478X mutation, retain residual enzymatic activity, and all patients carry at least one hypomorphic allele, confirming that the complete block of CoQ biosynthesis is lethal. These mutants are also partially stable and allow the assembly of the CoQ biosynthetic complex. In fact treatment with two hydroxylated analogues of 4-hydroxybenzoic acid, namely, vanillic acid or 3-4-hydroxybenzoic acid, restored the respiratory growth of yeast Δcoq6 cells expressing the mutant huCOQ6-isoa proteins. These compounds, and particularly vanillic acid, could therefore represent an interesting therapeutic option for COQ6 patients.
Abbreviations: COQ6, flavin-dependent monooxygenase; CoQ, coenzyme Q; CoQ10, coenzyme Q10; 4HB, 4-hydroxybenzoate; VA, vanillic acid; 3,4 diHB, 3,4 dihydroxybenzoic acid; SRNS, steroid resistant nephrotic syndrome; CYC1, cytochrome c1; pHBH, para-hydroxybenzoate hydroxylase; FAD, flavin adenine dinucleotide; COQ8-ADCK3, aarF domain containing kinase 3
Keywords: Coenzyme Q, Vanillic acid, COQ6, Steroid-resistant nephrotic syndrome
Highlights
-
•
Human COQ6 alleles are hypomorphic
-
•
Human COQ6 mutations are catalytically inactive but stable
-
•
4-Hydroxybenzoate analogues can bypass the CoQ deficiency due to COQ6 mutations
-
•
Vanillic acid could represent a potential therapeutic agent for this condition
1. Introduction
Coenzyme Q (CoQ) is a small lipophilic molecule comprised of a quinone ring and of a poly-isoprene tail of variable length: 6 units in the yeast Saccharomyces cerevisiae (CoQ6) and 10 in humans (CoQ10). CoQ is a key component of the respiratory chain, where it acts as an electron transporter between complexes I and II and complex III. CoQ also acts as an antioxidant and is a modulator of lipid β-oxidation and apoptosis [1].
In S. cerevisiae, synthesis of CoQ requires a set of at least nine genes (COQ genes), all of them with human orthologues. The precursor of quinone ring is 4-hydroxybenzoate (4HB), a catabolite of tyrosine, which is modified by a multienzymatic complex (CoQ complex) located on the mitochondrial inner membrane, on the matrix side [2]. This complex contains the majority of the enzymes involved in CoQ biosynthesis, although its exact composition is still under investigation.
Mutations in COQ genes cause primary CoQ deficiency, a clinically heterogeneous group of disorders. One peculiarity of CoQ deficiency is that patients respond to oral administration of CoQ10. However, treatment is still problematic for the relatively low bioavailability of CoQ10 due to its high hydrophobicity. This has stimulated different lines of research aimed at increasing the bioavailability of CoQ10 or at searching other molecules that could be beneficial in this disease [3].
Mutations in COQ6 cause a form of steroid-resistant nephrotic syndrome with a variable degree of neurologic involvement [4]. COQ6 encodes for a monooxygenase comprised in the CoQ complex, responsible for the C5-hydroxylation of the quinone ring. Vanillic acid (VA) and 3,4-dihydroxybenzoic acid (3,4 diHB), two analogues of 4HB that carry either a methoxyl or a hydroxyl group in position 5 of the ring, can bypass the Coq6 defect in yeast harboring catalytically inactive, but structurally stable, Coq6 alleles, which allow the formation of the CoQ complex [5].
2. Patients and methods
2.1. Patients and mutations
Patients and mutations have been reported previously [4]. In brief A353D and G255R were found in the homozygous state in two kindreds each. Q461fs478X and W447X were found in compound heterozygosity in a single family. We included in the analysis a novel variant, Y412C, which was found in the heterozygous state in a patient with SRNS in whom a second mutation could not be detected.
2.2. Construction of yeast expression vectors
The coding sequence of yeast COQ6 (yCOQ6) gene and the two human COQ6 isoforms were amplified from yeast genomic DNA or from cDNA (primers are available upon request) and cloned into the centromeric pCM189 yeast expression vector. Mutants were then generated using the QuikChange II site directed mutagenesis kit (Stratagene, La Jolla, CA, USA).
To replace the cytochrome c1 (CYC1) promoter of pCM189::yCOQ6 with the yCOQ6 endogenous promoter a fragment including the 5′UTR region of yCOQ6 and the first 844 bp was amplified from yeast genomic DNA (primers are available upon request) and cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA). A SacI–Msc I fragment from this construct was subcloned into the wild-type and mutated versions of pCM189::yCOQ6 after similar digestion. The correctness of all constructs was confirmed by direct sequencing.
2.3. Yeast strains, media, transformations and CoQ measurement
The Δcoq6 strain Y07287 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YGR255c::kanMX4) was purchased from the Euroscarf Consortium (Frankfurt, Germany). Rich YPD (1% yeast extract, 2% peptone and 2% glucose), YPG (1% yeast extract, 2% peptone and 3% glycerol) and selective SM GLU HLM (0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulphate, 2% glucose or galactose) and LG-pABA HLM medium were prepared as described [5]. Selective media were supplemented with amino acids to cover the yeast auxotrophies except for uracil which is carried by the plasmids. VA and 3,4 diHB were prepared as described [5]. All yeast DNA transformations were performed with the PEG-lithium acetate method as previously reported [6,7]. CoQ was measured as previously described [5,8].
2.4. Molecular modelling and bioinformatic analyses
Human COQ6-isoa sequence was aligned with a published alignment based on structure superpositions of different members of the aromatic hydroxylases family [9] using ClustalX and the alignment was curated by hand in order to favor gaps outside secondary structure elements. This alignment was then used in Modeler with Pseudomonas fluorescens para-hydroxybenzoate hydroxylase (Protein Data Bank code 1PBE) as target. Standard setup was used but for six regions where alpha-helices constraints were forced (45–60, 73–83, 220–227, 293–299, 319–328 and 397–401). These human COQ6 fragments overlap existing alpha helices found in pHBH and contiguous insertions. In silico analysis of the pathogenicity of the Y412C mutation was performed as described [10].
3. Results
3.1. COQ6 isoform a but not isoform b can partially complement Δcoq6 yeast
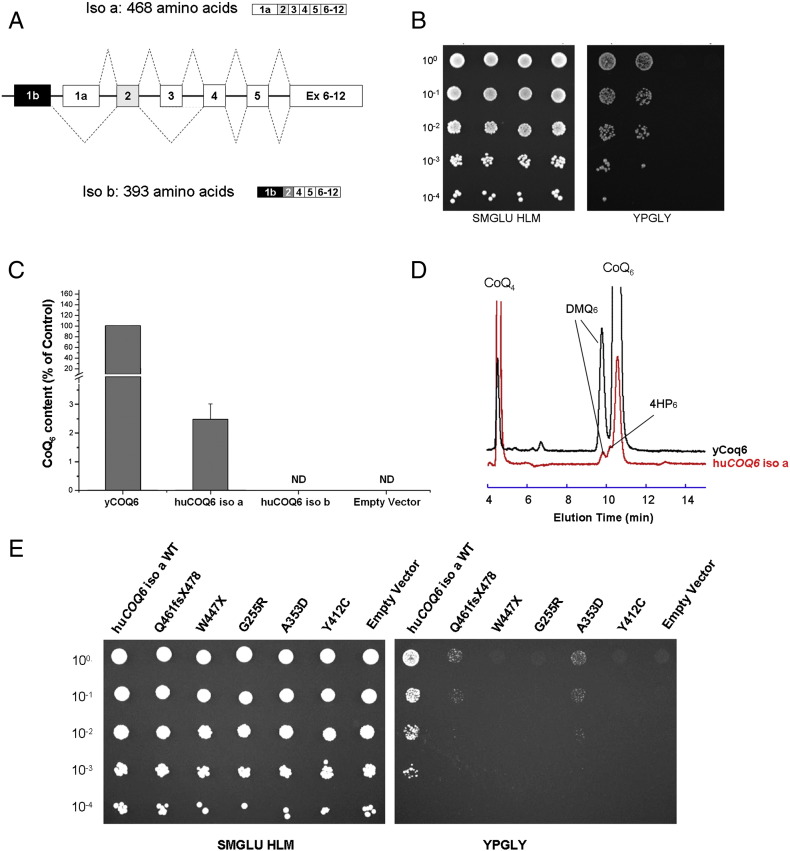
Human COQ6 encodes for at least two isoforms (huCOQ6-isoa and huCOQ6-isob) that differ for the transcription initiation site and for the splicing of exon 3 (Fig. 1A). huCOQ6-isoa is far more abundant than huCOQ6-isob [4]. In yeast there is only one isoform and its deletion causes the loss of the ability to synthesize CoQ and to grow on non-fermentable carbon sources [11]. We expressed the two human isoforms using a centromeric expression vector (pCM189) with the cytochrome c1 (CYC1) promoter, in S. cerevisiae BY4741Δcoq6 haploid strain, and checked the ability of the transformants to grow on a non-fermentable medium (YPG contains glycerol) which requires a functional respiratory chain. Only huCOQ6-isoa restored respiratory growth of the Δcoq6 yeast strain (Fig. 1B). The analysis of CoQ content of the different transformants grown in minimal medium revealed that the huCOQ6-isoa achieved levels of CoQ about 2.5% of those obtained with the yeast gene, while no detectable amount of CoQ was produced with huCOQ6-isob (Fig. 1C). This system is over 10 times more efficient than the one previously published, in which the wild type human gene could achieve levels of CoQ of just 0.2% of what could be obtained with the wild type yeast gene [4]. By the same analysis we detected only a very limited amount of 3-hexaprenyl-4-hydroxyphenol (4HP6) in huCOQ6-isoa transformants (Fig. 1D), indicating that the human protein is not rate limiting for C5-hydroxylation and that the low level of complementation is probably due to an inefficient stabilization of the CoQ complex [5].
Fig. 1.
(A) The two principal isoforms of human COQ6. Exon 2 is translated by a different reading frame in isob (in gray). (B) Growth assay of Δcoq6 cells transformed with either yCOQ6, huCOQ6-isoa, huCOQ6-isob or an empty pCM189 vector. Cells were plated onto respiratory rich medium (YPGLY) or on SM GLU–URA as positive control. (C) CoQ6 and demethoxy-coenzyme Q6 (DMQ6) amounts in Δcoq6 cells expressing either yCoq6, huCOQ6-isoa or huCOQ6-isob. Cells were grown in YNB–pABA 2% galactose containing 10 μM 4HB. The results are the average of 4 independent experiments. (D) Representative electrochromatogram of lipid extracts from Δcoq6 cells expressing either yCOQ6 (1 mg of cells) or huCOQ6-isoa (6 mg of cells). The elution position of the CoQ4 standard, of DMQ6, 4HP6 and CoQ6 are indicated. (E) Growth assay of Δcoq6 cells expressing huCOQ6-isoa wild type or carrying different mutations on the same media as in (B).
3.2. Missense mutations found in patients with SRNS affect COQ6 function
Because the complementation observed with this new huCOQ6-isoa construct is more efficient than the one obtained with the system employed in our previous work [4], we constructed plasmids expressing huCOQ6-isoa with the different mutations found in patients, including the novel Y412C, and repeated the growth assay. W447X, G255R, and Y412C mutants did not complement the respiratory growth defect of the Δcoq6 yeast strain (Fig. 1E). These data indicate that Y412C is a pathogenic COQ6 allele even though its role in determining the patient’s phenotype is still under investigation. Interestingly, the bioinformatic analysis could not reliably predict the consequences of this variant. Some residual growth was observed with A355D and, surprisingly, with the frameshift Q461fs478X allele (Fig. 1E).
3.3. Most human COQ6 mutations are hypomorphic
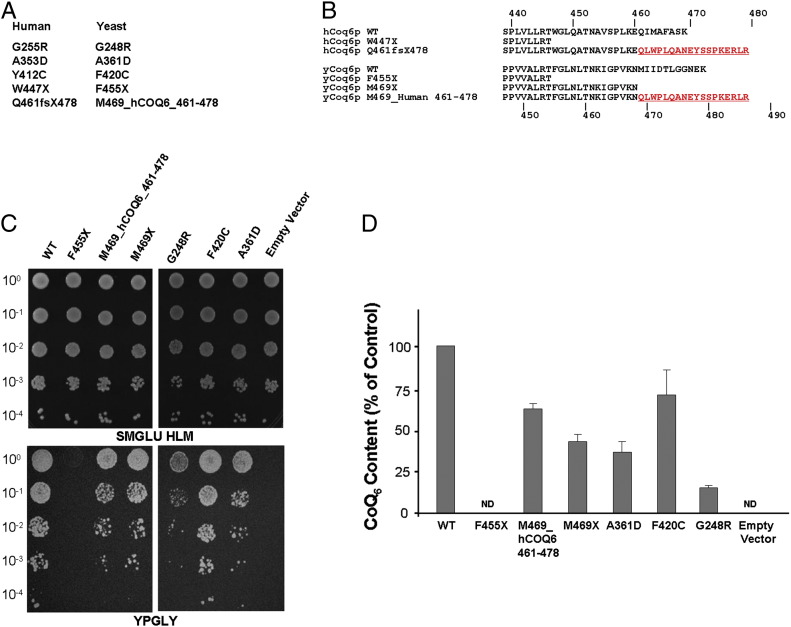
Since heterologous expression may fail to detect residual activity of mutated alleles due to the relatively low complementation efficiency observed with the human gene, we modeled the “human” mutations on the corresponding residues of the yeast gene (Fig. 2A and B). In the case of the Q461sf478X mutation, we expressed both a form with a truncation at position 469 (M469X) and a form with the 18 extra abnormal amino acids generated by the frameshift mutation in the human gene (Fig. 2B). In yeast Coq6, a phenylalanine (F420) is found in place of the corresponding human Y412. We therefore synthesized two different constructs: F420Y and F420C. Both wild type and mutated alleles were expressed either under the control of the strong CYC1 promoter or the physiological S. cerevisiae COQ6 promoter. With the CYC1 promoter, only the nonsense mutation F455X (corresponding to human W447X) failed to complement the respiratory growth defect of the Δcoq6 strain, while all the other alleles restored normal yeast growth and CoQ content (not shown). When the endogenous yeast COQ6 promoter drove the expression, the cells displayed a variable reduction in growth (Fig. 2C) and a more evident defect in CoQ content (Fig. 2D). As expected, the F420Y construct displayed normal growth and CoQ levels (not shown). These data suggest that the human G255R, A353D, Y412C and Q461sf478X mutations are hypomorphic since the corresponding yeast Coq6 mutants display residual activity.
Fig. 2.
(A) Missense mutations in huCOQ6-isoa and the corresponding residues in yCoq6. (B) Schematic representation of the huCOQ6-isoa constructs carrying the W447X and Q461fs478X mutations and their counterpart in yCOQ6. Growth assay (C) and CoQ6 levels (D) in mitochondria from Δcoq6 cells expressing different yCoq6 mutants under the control of the endogenous yeast promoter.
3.4. 4-Hydroxybenzoate analogues can restore growth in yeast Δcoq6 cells expressing human mutant alleles
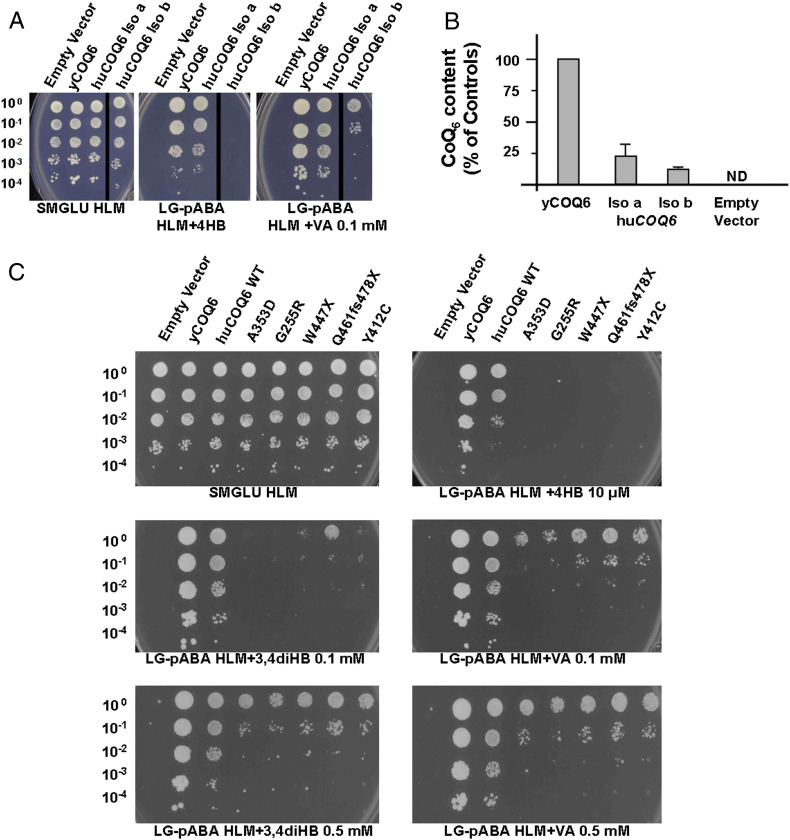
When added to the growth medium, two analogues of 4HB, VA and 3,4 diHB, can bypass the CoQ deficiency of yeast Δcoq6 cells expressing enzymatic inactive but structurally stable, Coq6 protein [5]. We first plated yeast Δcoq6 expressing huCOQ6-isoa and huCOQ6-isob on respiratory media containing either VA or 3,4 diHB. In the case of huCOQ6-isoa, VA did not further increase the growth, while it allowed partial recovery of growth in the case of huCOQ6-isob compared to medium with 4HB (Fig. 3A). A similar pattern was observed with 3,4 diHB (not shown). In agreement with previous data [5], no growth was observed for the strain transformed with the empty vector. We then measured the CoQ production after culturing in the presence of VA. Yeast expressing huCOQ6-isoa achieved CoQ levels about 20% of those obtained with yCOQ6, while strains expressing huCOQ6-isob produced about 11% of CoQ compared to the yeast gene (Fig. 3B).
Fig. 3.
(A) Growth assay of Δcoq6 cells expressing huCOQ6-isoa, huCOQ6-isob and yCoq6 on LG-pABA medium containing either 10 μM 4HB or 0.1 mM VA. (B) CoQ levels in Δcoq6 cells expressing huCOQ6-isoa, huCOQ6-isob and yCoq6 grown in YNB–pABA 2% galactose with 1 mM VA. (C) Growth assay in LG-pABA non-fermentable medium of Δcoq6 cells expressing human COQ6 mutants in the presence of VA or diHB.
Finally, we tested the effect of VA or 3,4 diHB in yeast expressing individual human mutant alleles. We detected a partial rescue of the phenotype in the presence of the analogues in all cases. VA seemed more efficient than 3,4 diHB, with effects already evident at 0.1 mM (Fig. 3C). These results indicate that the mutations impair the enzymatic activity of COQ6, but still allow formation of the CoQ complex. Interestingly this was observed also with the W447X truncating mutation, indicating that this should not be considered a null allele.
3.5. Molecular modeling of the human mutations
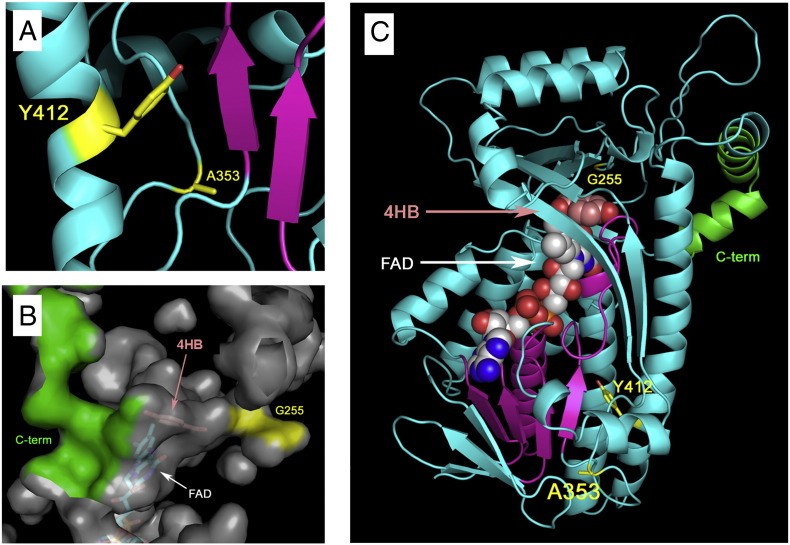
Because the structure of human COQ6 is not available, the protein was modeled on Pseudomonas fluorescens para-hydroxybenzoate hydroxylase (pHBH) with the exclusion of the N-terminal targeting sequence (that has no equivalent in pHBH). As depicted in Fig. 4A, residues Y412 and A353 interact with residues that belong to the classical Rossman fold which is implicated in FAD binding. Mutations probably affect the activity of huCOQ6-isoa by perturbing the binding/positioning of FAD. The substrate of Coq6 is prenyl-4HB. Based on the position of 4HB in the structure of pHBH [11], we find a similar pocket in huCOQ6-isoa (Fig. 4B). G255 is located in the active site pocket at about 1.7 nm from 4HB binding site and defines part of its surface (Fig. 4B). The substitution of the glycine with an arginine reduces significantly the volume of the pocket and adds positive charges that may hinder the binding of the hydrophobic substrate. W447 and Q461 residues are located in the C-terminus of the protein. As depicted in Fig. 4B and C, the C-terminal tail forms part of the surface of active pocket site. Alterations of this region are likely to cause a perturbation of the active site.
Fig. 4.
(A) A353 and Y412 and their side chains. In magenta the conserved residues that belong to the Rossman fold is implicated in FAD binding. (B) Contribution of G255 (yellow) and of the residues after W447 (green) to the surface of the active site pocket. Cofactor and substrate are shown in stick representation. (C) Overall representation of huCOQ6-isoa with 4HB and FAD, the different point mutants and of the C-terminal region comprising residues after W447 (green).
4. Discussion
Human COQ6 is transcribed in at least two isoforms which differ for the N-terminal portion. We have previously shown that huCOQ6-isoa can restore the ability of Δcoq6 yeast to grow on a nonfermentable carbon source, although the amount of CoQ synthesized by the cells was very low [4]. We have now developed a more efficient system based on pCM189 vectors which allows a higher CoQ production (2.5% of yeast versus 0.2%). It should be noted that often complementation of yeast deletion mutant with their human orthologues results in low levels of CoQ6 production compared to the human gene. Human COQ8-ADCK3 yielded less than 0.5% CoQ6 compared to the yeast gene when expressed from a multicopy plasmid [12].
Yeast expressing huCOQ6-isoa do not display significant 4HP6 accumulation, indicating a reduced ability to stabilize the CoQ complex rather than a catalytic problem. In fact, expression of a stable but catalytically inactive Coq6 or overexpression of Coq8 results in significant accumulation of 4HP6 [5].
As expected (it lacks a significant portion of the FAD binding domain) huCOQ6-isob is enzymatically inactive; nevertheless, it appears to be able partially stabilize the CoQ biosynthetic complex, as both VA and 3,4 diHB can restore CoQ synthesis in Δcoq6 yeast expressing this isoform. These data imply that huCOQ6-isob is transported into mitochondria. We speculate that in human cells it could compete with huCOQ6-isoa for assembly into the CoQ complex and could therefore play a regulatory/inhibitory role in CoQ biosynthesis. Further work is needed to address this issue.
Mutations in human COQ6 cause a multisystem disorder with a predominant renal involvement and variable neurological manifestations. Our data show that with the exception of the W447X mutant, all other alleles retain residual activity, including the frameshift mutant Q461sf478X, which displayed significant residual growth as documented in Figs. 1E and 2C and D. This is consistent with the fact that the clinical phenotype in the patient who harbored this mutation in trans with W447X was not significantly different from patients who carried missense mutations. In fact, all patients harbored at least one hypomorphic allele, in agreement with the notion that a complete block in CoQ biosynthesis is incompatible with life [13,14]. Interestingly, when we introduced the mutation into the yeast gene, the CoQ content was only slightly lower in the M469X allele compared to the form with the 18 abnormal C-terminal amino acids, suggesting that despite being important for the formation of the catalytic site the eight C-terminal amino acids of COQ6, which are conserved only in higher vertebrates are not that crucial for the stability and the functionality of the enzyme. Yeast proves to be a simple but powerful model to analyze the pathogenicity of COQ gene variants, which will be particularly useful in the upcoming years when exome sequencing will become routine clinical practice.
We finally tested if VA and 3,4 diHB could rescue CoQ deficiency in yeast expressing the human COQ6 mutants. In all cases (including the W447X) we noted partial recovery of growth in non-fermentable media, indicating that 3,4 diHB and especially VA can bypass the CoQ biosynthetic defect caused by these mutations. This result suggests that these mutations impair mostly COQ6 catalytic activity rather than its structural stability. In fact, Y412C and A353D seem to perturb the binding of the FAD, while G255R, W447X and Q461fs478X are likely to cause a rearrangement of the active site pocket. Direct proof of this by Western blot could not be achieved because of the poor quality of the available COQ6 antibodies that recognize multiple aspecific signals in yeast.
The response of these mutants to VA could have important implications for patient therapy. In fact, although patients respond to CoQ supplementation, very high doses of the compound are required, because of its low bioavailability and its limited delivery to mitochondria. VA is non-toxic (it is commonly used as a flavoring agent by the food industry), has a good oral bioavailability [15], and acts by reactivating endogenous CoQ biosynthesis. This has critical therapeutic implications since it has been shown that in C. elegans coq-3 mutants reactivation of endogenous biosynthesis was much more effective than exogenous CoQ supplementation [16]. In fact endogenously produced CoQ will be delivered to the appropriate subcellular compartments, while with exogenous supplementation the distribution of CoQ is not controlled and CoQ may have difficulties to efficiently reach the mitochondrial inner membrane. We still have no idea of what could be the potential dangers in the long or very long term (the treatment must be continued lifelong) of having an excess of CoQ in other cellular compartments. Furthermore, the QH2/Q ratio can be changed in these cells contributing to a new redox environment that can produce higher reactive oxygen species [17]. Restoration of endogenous biosynthesis, but not CoQ supplementation, would contribute to rescue the original balance. Although future studies are clearly needed to address the safety and efficacy of VA in patients, this approach could represent a major improvement in the treatment of patients with CoQ deficiency due to COQ6 mutations.
Competing interests statement
None of the authors have competing interests to declare.
Acknowledgments
This work has been supported by grants from Telethon Italy, Fondazione CARIPARO, and University of Padova (CPDA123573/12) (to L.S.), the Italian Ministry of Health (GR-2009-1578914) (to E.T.), Région Rhônes-Alpes CIBLE 2009 (to F.P.), Spanish FIS grant PI11-00078 (to P.N.) and Proyecto Excelencia P08-CTS-03988 (to P.N.). We are grateful to Mohammad Ozeir for performing preliminary experiments.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Turunen M., Olsson J., Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Tran U.C., Clarke C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;(7 Suppl.):S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trevisson E., DiMauro S., Navas P., Salviati L. Coenzyme Q deficiency in muscle. Curr. Opin. Neurol. 2011;24:449–456. doi: 10.1097/WCO.0b013e32834ab528. [DOI] [PubMed] [Google Scholar]
- 4.Heeringa S.F., Chernin G., Chaki M., Zhou W., Sloan A.J., Ji Z., Xie L.X., Salviati L., Hurd T.W., Vega-Warner V., Killen P.D., Raphael Y., Ashraf S., Ovunc B., Schoeb D.S., McLaughlin H.M., Airik R., Vlangos C.N., Gbadegesin R., Hinkes B., Saisawat P., Trevisson E., Doimo M., Casarin A., Pertegato V., Giorgi G., Prokisch H., Rotig A., Nurnberg G., Becker C., Wang S., Ozaltin F., Topaloglu R., Bakkaloglu A., Bakkaloglu S.A., Muller D., Beissert A., Mir S., Berdeli A., Varpizen S., Zenker M., Matejas V., Santos-Ocana C., Navas P., Kusakabe T., Kispert A., Akman S., Soliman N.A., Krick S., Mundel P., Reiser J., Nurnberg P., Clarke C.F., Wiggins R.C., Faul C., Hildebrandt F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozeir M., Muhlenhoff U., Webert H., Lill R., Fontecave M., Pierrel F. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 2011;18:1134–1142. doi: 10.1016/j.chembiol.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Trevisson E., Burlina A., Doimo M., Pertegato V., Casarin A., Cesaro L., Navas P., Basso G., Sartori G., Salviati L. Functional complementation in yeast allows molecular characterization of missense argininosuccinate lyase mutations. J. Biol. Chem. 2009;284:28926–28934. doi: 10.1074/jbc.M109.050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassandrini D., Cilio M.R., Bianchi M., Doimo M., Balestri M., Tessa A., Rizza T., Sartori G., Meschini M.C., Nesti C., Tozzi G., Petruzzella V., Piemonte F., Bisceglia L., Bruno C., Dionisi-Vici C., D'Amico A., Fattori F., Carrozzo R., Salviati L., Santorelli F.M., Bertini E. Pontocerebellar hypoplasia type 6 caused by mutations in RARS2: definition of the clinical spectrum and molecular findings in five patients. J. Inherit. Metab. Dis. 2013;36:43–53. doi: 10.1007/s10545-012-9487-9. [DOI] [PubMed] [Google Scholar]
- 8.Salviati L., Trevisson E., Rodriguez Hernandez M.A., Casarin A., Pertegato V., Doimo M., Cassina M., Agosto C., Desbats M.A., Sartori G., Sacconi S., Memo L., Zuffardi O., Artuch R., Quinzii C., Dimauro S., Hirano M., Santos-Ocana C., Navas P. Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J. Med. Genet. 2012;49:187–191. doi: 10.1136/jmedgenet-2011-100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koskiniemi H., Metsa-Ketela M., Dobritzsch D., Kallio P., Korhonen H., Mantsala P., Schneider G., Niemi J. Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis. J. Mol. Biol. 2007;372:633–648. doi: 10.1016/j.jmb.2007.06.087. [DOI] [PubMed] [Google Scholar]
- 10.Sacconi S., Feasson L., Antoine J.C., Pecheux C., Bernard R., Cobo A.M., Casarin A., Salviati L., Desnuelle C., Urtizberea A. A novel CRYAB mutation resulting in multisystemic disease. Neuromuscul. Disord. 2012;22:66–72. doi: 10.1016/j.nmd.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Schreuder H.A., Prick P.A., Wierenga R.K., Vriend G., Wilson K.S., Hol W.G., Drenth J. Crystal structure of the p-hydroxybenzoate hydroxylase-substrate complex refined at 1.9 A resolution. Analysis of the enzyme-substrate and enzyme-product complexes. J. Mol. Biol. 1989;208:679–696. doi: 10.1016/0022-2836(89)90158-7. [DOI] [PubMed] [Google Scholar]
- 12.Xie L.X., Hsieh E.J., Watanabe S., Allan C.M., Chen J.Y., Tran U.C., Clarke C.F. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta. 2011;1811:348–360. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng M., Falk M.J., Haase V.H., King R., Polyak E., Selak M., Yudkoff M., Hancock W.W., Meade R., Saiki R., Lunceford A.L., Clarke C.F., Gasser D.L. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 2008;4:e1000061. doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levavasseur F., Miyadera H., Sirois J., Tremblay M.L., Kita K., Shoubridge E., Hekimi S. Ubiquinone is necessary for mouse embryonic development but is not essential for mitochondrial respiration. J. Biol. Chem. 2001;276:46160–46164. doi: 10.1074/jbc.M108980200. [DOI] [PubMed] [Google Scholar]
- 15.Gitzinger M., Kemmer C., Fluri D.A., El-Baba M.D., Weber W., Fussenegger M. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 2012;40:e37. doi: 10.1093/nar/gkr1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez F., Saiki R., Chin R., Srinivasan C., Clarke C.F. Restoring de novo coenzyme Q biosynthesis in Caenorhabditis elegans coq-3 mutants yields profound rescue compared to exogenous coenzyme Q supplementation. Gene. 2012;506:106–116. doi: 10.1016/j.gene.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]