Abstract
Vitrification is the state-of-the-art specimen preparation technique for molecular electron microscopy (EM) and therefore negative staining may appear to be an outdated approach. In this paper we illustrate the specific advantages of negative staining, ensuring that this technique will remain an important tool for the study of biological macromolecules. Due to the higher image contrast, much smaller molecules can be visualized by negative staining. Also, while molecules prepared by vitrification usually adopt random orientations in the amorphous ice layer, negative staining tends to induce preferred orientations of the molecules on the carbon support film. Combining negative staining with image classification techniques makes it possible to work with very heterogeneous molecule populations, which are difficult or even impossible to analyze using vitrified specimens.
Keywords: Negative staining; Microscopy, Electron; Protein Conformation
Introduction
EM has proved to be an exceptionally versatile tool to study the structure of proteins and macromolecular complexes. Biological molecules are problematic specimens for EM, because of their susceptibility to radiation damage, their poor capacity to scatter electrons, and their proneness to dehydration in the vacuum of the electron microscope. A main requirement for EM specimen preparation is to prevent the structural collapse upon sample dehydration, and ideally also to increase specimen contrast.
Negative staining, the embedding of a specimen in a layer of dried heavy metal solution, was introduced early on as a quick and easy specimen preparation technique that significantly increases the specimen contrast. Images of negatively stained molecules are amenable to image averaging techniques that increase the signal-to-noise ratio (SNR) and thus allow finer details of the molecule to be visualized. Using computational tools to combine different views, images of negatively stained specimens can also be used to produce threedimensional (3D) reconstructions of the molecule under investigation (e.g. (1)). Embedding of the sample in a layer of dried staining solution provides some protection against the collapse of the specimen due to dehydration, but 3D reconstructions from specimens prepared in this manner are usually still significantly flattened. Moreover, microcrystals formed by the heavy metals upon drying of the negative stain solution limit the resolution of the 3D map that can be achieved to about 20 Å.
In 1984, Dubochet and colleagues revolutionized single particle EM by introducing specimen vitrification, in which the sample is applied to a grid covered with holey carbon film and quickly frozen by plunging the grid into liquid ethane (2). The rapid freezing prevents the water from forming ice crystals and embeds the molecules in a layer of vitrified (or amorphous) ice. Vitrification preserves the specimen in a near-native environment, eliminating not only specimen distortions due to dehydration and adsorption but also the limitation in achievable resolution associated with negative staining. A drawback of vitrification is the poor SNR in images of vitrified specimens, which poses severe difficulties especially for the study of small molecules. Nevertheless, under favorable circumstances, high-resolution structures can be obtained even with rather small molecules in vitrified ice (e.g. (3-5)).
Molecules usually adopt more or less random orientations in an amorphous ice layer providing many different views of the molecule. These views can be exploited to calculate a 3D reconstruction with the angular reconstitution approach (6), making it unnecessary to record images of tilted specimens. While random orientations of the molecules are advantageous for homogeneous samples, they create a severe problem for heterogeneous samples. To obtain a reliable 3D reconstruction of a molecule it is essential that only images of identical molecules are combined. However, from projection views alone, it is usually not possible to distinguish between molecules in different orientations and molecules in different conformations.
Vitrification has proved to be such a powerful technique to determine the structure of macromolecules that many scientists have begun to consider negative staining to be old-fashioned and not worth pursuing. If applicable, vitrification is undoubtedly the best technique to obtain an undistorted 3D map of a molecule by EM. However, negative staining has unique advantages and can provide important information on biological molecules that is not easily obtained or, in the case of very heterogeneous samples, not even possible to obtain with vitrified specimens. In this paper we describe different negative staining protocols. We also demonstrate the unique information that can be gathered by applying classification methods to images of heterogeneous samples prepared by the conventional negative staining protocol. To illustrate these points, we will use mainly recent examples from our own work.
Materials and Methods
Proteins
Recombinant human transferrin receptor (TfR) and transferrin (Tf) were prepared as described in (5). S. cerevisiae Sec23p/Sec24p was prepared as described in (7). Recombinant bacteriophage T7 primase-helicase was prepared as described in (8). Recombinant integrin aVb3 was prepared as described in (9). Recombinant integrin a5b1 headpiece and fibronectin fragments 7-10 (Fn7-10) and 9-10 (Fn9-10) were prepared as described in (10). S. cerevisiae proteasome was prepared as described in (11). Rabbit spleen 20S and 26S immunoproteasome and recombinant mouse PA26a were prepared as described in (12). Rhodobacter sphaeroides reaction center (RC) – light-harvesting complex 1 (LH1) photounits were prepared as described in (13).
Electron microscopy
Unless stated otherwise, specimens were prepared for EM using the conventional negative staining procedure. Briefly, a 2.5 ml drop of sample solution was adsorbed to a glow-discharged carbon-coated copper grid, washed with two drops of deionized water, and stained with two drops of freshly prepared 0.75% uranyl formate.
Unless stated otherwise, samples were imaged at room temperature using a Philips Tecnai T12 electron microscope equipped with an LaB6 filament and operated at an acceleration voltage of 120 kV. Images were taken at a magnification of 52,000x and a defocus value of 1.5 mm on Kodak SO-163 film using low-dose procedures. Films were developed for 12 minutes with fullstrength Kodak D-19 developer at 20°C. All micrographs were visually inspected with a laser diffractometer, and only drift-free images were selected for digitization with a Zeiss SCAI scanner using a step size of 7 mm. Micrographs were binned over 3 ¥ 3 pixels to yield a pixel size of 4.04 Å on the specimen level.
Image processing
Particles were selected interactively from images using the display program WEB associated with SPIDER (14), the program used for all subsequent image processing steps. Selected particles were windowed into individual images with a size depending on the molecule under investigation. The side length of the images was typically chosen to be approximately twice the length of the longest particle dimension. Particle images were first subjected to 10 rounds of alignment and classification, specifying a number of output classes depending on the heterogeneity of the particular sample. Unique averages were selected from the resulting class averages and used as references for 8 cycles of multi-reference alignment.
To compare projection averages of the S. cerevisiae proteasome with the corresponding crystal structure, the crystal structure was converted into a density map and resolution-filtered to 25 Å. Projections from the density map were calculated at angular intervals of 2º and cross-correlated with the respective projection averages. The projections with the highest cross-correlation coefficients are shown in Figure 4a.
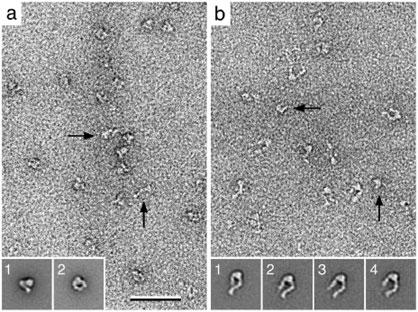
Fig. 4. Apparent sample heterogeneity due to different particle orientations.
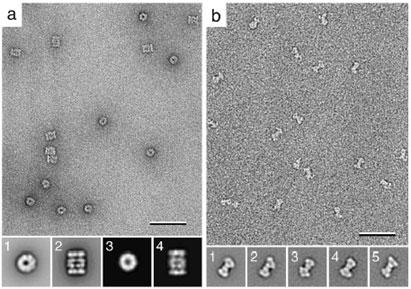
a: Image of a negatively stained yeast proteasomes. Classification of the particle images yielded two class averages (insets 1 and 2). Comparison of the two class averages with projections from a resolution-limited model generated from the crystal structure (26) identified the two averages to correspond to a top (inset 3) and a side view (inset 4) of the proteasome. b: Image of negatively stained yeast Sec23p/Sec24p complexes. Classification of the particle images yielded a variety of slightly different class averages. 3D reconstructions of the classes shown in insets 1 to 5, calculated using images of tilted specimens, looked identical, demonstrating that the variations in the class averages are due to slightly different orientations, in which the complexes adsorbed to the carbon film. The scale bars correspond to 50 nm and the inset panels have a side length of 34 nm in a and 32 nm in b. Figure 4b modified and reprinted from (7). Copyright 2001 with permission from the National Academy of Sciences, USA.
Results and Discussion
Negative staining protocols
The conventional negative staining protocol involves the adsorption of the specimen to a glow-discharged carbon-coated EM grid, which is washed with two drops of deionized water and subsequently stained with two drops of heavy metal solution. To obtain thinner stain embedding, excess stain solution can be removed from the grid by vacuum aspiration. This basic protocol can easily be adapted if required. Buffer solution can be used instead of water to wash the grid if this is necessary for sample stability, although this generally results in a somewhat higher background. If a membrane protein is to be visualized, at least five drops of water should be used to remove the detergent from the grid, since detergents can interfere with staining.
The conventional negative staining protocol normally induces specimens to adsorb to the carbon support film in one or a limited number of preferred orientations. For example, a complex between a construct of the transferrin receptor containing only the extracellular domains but lacking the transmembrane and cytoplasmic domains (TfR) with transferrin (Tf) adsorbs to the carbon film predominantly in two orientations (Fig. 1b, insets 1 and 2). It is characteristic for this method to create stain clouds that surround the molecules, producing a strong contrast between the background and the particle (Fig. 1a). When images are tilted to provide the different views of the specimen needed to calculate a 3D map by the random conical tilt approach (1), these stain clouds are particularly evident (Fig. 1b).
Fig. 1. Images of the TfR-Tf complex obtained with different negative staining protocols.
a and b: Images of an untilted (a) and a 60° tilted sample (b) prepared by the conventional negative staining protocol using uranyl formate. The particles are surrounded by a dark stain cloud, which is particularly evident in the image of the tilted specimen. Class averages (insets) show the two predominant orientations, in which the complex adsorbs to the carbon film, revealing a side view (inset 1) and a top view (inset 2) of the complex. c and d: Images of an untilted (c) and a 60° tilted sample (d) prepared by the carbon sandwich technique using uranyl formate. The negative stain forms a continuous layer and no stain cloud is apparent in images of untilted or tilted specimens. Class averages show that the complexes are seen in the same orientations as in the conventional negative staining protocol (insets 1 and 2). Due to the additional carbon layer, a significant number of complexes are being squashed upon drying and therefore can not be used for structure determination (insets 3 and 4). e: Image obtained with an untilted sample embedded in a mixture of glucose and ammonium molybdate, showing the image contrast to be much weaker than in the case of uranyl formate staining. Classification of particles selected from such images reveals that the molecules adsorb to the grid in random orientations. Some of the resulting class averages are shown in insets 1 to 10. The lines in panels a to d indicate the tilt axis. The scale bar corresponds to 50 nm and the inset panels have a side length of 26 nm.
The conventional negative staining protocol is quick and easy, but 3D maps calculated from specimens prepared in this way show severe deformations due to flattening and incomplete stain embedding. To overcome incomplete stain embedding, Frank and colleagues have developed a carbon sandwich technique, in which the specimen is embedded in a continuous layer of stain in between two carbon films. While specimens prepared using this technique usually still adsorb in preferred orientations (Fig. 1d, insets 1 and 2), images do not have stain clouds surrounding the molecules (Fig. 1c and d). While this preparation technique prevents artifacts due to incomplete stain embedding, a fraction of the molecules can become significantly squashed, leading to a spread-out appearance of the molecules (Fig. 1d, insets 3 and 4). Such particles should be excluded from structure determination. To reduce dehydration-induced flattening of the specimen or the squashing of the molecules in the carbon sandwich technique, glycerol or glucose can be added to the specimen or staining solution. If sugar or glycerol is added directly to the specimen solution, this often causes the specimen to adsorb to the carbon film in random orientations (Fig. 1e). Glycerol and sugars are also very sensitive to radiation damage. Therefore, specimens prepared with such additives need to be imaged at liquid nitrogen temperature.
In all the negative stain protocols described above the specimen is dried. The best specimen preservation is however achieved with cryo-negative staining techniques, which avoid drying of the specimen. In a protocol pioneered by Adrian and co-workers, the specimen is vitrified in a saturated ammonium molybdate solution (15). This technique, however, exposes the specimen to high ionic strength, which can cause the dissociation of many macromolecular complexes. In an alternative, gentler approach, developed in the Stark laboratory, the sample is mixed with glycerol, stained in a carbon layer sandwich and then frozen (16). Both techniques provide high contrast due to the heavy metal stain while avoiding dehydration of the specimen. Molecules prepared in either way usually adopt random orientations, so that the angular reconstitution approach must be used for 3D reconstruction (6).
The carbon sandwich technique, the addition of glycerol or glucose, and the cryo-negative staining approaches all improve the quality of 3D structures by reducing artifacts due to dehydration, adsorption and incomplete stain embedding. Conventional negative staining is however perfectly adequate when visualizing the 3D structure of small molecules (< 250 kDa), where the above problems are less severe, and when only projection structures are being determined. The thin layer of stain produced by conventional staining is actually an advantage when visualizing very small molecules. Moreover, the adsorption of the molecules to the carbon film in one or only a limited number of preferred orientations, which is usually observed with samples prepared by this technique, is beneficial in the analysis of heterogeneous samples. The use of projection images of negatively stained samples to analyze heterogeneous samples is the focus of this paper.
Choice of negative stains
A variety of heavy metal compounds are available for conventional negative staining. Among the most commonly used stains are uranyl and tungstate stains, ammonium molybdate and aurothioglucose (for a more complete list of stains, see (17)). It is important to note that stains are usually not inert, but have different characteristics that can lead to different staining of the specimen (e.g., (18-20)).
Tungstate stains and ammonium molybdate are negatively charged metal ions and have the advantage that the pH of the stain solutions can be neutralized. Aurothioglucose carries no charge and preserves the specimen particularly well due to its sugar component. However, aurothioglucose produces only poor image contrast and is very sensitive to radiation damage, requiring images to be taken at liquid nitrogen temperature. The positively charged uranyl stains often generate the highest contrast and have a fixative effect, but they require the stain solution to be acidic (pH ~ 4.5) in order to prevent precipitation of the stain. This is problematic when studying proteins that undergo pH-dependent conformational changes, such as viral fusion proteins. Recent studies have demonstrated, however, that a uranyl acetate solution fixes protein structure on the millisecond timescale (21), alleviating this problem.
For most applications uranyl stains are the best choice. A solution of the more commonly used uranyl acetate is stable over many months. A uranyl formate solution is only stable over a few days, but it yields better staining of the specimen due to a finer grain size. This can be important when visualizing very small molecules (< 100 kDa). In our hands, visualizing the 40 kDa fibronectin Fn7-10 fragment (Fig. 2a) and the 75 kDa Tf molecule (Fig. 2b) was only possible by conventional negative staining with uranyl formate. We could not see these molecules using any other negative stain or negative staining procedure.
Fig. 2. Visualizing small molecules (< 100 kDa) prepared by the conventional negative staining protocol using uranyl formate.
a: Image of a mixture of integrin α5β1 headpieces and a fibronectin (Fn) fragment containing Fn domains 7 to 10 (Fn7-10, MW ~40 kDa). The image not only visualizes the Fn7-10 fragment bound to the α5β1 headpiece (asterisks), but also unbound Fn7-10 fragment (arrows). Class averages of the unbound Fn7-10 fragment obtained from such images resolve the four individual 10-kDa domains in the flexible Fn7-10 fragment (insets 1 and 2). b: Individual molecules can clearly be seen in images of negatively stained Tf molecules (MW~ 70 kDa). Class averages show a top view (inset 1) and a side view of the molecule (inset 2). The top view resolves the two lobes of Tf (MW ~35 kDa) as well as the two domains of each lobe (MW ~17 kDa). The scale bar corresponds to 50 nm and the inset panels have a side length of 26 nm. Insets in Figure 4a modified and reprinted from (10). Copyright 2003 with permission from EMBO Journal.
The need for image classification
Only in the most favorable cases is the sample to be studied truly homogeneous. Even if a protein appears pure and shows only a single band on an SDS PAGE, gel, the particles are likely to appear heterogeneous when viewed by negative stain EM. A minor degree of heterogeneity can be introduced by the negative staining procedure itself due to distortions upon adsorption and/or a variable degree of stain embedding. These are usually only significant problems with larger molecules. More significant heterogeneity in the molecule population may however occur because of (i) particles adsorbing to the grid in different orientations (resulting in identical molecules having a different appearance), (ii) particles assembling into different oligomeric states, and (iii) particles adopting different conformational states. While heterogeneity is usually already present in pure protein preparations, it is even more pronounced when macromolecular complexes are studied, which in many cases can fall apart. Image classification is the computational method that allows one to obtain meaningful structural information from heterogeneous specimens.
As a general rule, biochemical data such as SDS PAGE and gel filtration chromatography do not suffice to rule out sample heterogeneity. Due to the more or less random orientation of the molecules and the poor SNR of the images, it is virtually impossible to assess sample heterogeneity by cryo-EM of vitrified samples. Structure determination using vitrified samples therefore always carries the risk of producing a distorted image of the molecule under investigation due to averaging images of non-identical molecules. Negative staining yields a much better SNR in the images and usually induces molecules to adsorb to the support film in only one or a few preferred orientations. This makes it possible to assess sample heterogeneity by image classification. When working with a structurally uncharacterized molecule, it is therefore good practice to first perform a negative stain analysis prior to any attempt of working with vitrified specimens.
We illustrate this point with the example of the complex formed by the headpiece of integrin α5β1 with the fibronectin Fn9-10 fragment (10). Gel filtration of the complex showed only two peaks corresponding to the complex and excess Fn9-10 used for complex formation, suggesting no unliganded integrin α5β1 headpieces to be present (Fig. 3a). Images of the peak fraction containing the complex prepared by negative staining immediately revealed, however, heterogeneity in the sample (Fig. 3b) due to dissociation of the ligand from the integrin headpiece, which is accompanied by a conformational change in the headpiece (Fig. 3c and 3d). This heterogeneity would not have been seen in vitrified preparations and without doubt cryo-EM of vitrified samples would have produced a flawed 3D reconstruction. By contrast, using negative stain EM in combination with image classification techniques it was straightforward to separate images of the two different molecules. Moreover, it was possible to obtain meaningful 3D reconstructions from this heterogeneous sample for both the unliganded integrin headpiece (Fig. 3e) and its complex with the Fn9-10 fragment (Fig. 3f).
Fig. 3. Negative stain electron microscopy of the integrin α5β1 headpiece with and without a bound fibronectin (Fn) fragment containing Fn domains 7 to 10 (Fn9-10).
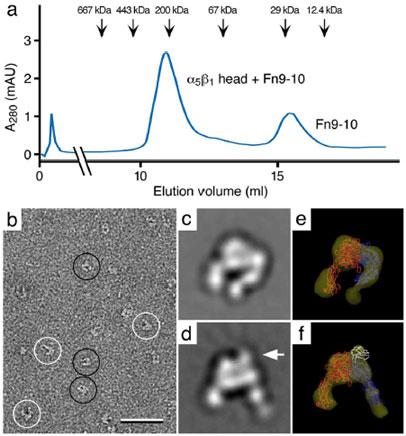
a: Elution profile from a gel filtration column used to purify the complex of α5β1 headpiece with an Fn9-10 fragment. The elution profile shows two peaks that correspond to the α5β1-Fn9-10 complex (~200 kDa) and unbound Fn9-10 fragment (~30 kDa). b: Negative stain electron microscopy reveals that the α5β1 headpiece adopts two conformations, namely a closed (black circles) and an open conformation (white circles). c and d: Class averages representing the closed (c) and the open conformation (d). Binding of Fn9-10 fragment (arrow in d) induces the open conformation of the headpiece, while the unliganded is in the closed conformation (c). e and f: 3D reconstructions of an unliganded (e) and an Fn9-10-liganded α5β1 headpiece (f) with the fit atomic structures of the αV and β3 subunit (33) in red and blue, respectively, and of the Fn9-10 fragment (34) in white. The scale bar corresponds to 50 nm and panels c to f have a side length of 22 nm. Figure panels modified and reprinted from (10). Copyright 2003 with permission from EMBO Journal.
Principle of image classification
A mathematical description of classification algorithms is beyond the scope of this paper and the interested reader is referred to (22) as an excellent introduction to the subject. Briefly, classification algorithms provide mathematical tools to quantify the similarity among different images. Variants of the K-means and hierarchical classification method are the most commonly used algorithms in single particle EM, but other algorithms, e.g. the Baysean method (23), are still being explored for the application to sets of images. All classification methods are in essence based on a comparison of the intensities of all pixels in one image with those of all the corresponding pixels in another image. For the best result of classification it is therefore of crucial importance that all particle images be aligned to each other as precisely as possible.
The details of how a set of images is aligned and classified strongly depend on both the software package and the classification algorithm used, and we therefore only provide a very general outline. The first step has to be a translational and rotational alignment of the particle images, for which reference-free or multi-reference alignment procedures can be used (22). So not to interfere with the alignment and classification algorithms, each image ideally contains only one particle. The alignment procedure is done iteratively over many cycles and is considered completed when overall image shifts and rotations no longer decrease upon further alignment cycles.
The aligned images are then subjected to classification. After specifying into how many classes the images should be sorted, the images in each class are averaged to create class averages with improved SNR. The number of required classes depends on the heterogeneity in the sample and cannot be predicted. It is therefore good practice to run multiple classifications specifying different numbers of output classes, e.g. 20, 50, and 100 classes. One good indication that sufficient output classes were selected is that a number of projection structures are represented by more than one class average. The more heterogeneous the population, the more classes are required, and accordingly the larger the image set has to be to obtain a sufficient number of images in each class to calculate meaningful averages.
To assess whether the classification procedure was successful, the images in each class can be visually compared to the corresponding class average. This is usually easy when working with images of negatively stained samples, where the particles are clearly seen due to the high image contrast. The same assessment is substantially harder for images of vitrified specimens, where the poor SNR of the images can make it difficult to even see the particle. At the same time the poor SNR of images taken from vitrified specimens reduces the probability that the images are being assigned to the correct class. This is of particular concern with heterogeneous samples and emphasizes the notion that a structurally uncharacterized protein should always first be analyzed in negative stain prior to analysis of vitrified specimens.
If desired, the unique classes can be selected from the best classification result and used as references for multi-reference alignment. Here, all the images of a data set are cross-correlated with all the references and assigned to the reference that yielded the highest correlation coefficient. All the images assigned to the various references are again averaged. Multi-reference alignment is also performed iteratively with the averages of each cycle being used as the references for the next cycle. The procedure is considered completed when the averages show no significant changes upon further cycles. No further improvement in the resolution of the averages upon further cycles, as assessed by the Fourier ring correlation or the spectral SNR criterion, is also an indication that the multi-reference alignment procedure is completed.
Various software packages are available that contain modules to perform image alignment, classification, and multi-reference alignment. IMAGIC (24) is commercially available, whereas SPIDER (14) and EMAN (25) are academic packages. Of these three programs, we consider SPIDER the most versatile one, allowing various classification algorithms to be applied to a data set.
Examples for the use of image classification
At this point we emphasize again that if the goal is to determine the 3D structure of a protein or a macromolecular complex, cryo-EM of vitrified specimens is the method of choice. If the sample is however too heterogeneous to use vitrified specimens, much care should be taken to find the negative stain and the preparation protocol that preserve the specimen with as little preparation artifacts as possible in order to obtain a reliable 3D map. The appropriate combination depends on the molecule that is being studied. This paper, however, does not focus on the determination of 3D structures. It is our intention to show that many biological questions can be addressed by simply taking projection images of negatively stained specimens. For this purpose it is in most cases sufficient to use the conventional negative stain approach with uranyl formate as the stain. All specimens used as examples for classification in the next paragraphs were prepared in this way.
Apparent heterogeneity due to different orientations. A fundamental problem in EM is the difficultly in distinguishing between molecules viewed from different orientations and molecules in different conformational states from projection views alone. There are two ways to determine whether differences in projection averages arise from different views of the molecule or from conformational variability.
If the crystal structure of the molecule is available, it can be used to create a resolution-limited model, typically using a resolution cut-off of about 25 Å. Projection views can then be generated from this model at regular angular intervals, e.g. 2°. If comparison by cross-correlation identifies a highly similar projection view of the molecule for all the experimental class averages, the structural heterogeneity is most likely due to different orientations of the molecules on the carbon film. This situation is illustrated by a negative stain preparation of yeast proteasomes (Fig. 4a), where classification produced two projection averages (Fig. 4a, insets 1 and 2). Comparison with projections from a 25 Å resolution-limited model generated from the crystal structure (26) identified the two projection averages to correspond to a top (Fig. 4a, inset 3) and a side view (Fig. 4a, inset 4) of the proteasome.
The situation is more difficult if no structural information is available on the molecule under investigation. In this case 60°/0° image pairs have to be collected to calculate 3D maps by the random conical tilt approach. The particle images from the untilted specimen are subjected to classification and individual 3D reconstructions are calculated for all or at least some of the classes using the corresponding particle images from the tilted specimen. If the 3D maps look the same, the classes represent different views rather than different conformations. This is illustrated by the Sec23p/24p complex. Visualized by negative stain EM, this complex has a bone-like appearance (Fig. 4b), but classification of the particle images produced averages with notable variations in the shape of the molecules (Fig. 4b, insets 1 to 5). Since there was no crystal structure available at the time of our EM analysis, individual 3D reconstructions were calculated for several classes, which looked essentially the same. Therefore, all images were combined to calculate a single 3D reconstruction (7) and the accuracy of our structure was later confirmed by a crystal structure of the Sec23p/24p complex (27). Because the sample was homogenous, this complex would also have been amenable to structure determination by cryo-EM of vitrified specimens.
If 3D maps obtained from different classes are different, experience is required to make the decision whether the differences arise from true conformational changes in the molecule or from preparation artifacts such as specimen flattening or incomplete stain embedding. A certain degree of variation in the 3D structures is indeed expected, because the staining pattern and the deformations due to adsorption interactions and sample dehydration all depend on the specific orientation of the particle on the carbon film. As discussed above, addition of glucose or glycerol to the sample and the use of the carbon sandwich technique minimize such variations induced by the negative stain preparation.
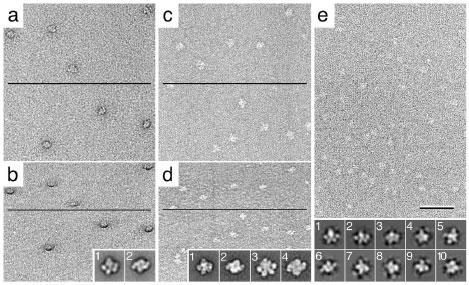
Heterogeneity due to different oligomeric states. Sample heterogeneity arises when a protein can exist in different oligomeric states as exemplified by the bifunctional primase-helicase of bacteriophage T7, the crystal structure of which has recently been solved (8). This protein oligomerizes into a ring-shaped structure and appears rather homogeneous when visualized by negative stain EM (Fig. 5). Upon image classification it becomes evident, however, that mixed populations of six- (Fig. 5, inset 1) as well as seven-membered rings (Fig. 5, inset 2) are present in the sample. Currently, it is not known what drives the equilibrium between the two oligomeric states, and an efficient way to separate the two forms has yet to be found. Such heterogeneity may not have been discernable in vitrified specimens and may have produced unreliable features in a 3D reconstruction determined by cryo-EM of vitrified specimens.
Fig. 5. Sample heterogeneity due to different oligomeric states.
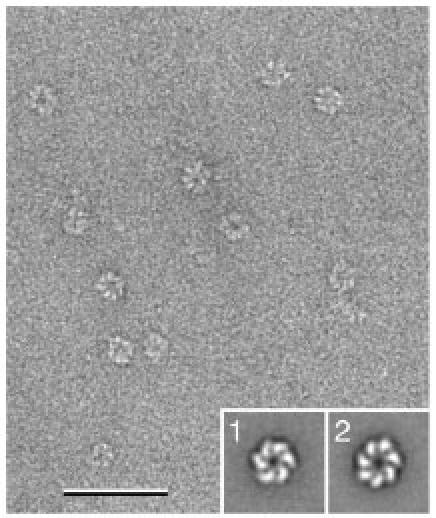
Image of negatively stained T7 helicase/primase in the presence of dTDP. While the particles appear rather homogeneous in the micrograph, image classification revealed the protein formed six- (inset 1) as well as seven-membered rings. The scale bar corresponds to 50 nm and the inset panels have a side length of 30 nm.
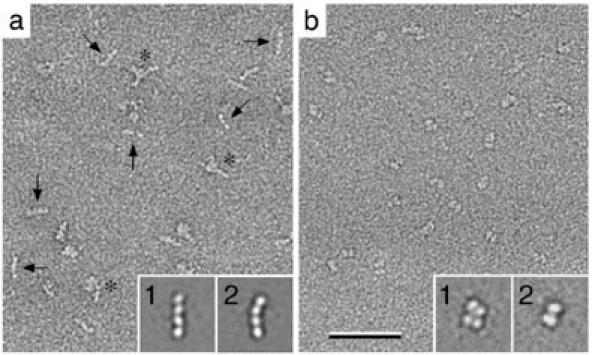
Heterogeneity due to different conformations. Like many proteins, integrin αVβ3 undergoes an extensive conformational change upon activation (9). For the presented analysis a construct of integrin αVβ3 containing only the extracellular domains but lacking the transmembrane and cytoplasmic domains was used. In the presence of inactivating Ca2+ ions αVβ3 adopts a compact conformation (Fig. 6a), while activating Mn2+ ions induce an extended conformation (Fig. 6b). It is rare to find experimental conditions that shift a conformational equilibrium completely to one side or the other, leaving at least some residual conformational heterogeneity in the particle population. In the case of the αVβ3 construct, about 14% of the molecules were in the extended conformation even in the presence of Ca2+, whereas about 20% of the molecules remained in the compact conformation in the presence of Mn2+. Addition of the strong ligand mimetic cyclic peptide cyclo-RGDfV induced more than 98% of the molecules to adopt the extended conformation irrespective of the cation present (9). These results demonstrate that by simple quantification of the subpopulations, classification can provide quantitative information for example on the potency of activating agents.
Fig. 6. Conformational equilibrium of integrin αVβ3.
a: Image of αVβ3 in the presence of inhibiting Ca2+ ions, where most of the molecules adopt a compact, closed conformation (insets 1 and 2 show representative class averages). Some of the molecules however can be seen in an extended, open conformation (arrows). b: In the presence of activating Mn2+ ions, the situation is reversed and most molecules are in the extended conformation (insets 1 to 4 show representative class averages), while only few molecules adopt the compact conformation (arrows). The scale bar corresponds to 50 nm and the inset panels have a side length of 40 nm. Figure 4 modified and reprinted from (9). Copyright 2002 with permission from Elsevier.
Another example for the use of quantitative classification is the determination of binding constants.
Figure 7a shows an image, where TfR at a concentration of 25 nM was mixed with Tf at a 1:1 molar ratio. The mixture was directly applied to a grid and negatively stained. Figure 7b shows the classes resulting from classification into 30 classes. Five unique classes representing unliganded TfR and Tf and TfR with one or two Tf molecules bound were selected for multi-reference alignment (Fig. 7c). By determining the percentage of the TfR-containing complexes formed at varying mixing ratios, it is possible to obtain a binding curve and thus the binding constant. The high contrast and the few preferred orientations obtained by negative staining are crucial for this approach to work, which was indeed used to determine the association constant for the binding of the 19S regulatory particle to the 20S proteasome (28).
Fig. 7. Quantitative classification of TfR-Tf complexes.
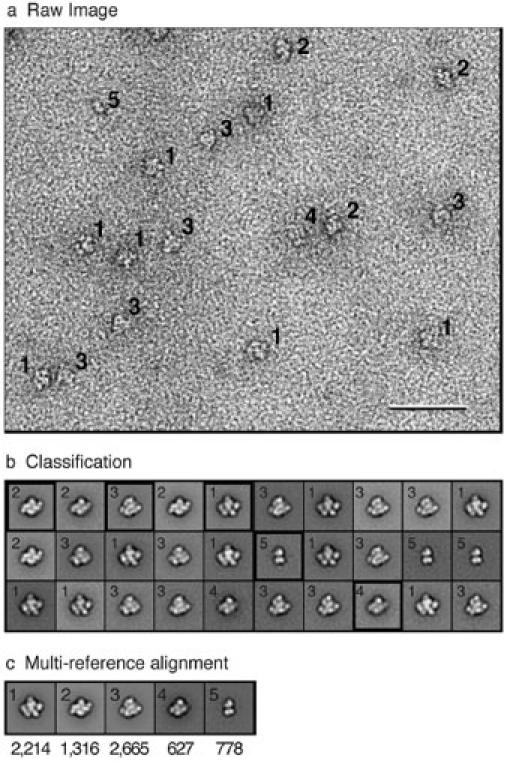
a: Image of a 1:1 mixture of Tf and TfR, revealing five different particle types. b: Classification of the particle images into 30 classes yielded five unique projection averages corresponding to TfR with two Tf molecules bound (label 1: side view; label 2: top view), TfR with one Tf molecule bound (label 3), TfR by itself (label 4), and Tf by itself (label 5). One projection average for each unique class was selected for multi-reference alignment (black frames). c: Final projection averages of the five classes with the number of particle images in each class noted below. The scale bar corresponds to 50 nm and the panels in b and c have a side length of 30 nm.
Analysis of mixed complexes. Complexes can be unstable, making it impossible to purify them to homogeneity. In these cases it is possible to simply mix the proteins and adsorb the mixture to an EM grid. Although such preparations are intrinsically very heterogeneous, this is sometimes the only practical approach for obtaining structural data for an unstable complex.
This mixing approach was successfully used to determine the structure of the ternary complex formed by the 20S proteasome with the 19S regulatory particle and the proteasome activator PA26 (12). Since only the α subunit of the PA26 complex was available, which has a low binding affinity for the proteasome, a large excess of PA26α had to be mixed with 26S proteasome to obtain a ternary complex, and the mixture had to be adsorbed to an EM grid without any further purification. Images of this preparation were therefore dominated by small ring-shaped molecules in the background formed by unbound PA26α, but various proteasome-containing complexes could also be seen (Fig. 8). Classification revealed all the expected complexes, namely unliganded 20S proteasome, 20S proteasome with one or two 19S regulatory particles bound, 20S proteasome with one or two PA26 bound, and 20S proteasome with one 19S and one PA26 bound (Fig. 8, insets 1 to 6). No other approach would have allowed structural analysis of such a heterogeneous sample. If required even 3D reconstructions could have been produced for all six complexes by recording images of tilted specimens.
Fig. 8. Complexes formed in a mixture of 20S proteasome, 19S regulatory particle and the α subunit of proteasome activator PA26.
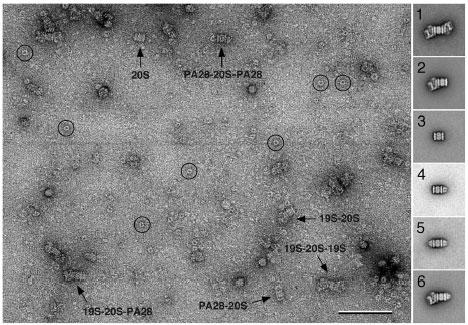
Image of a mixture of 26S proteasome with an excess of PA26α. Due to its low binding affinity for the proteasome, many unbound PA26α rings are present in this preparation (circles). Classification of images of proteasome-containing particles yielded projection averages of all the expected complexes, namely 20S proteasome with two (inset 1) or one 19S regulatory particles bound (inset 2), unliganded 20S proteasome (inset 3), 20S proteasome with one (inset 4) or two PA26α rings bound (inset 5) as well as the ternary complex of a 20S proteasome with a 19S regulatory particle and a PA26α ring (inset 6). The scale bar corresponds to 50 nm and the inset panels have a side length of 48 nm. Figure modified and reprinted from (10). Copyright 2003 with permission from EMBO Journal.
Use of classification with images of 2D crystals. Image classification is typically associated with single particle EM, but it can also be helpful in the analysis of 2D crystals formed by structurally distinct unit cells. In this approach the unit cells are extracted from the 2D array and classified as individual particles.
An early application of classification to 2D crystals was the analysis of the binding of maltose-binding protein (MBP) to maltoporin 2D crystals (29). MBP bound to only one out of three symmetry-related binding sites per maltoporin trimer. Classification was therefore used to select similar unit cells of the decorated maltoporin array and a projection map could be generated showing the outline of an MBP molecule interacting with a maltoporin trimer.
A different approach was used for the analysis of 2D crystals formed by reaction center (RC) – light-harvesting complex 1 (LH1) photounits from Rhodobacter sphaeroides (Fig. 9a and b) (13). The crystal contacts in these arrays were mediated by the 16-membered ring formed by the LH1 molecules (Fig. 9d). The RC in the center of the LH1 ring adopted a random orientation and was smeared out upon crystallographic (Fig. 9d) or single-particle averaging without prior alignment of the unit cells (Fig. 9e). Only by rotational alignment of the individual unit cells was it possible to resolve the projection structure of the RC in the center of the LH1 ring (Fig. 9f). A similar strategy was also used for 2D crystals formed by RC-LH1 photounits from Rhodospirillum rubrum (30).
Fig. 9. Single-particle processing of 2D crystals formed by RC-LH1 photounits from Rhodobacter sphaeroides.
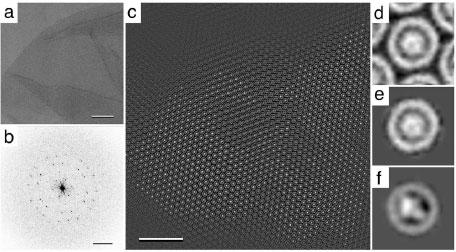
a and b: Image of a negatively stained RC-LH1 2D crystal (a) and the corresponding calculated power spectrum (b). c: The same crystal area as in panel a after Fourier-peak filtration, revealing the individual RC-LH1 complexes. d: Projection structure of the RC-LH1 complex obtained by crystallographic averaging of the image shown in a. e: Single particle average of the unit cells marked in panel c without rotational alignment. f: Single particle average of the same unit cells used to generate the average in panel e after rotational alignment. While the RC in the center of the LH1 ring has no features in averages d and e, it has a distinct shape in average f. The scale bars in a and c correspond to 100 nm, the scale bar in b to (6 nm)-1, and panels d to f have a side length of 18 nm. Figure modified and reprinted from (13). Copyright 1998 with permission from Elsevier.
The most vigorous single particle processing of a 2D crystal was applied to vitrified ordered arrays of the Na+/K+-ATPase from dog kidney. The resolution of the projection map improved from about 20 Å obtained with conventional electron crystallographic image processing (31) to 11 Å by the single particle/classification approach (32). As the borders between single particle and electron crystallographic processing are starting to blur, software packages are being developed that include classification modules that will be equally easy to apply to images of single particles and 2D crystals. One example is the Zephyr package that is currently being developed in the groups of David DeRosier and Nikolaus Grigorieff at Brandeis University.
Acknowledgments
The authors would like to thank Nikolaus Grigorieff, William Brieher, and Margaret Coughlin for critical reading of the manuscript. The molecular EM facility at Harvard Medical School was established by a generous donation from the Giovanni Armenise Harvard Center for Structural Biology. This work was supported by NIH grant GM62580 (to TW), which also provides funds to maintain the EM facility. Melanie Ohi thanks The Jane Coffin Childs Memorial Fund for Medical Research for a fellowship award and Ying Li is supported by an NRSA fellowship from the NIH. The authors have no conflicts of interest to declare related to this publication.
Appendix
Protocols
Preparation of a 0.75% uranyl formate solution
Attention: uranyl formate is radioactive, toxic and light-sensitive
Weigh out 37.5 mg of uranyl formate into a small beaker
Add 5 ml of boiling deionized water and stir for 5 min in the dark
Add drops of 5 M NaOH until the stain solution becomes slightly darker yellow (too much NaOH will precipitate the stain) and stir for another 5 min in the dark
Filter the solution with a 0.2 μm syringe filter into a Falcon tube wrapped with aluminum foil and add deionized water to a final volume of 5 ml
Conventional negative staining protocol
Place two 50 μl drops of deionized water and two 50 μl drops of uranyl formate stain on a piece of parafilm
Apply 2.5 μl of sample to a glow-discharged EM grid covered with a continuous carbon film and let the sample adsorb for 30 sec
Blot the grid from the side with a piece of filter paper, briefly touch the first drop of water with the grid, blot with filter paper, briefly touch the second drop of water, blot with filter paper, briefly touch the first drop of uranyl formate, blot with filter paper, touch the second drop of uranyl formate for 20 sec, and blot with filter paper (avoid complete drying of the grid in between the drops)
Completely dry the grid by vacuum aspiration touching only the rim of the grid
The particles on the carbon film should be well separated (to allow for their extraction into individual images for computational processing) but not too sparse (to avoid having to collect too many images). The particle concentration on the grid is best adjusted by dilution of the sample solution. It is also possible to vary the time for glow discharging and for sample adsorption.
Useful modifications to the protocol
For sensitive specimens, distilled water can be replaced by buffer solution for the washing steps
The number of washing drops can be increased to remove detergent or omitted to induce immediate fixation of the specimen
Glycerol or glucose can be added to the sample solution or the staining solution to minimize specimen flattening (only recommended for calculating 3D reconstructions)
Carbon sandwich technique
Place two 50 μl drops of deionized water and one 50 μl drop of uranyl formate stain on a piece of parafilm
Float thin piece of carbon in a small container of uranyl formate stain
Apply 2.5 μl of sample to a glow-discharged EM grid covered with a continuous carbon film and let the sample adsorb for 30 sec
Blot the grid from the side with a piece of filter paper, briefly touch the first drop of water with the grid, blot with filter paper, briefly touch the second drop of water, blot with filter paper, briefly touch the first drop of uranyl formate, blot with filter paper, touch the second drop of uranyl formate for 20 sec
With the sample side facing up, plunge the grid into the container holding the uranyl formate with the floating piece of carbon. Position the grid under the carbon and then lift the grid out of the container picking up the floating piece of carbon in the process. Gently blot the grid from the side using filter paper
Useful modifications to the protocol
Uranyl formate can be substituted with any other stain
Glycerol or glucose can be added to the sample solution and the grid can be frozen in liquid nitrogen to minimize specimen flattening
Cryo-negative stain using uranyl formate (Walz lab method adapted from Golas et al.)
Float thin piece of carbon in a small container of deionized water. Pick up piece of carbon with a holey carbon grid (QUANTIFOIL, Germany). The thin piece of carbon should rest on the carbon side of the holey grid. Allow the grid to air dry overnight or for several hours. Do not blot the grid after picking up the carbon
Sample should contain 5-10% glycerol or sucrose
Glow discharge the thin layer of carbon supported by the holey carbon film
Place one 50 μl drop of deionized water and one 50 μl drop of uranyl formate stain on a piece of parafilm
Float thin piece of carbon in a small container of uranyl formate stain
Apply 5.0 μl of sample to a glow discharged holey carbon grid prepared as described above and let the sample adsorb for 30 sec
Blot the grid from the side with a piece of filter paper, briefly touch the drop of water with the grid, blot with filter paper and then touch the drop of uranyl formate for 30 sec
With the sample side facing up, plunge the grid into the container holding the uranyl formate with the floating piece of carbon. Position the grid under the carbon and then lift the grid out of the container picking up the floating piece of carbon in the process. Dry for 20-30 sec and then carefully blot the grid from the side using filter paper. Dry at room temperature for approximately 2 minutes and then freeze the sample in liquid nitrogen
Cryo-negative staining (according to Adrian et al.)
Place a 100 μl droplet of 16% ammonium molybdate (pH range 7.0 to 8.0) on a piece of parafilm
Apply 4.0 μl of sample to a holey carbon film (QUANTIFOIL, Germany) that has not been glow discharged. Adsorb for 30 sec
Float the grid in the 16% ammonium molybdate droplet (sample side facing droplet) for 60 sec
Blot with filter paper, air dry 1-3 sec, and plunge into liquid ethane
Both the length of blotting and the amount of time the sample is allowed to dry before plunge freezing is critical for obtaining the optimal thickness of vitrified ice. These times must be experimentally determined.
References
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Rahman MA, Gowen B, Volynski KE, Ashton AC, Manser C, van Heel M, Ushkaryov YA. Structure of alpha-latrotoxin oligomers reveals that divalent cation-dependent tetramers form membrane pores. Nat Struct Biol. 2000;7:48–53. doi: 10.1038/71247. [DOI] [PubMed] [Google Scholar]
- Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ, Cheng Y, Petre BM, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc Natl Acad Sci USA. 2001;98:10704–10709. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz T, Jamieson SJ, Bowers CM, Bullough PA, Hunter CN. Projection structures of three photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6 Å, LH1 and RC-LH1 at 25 Å. J Mol Biol. 1998;282:833–845. doi: 10.1006/jmbi.1998.2050. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Adrian M, Dubochet J, Fuller SD, Harris JR. Cryo-negative staining. Micron. 1998;29:145–160. doi: 10.1016/S0968-4328(97)00068-1. [DOI] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- Bremer A, Henn C, Engel A, Baumeister W, Aebi U. Has negative staining still a place in biomacromolecular electron microscopy? Ultramicroscopy. 1992;46:85–111. doi: 10.1016/0304-3991(92)90008-8. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Dahlmann B, Hegerl R, Kopp F, Kuehn L, Pfeifer G. Electron microscopy and image analysis of the multicatalytic proteinase. FEBS Lett. 1988;241:239–245. doi: 10.1016/0014-5793(88)81069-X. [DOI] [PubMed] [Google Scholar]
- Brink J, Van Breemen JF, Keegstra W, Van Bruggen EF. Computer image analysis of two-dimensional crystals of beef heart NADH: ubiquinone oxidoreductase fragments. I. Comparison of crystal structures in various negative stains. Ultramicroscopy. 1989;27:79–90. doi: 10.1016/0304-3991(89)90202-7. [DOI] [PubMed] [Google Scholar]
- Walz T, Haner M, Wu XR, Henn C, Engel A, Sun TT, Aebi U. Towards the molecular architecture of the asymmetric unit membrane of the mammalian urinary bladder epithelium: a closed "twisted ribbon" structure. J Mol Biol. 1995;248:887–900. doi: 10.1006/jmbi.1995.0269. [DOI] [PubMed] [Google Scholar]
- Zhao FQ, Craig R. Capturing time-resolved changes in molecular structure by negative staining. J Struct Biol. 2003;141:43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. 1st ed. San Diego: Academic Press, Inc.; 1996.
- Samso M, Palumbo MJ, Radermacher M, Liu JS, Lawrence CE. A Bayesian method for classification of images from electron micrographs. J Struct Biol. 2002;138:157–170. doi: 10.1016/s1047-8477(02)00001-1. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Adams GM, Crotchett B, Slaughter CA, DeMartino GN, Gogol EP. Formation of proteasome-PA700 complexes directly correlates with activation of peptidase activity. Biochemistry. 1998;37:12927–12932. doi: 10.1021/bi981482i. [DOI] [PubMed] [Google Scholar]
- Stauffer KA, Hoenger A, Engel A. Two-dimensional crystals of Escherichia coli maltoporin and their interaction with the maltose-binding protein. J Mol Biol. 1992;223:1155–1165. doi: 10.1016/0022-2836(92)90266-m. [DOI] [PubMed] [Google Scholar]
- Stahlberg H, Dubochet J, Vogel H, Ghosh R. Are the light-harvesting I complexes from Rhodospirillum rubrum arranged around the reaction centre in a square geometry? J Mol Biol. 1998;282:819–831. doi: 10.1006/jmbi.1998.1975. [DOI] [PubMed] [Google Scholar]
- Tahara Y, Ohnishi S, Fujiyoshi Y, Kimura Y, Hayashi Y. A pH induced two-dimensional crystal of membrane-bound Na+,K+-ATPase of dog kidney. FEBS Lett. 1993;320:17–22. doi: 10.1016/0014-5793(93)81648-J. [DOI] [PubMed] [Google Scholar]
- Tahara Y, Oshima A, Hirai T, Mitsuoka K, Fujiyoshi Y, Hayashi Y. The 11 Å resolution projection map of Na+/K+-ATPase calculated by application of single particle analysis to two-dimensional crystal images. J Electron Microsc. 2000;49:583–587. doi: 10.1093/oxfordjournals.jmicro.a023847. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αvβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]


