Summary
Posttranslational modification with ubiquitin (Ub) controls many cellular processes, and aberrant ubiquitination can contribute to cancer, immunopathology, and neurodegeneration. The versatility arises from the ability of Ub to form polymer chains with eight distinct linkages via lysine side chains and the N terminus. In this study, we engineered Di-Ub probes mimicking all eight different poly-Ub linkages and profiled the deubiquitinating enzyme (DUB) selectivity for recognizing Di-Ub moieties in cellular extracts. Mass spectrometric profiling revealed that most DUBs examined have broad selectivity, whereas a subset displays a clear preference for recognizing noncanonical over K48/K63 Ub linkages. Our results expand knowledge of Ub processing enzyme functions in cellular contexts that currently depends largely on using recombinant enzymes and substrates.
Graphical Abstract
Highlights
-
•
Synthesis of Di-ubiquitin-based active site probes representing all eight linkages
-
•
Mass spectrometric profiling of DUB-Ub linkage preference in whole cell extracts
-
•
Activity-based Di-Ub probe screen for DUB specificity toward poly-Ub linkages
-
•
DUBs detected with a preference for noncanonical linkages over K48/K63-linked Ub
Ubiquitination controls many cellular processes. McGouran et al. engineered Di-Ub probes mimicking all eight different poly-Ub linkages and profiled the deubiquitinating enzyme (DUB) selectivity. Most DUBs examined have broad selectivity while a subset displays a preference for recognizing noncanonical linkages.
Introduction
Many biochemical processes in the eukaryotic cell are controlled by the posttranslational modification of proteins with ubiquitin (Ub), a 76 amino acid polypeptide that is highly conserved among eukaryotes (Kerscher et al., 2006). Ub adopts a globular structure in which all seven lysine residues and the N terminus are solvent exposed and can therefore form linkages with the C terminus of another Ub to form poly-Ub chains (Ye et al., 2012). The Ub linkage type and the chain length encode the fate of a ubiquitinated substrate. Protein modification by K48-linked poly-Ub chains is a well-established signal for recognition and initiation of degradation by the 26S proteasome complex, whereas proteins tagged with K63-linked Ub chains are known to be involved in nonproteolytic events such as in DNA damage responses and immune signaling (Komander and Rape, 2012). The roles of “atypical” poly-Ub chains linked through the Ub N terminus, K6, K11, K27, K29, or K33 are less well understood, but proteins tagged in such a way also appear to be directed toward proteasomal degradation (Kulathu and Komander, 2012, Xu et al., 2009). The molecular basis for the biological diversity of poly-Ub chains stems from their distinct structural features. Some Ub-linkages such as K6, K11, and K48 adopt compact structures where adjacent moieties interact with each other whereas linear and K63-linked chains display more open conformations where the linkage site is the only interface, although all of them show a degree of flexibility and dynamics (Ye et al., 2012). This implies that enzymes involved in ubiquitin conjugation (E1/E2/E3 ligases) and deconjugation (deubiquitinating enzymes [DUBs]) must bear specific recognition features to create and process different poly-Ub chains in their biological context, often involving direct interactions with cognate protein substrates and perhaps with other components of multiprotein complexes they are embedded in (Reyes-Turcu et al., 2009). DUBs are proteases that mediate the removal or processing of Ub chains or linked Ubl proteins by hydrolyzing the isopeptide bond between the C-terminal glycine of the distal molecule and the lysine residue in either the proximal Ub or the protein substrate. DUBs have recently emerged as key regulators in a multitude of processes from gene transcription to protein degradation and from cell division to cell death (Nijman et al., 2005, Tsou et al., 2012). They consist of a protease family comprising ∼90 members in human cells that can be grouped into five distinct families including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor domain proteases (OTUs), and Machado-Joseph disease (MJD) protein family, all of which are cysteine proteases. The fifth DUB family JAB1/MPN/MOV34 metalloenzymes (JAMMs) are metalloproteases. Available knowledge of DUB specificity or mode of regulation in cells is scant due to challenges in isolating DUB containing protein complexes or expressing full-length recombinant enzymes (Reyes-Turcu et al., 2009). Current efforts to study DUBs in vitro generally focus on the production of recombinant enzymes and the use of artificial substrates (e.g., Ub-AMC). Such investigations may not always reflect their substrate specificity and selectivity in a cellular environment.
Although K48- and K63-linked ubiquitin dimers and oligomers have been accessible through chemoenzymatic synthesis for a number of years (Pickart and Raasi, 2005, Piotrowski et al., 1997), synthetic access to the less well studied atypical linkages has only been established more recently. Selectively functionalized Ub variants have been assembled from peptide building blocks using native chemical ligation-based strategies (Kumar et al., 2010, Yang et al., 2010) as well as by optimized solid phase peptide synthesis of full length Ub (El Oualid et al., 2010). In each of these strategies, a mercapto-lysine residue was utilized for ligation with a C-terminal Ub thioester followed by desulfurization to obtain an isopeptide linkage containing a native lysine residue. Genetic incorporation of protected Boc-lysine in Escherichia coli followed by orthogonal amine protection schemes and Ag(I)-mediated thioester to amine ligation represents an elegant alternative approach (Castañeda et al., 2011, Virdee et al., 2010) that does not require chemical peptide synthesis. The successful genetic incorporation of protected mercapto-lysine derivatives in E. coli has now also enabled the combination of unnatural amino acid incorporation with the facile ligation methodology enabled by mercapto-lysine derivatives (Virdee et al., 2011). In addition to methods for the generation of native isopeptide-linked Ub dimers, there has also been an increased interest in novel straightforward methods for ligation of Ub by non-native linkages. Cu(I)-catalyzed triazole formation has been successfully employed for the generation of Ub-protein conjugates (Weikart and Mootz, 2010) as well as Ub dimers (Eger et al., 2010, Weikart et al., 2012). It was further demonstrated that the obtained conjugates are resistant to cleavage by DUBs (Weikart and Mootz, 2010).
As an addition to the repertoire of conventional in vitro assays, Ub-based active site probes, targeting DUB catalytic sites in crude cell extracts were successfully used to discover and profile active enzyme species (Borodovsky et al., 2001, de Jong et al., 2012, Love et al., 2009) and small molecule DUB inhibitors (Altun et al., 2011). The latter approach does not require biochemical purification of DUBs and can report on their activity and inhibition within their biological context, but so far was restricted to single Ub or Ub linked to a branched peptide as recognition scaffolds (Iphöfer et al., 2012).
In this study, we have engineered active site probes on a dimeric ubiquitin (Di-Ub) scaffold representing all eight different Ub-linkages, using biochemical methods coupled with discovery proteomics and quantitative mass spectrometry to define DUB poly-Ub linkage specificity in a cellular environment.
Results and Discussion
Synthesis, Reactivity, and Purification of Di-Ub Activity-Based Probes
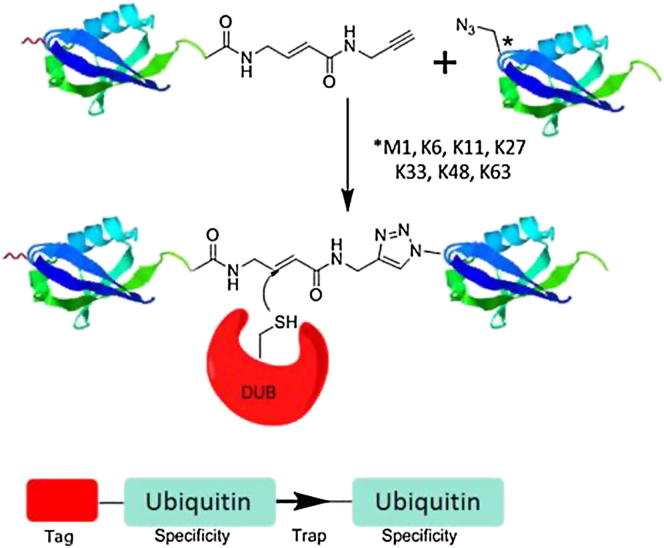
Activity-based Di-Ub probes formed of two recombinant Ub proteins were designed (Figure 1) with the following consideration in mind: the isopeptide bond present in the natural Ub dimer was to be replaced by a reactive electrophile for the covalent capture of cysteine protease DUBs. The linkage position was controlled by site-directed mutagenesis and incorporation of the unnatural amino acid azidohomoalanine (Aha) into the proximal Ub using the methionine analog incorporation approach (Kiick et al., 2002). Subsequently, the proximal Ub was linked by Cu(I)-catalyzed 1,4-triazole formation to the C terminus of the distal Ub. Molecular modeling indicated similarities in spatial requirements of the linker region between the natural linkage containing a Gly to Lys isopeptide bond and the reactive electrophilie linked via a 1,4-triazole (Figure S1 available online). A recent computational study using quantum mechanical/molecular mechanical (QM/MM) methods confirmed that K48-linked Ub-dimer contains similar geometric features in both native Ub dimer and a synthetic triazole-linked Ub dimer (Dresselhaus et al., 2013). The comparison was carried out between K48-linked Ub dimer with a native isopeptide linkage and a triazole containing dimer based on click conjugation of a K48AzPhe containing Ub.
Figure 1.
Schematic Illustration of Di-Ub Probes and Applications
Di-Ub probes were designed, consisting of two linked Ub moieties representing all Ub-linkages determining the specificity for DUBs that bind to the probe. Linkage specificity is achieved by incorporating Aha at the N-terminal methionine or in the positions of lysine residues in the proximal Ub. An electrophilic moiety in the “warhead” linking the two Ub molecules allowed covalent trapping of DUBs/Ub processing enzymes with a cysteine in the active site. DUBs/Ub processing enzymes bound to the probe could be characterized by gel electrophoresis and immunoblotting or identified after an immunoprecipitation (IP) by tandem mass spectrometry (LC-MS/MS) analysis.
See also Figure S1.
With this approach, Di-Ub probes mimicking linkages to each of the seven lysine residues or the N-terminal methionine were synthesized. The HA-tag present on the N-terminal of the distal Ub (derived from HA-Ub(1-75)-alkyne) allowed visualization and retrieval for identification using tandem mass spectrometry (LC-MS/MS).
Introduction of an alanine residue at position two of ubiquitin (I2A) to facilitate N-terminal Met/Aha cleavage by methionine aminopeptidase (Wang et al., 2008) was followed by individual K to M point mutations at each of the lysine residues to afford the seven Ub mutant plasmids. The Ub mutants and WT Ub were expressed in a methionine auxotrophic E. coli strain for azidohomoalanine incorporation (M1Aha, K6Aha, K11Aha, K27Aha, K29Aha, K33Aha, K48Aha, K63Aha) and purified (Pickart and Raasi, 2005). The purified proteins were characterized by SDS-PAGE and silver staining. MALDI-TOF MS of the intact proteins displayed efficient N-terminal Met/Aha cleavage as determined by the strength of signal at 8,504 m/z (corresponding to the mass of N-terminal cleaved Ub) versus a negligible to minor signal detected at 8,639 m/z (corresponding to uncleaved Ub). LC-MS/MS analysis after digestion with trypsin was also carried out to verify site-specific Aha incorporation (see Figures 2, S2, and DNA Sequences for Wild-Type and Mutant Ubiquitin). HA-Ub-alkyne and VME probes were prepared and purified according to literature procedures (Borodovsky et al., 2001, Borodovsky et al., 2002, McGouran et al., 2012). The HA-Ub-alkyne was ligated to the relevant Ub Aha mutant by Cu(I) catalyzed triazole formation (Rostovtsev et al., 2002) to afford the corresponding HA-tagged Di-Ub probes.
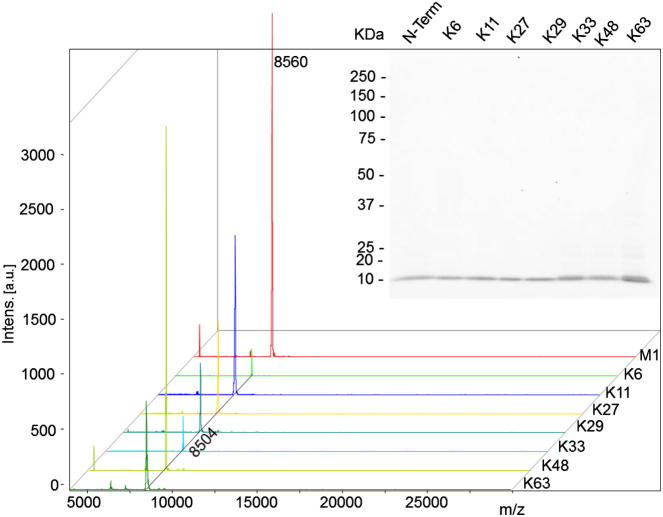
Figure 2.
MALDI-TOF Spectra and SDS-PAGE Analyses of Purified Aha Incorporated Ubiquitin Wild-Type and Mutant Proteins
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis revealed a distinct peak of the expected m/z for the Ub WT and mutants. The Aha incorporated mutants with N-terminal methionine cleavage were expected for [M+H]+ at 8,504 m/z and were detected between 8,500 and 8,508 m/z. For M1Aha, a [M+H]+ peak was expected at 8,561 m/z and detected at 8,560 m/z. The dialyzed Ub Aha mutants were separated by an 18% SDS-PAGE and visualized by silver staining (inset).
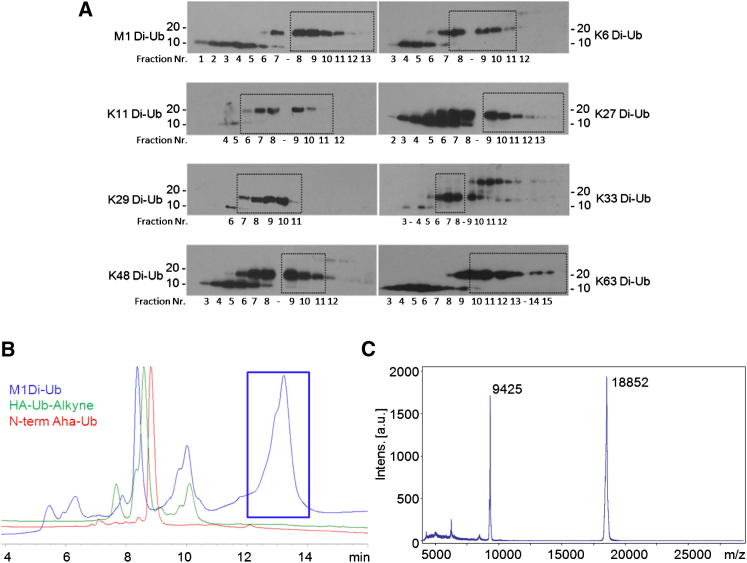
HPLC purification of all Di-Ub probes was performed using strong cation exchange chromatography to obtain high purity preparations removing any unconjugated HA-Ub-alkyne. The separation of Ub material of HPLC fractions was monitored by SDS-PAGE and anti-HA Western blotting (Figure 3A). The fractions marked with a dotted box were pooled, concentrated, and used for subsequent Ub processing enzyme profiling experiments. Representative HPLC chromatograms and intact protein MALDI-TOF MS spectra are shown for the M1 Di-Ub probe (Figures 3B and 3C).
Figure 3.
Purification and Characterization of Di-Ub Probes
(A) HPLC fractions of Di-Ub probe formation reactions analyzed by SDS-PAGE and anti-HA western blotting. Fractions containing high amounts of Di-Ub probe were pooled and concentrated (marked with dotted boxes).
(B) Overlay of Aha-Ub (red), HA-Ub-alkyne (green), and M1 Di-Ub probe (blue) chromatograms after HPLC purification. Peaks at 9 min correspond to Aha Ub and hydrolyzed HA-Ub-thioester, whereas the peak at 10 min corresponds to HA-Ub-Alkyne probe and the peak at 13 min represents the M1 Di-Ub probe, as verified by MALDI-TOF MS.
(C) MALDI-TOF MS spectrum of purified intact M1 Di-Ub probe with an observed average mass [M+2H]2+ 9,425 m/z, calculated 9,430 m/z and [M+H]+ 18,852 m/z, calculated 18,859 m/z.
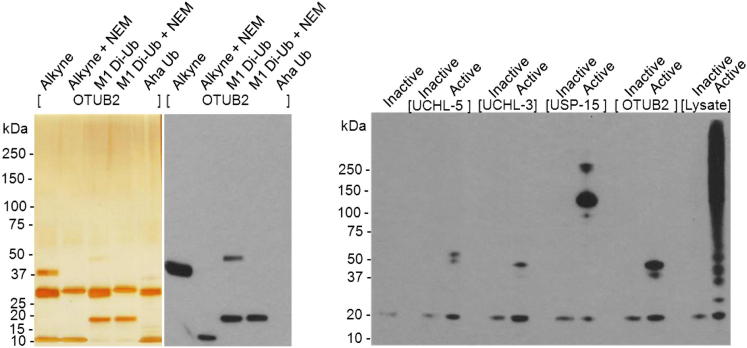
The reactivity of the purified M1 Di-Ub probe was tested by incubation with the deubiquitinating enzyme otubain 2 (OTUB2) alongside the monomeric HA-Ub-alkyne probe and visualized by both silver stain and western blotting (Figure 4A). Covalent trapping of OTUB2 with the probes was observed as a characteristic mass shift and shown to be inhibited by the cysteine alkylating agent N-ethylmaleimide (NEM). In order to demonstrate the requirement for the presence of a reactive electrophile, a proportion of the material was deactivated by reaction with sodium 2-mercaptoethanesulfonate (MESNa). Inactivated and active M1 Di-Ub probes were incubated with four different recombinant DUBs (OTUB2, UCH-L3, UCH-L5, and USP15) and also a cellular extract prepared from HEK293T cells (Figure 4B). All DUBs were successfully labeled with the active M1 Di-Ub probe, but did not react with the inactivated counterpart. Incubating the active probe with lysate resulted in strong labeling of proteins whereas lysate incubation with the inactive probe did not lead to any labeling. This demonstrates that the electrophilic trap is required for covalent capture of cellular DUBs (Figure 4B). Furthermore, conjugation through the native C terminus of the proximal Ub of the Di-Ub probe by E3 ligases does not occur to any significant extent under these labeling conditions.
Figure 4.
Di-Ub Probe Activity-Based Labeling of Recombinant and Lysate-Derived DUBs
(A) OTUB2-labeling with purified M1 Di-Ub and HA-Ub-alkyne probes. Reactions were separated by SDS-PAGE and visualized by silver staining and anti-HA western blotting.
(B) Active or inactive M1 Di-Ub probe was incubated with recombinant DUB or lysate, followed by SDS-PAGE separation and visualization by anti-HA western blotting.
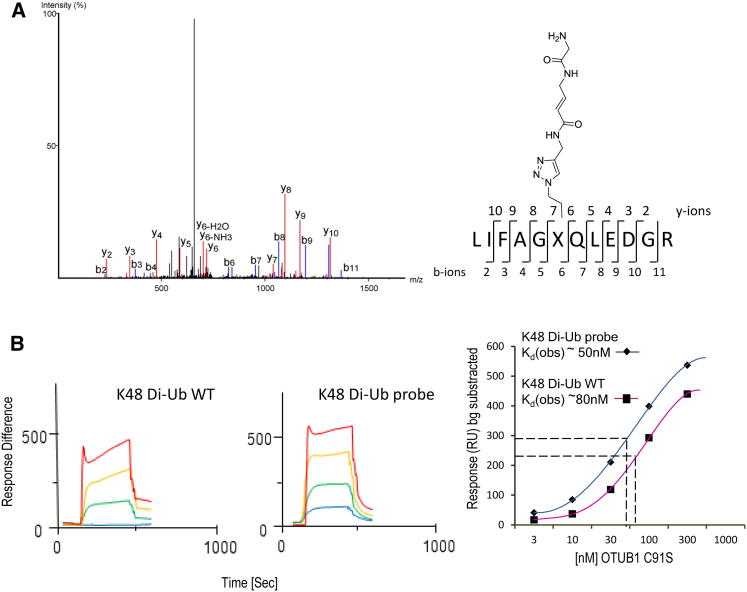
To verify site-specific linkage of the Ubiquitin moieties within the Di-Ub probes, LC-MS/MS analyses after digestion with trypsin were performed (Figures 5A and S3A–S3F). To assess the effect of introducing a nonnative triazole linker on the conformation of di-Ubiquitin, surface plasmon resonance (SPR) was carried out to compare the K48 Di-Ub probe to wild-type (WT) K48 Di-Ubiquitin with regards to its binding affinity for OTUB1, a DUB selective for K48-linked poly-Ub chains (Edelmann et al., 2009) (Figure 5B, left and middle panel). A catalytically inactive mutant (OTUB1 C91S) was used to avoid covalent binding of the probe. SPR analysis indicated KD values of ∼50 nM (Di-Ub WT) versus ∼80 nM (Di-Ub probe), respectively (Figure 5B, right panel). The OTUB1 inactivation mutation may further stabilize the substrate interaction as compared to its wild-type enzyme (Wiener et al., 2013), but appears to be similar between natural Di-Ub and the Di-Ub probe.
Figure 5.
Comparison of K48 Di-Ub Probe Linkage to Native Isopeptide-Linked Ub Dimer
(A) MS/MS spectrum of the peptidic fragment of trypsin-digested K48 Di-Ub probe bearing the electrophilic trap-derived covalent adduct (marked as X) in position 48. The proximal Ubiquitin-derived peptide (43–54) is shown with addition of the distal Ubiquitin C-terminal fragment as a modification. The b and y fragment ions are indicated in blue and red, respectively. See also Figures S3A–S3F.
(B) K48 Di-Ub wild-type (WT) (left panel) and K48 Di-Ub probe (middle panel) surface plasmon resonance data exposed to catalytically inactive OTUB1 C91S in 300 s cycles (10 nM: blue, 30 nM: green, 100 nM: yellow, 300 nM: red, respectively). Response units are shown after background subtraction (Response Difference). Right panel: Equilibrium analysis of the Di-Ub probe and native Di-Ub/OTUB1 interaction to determine the dissociation constants.
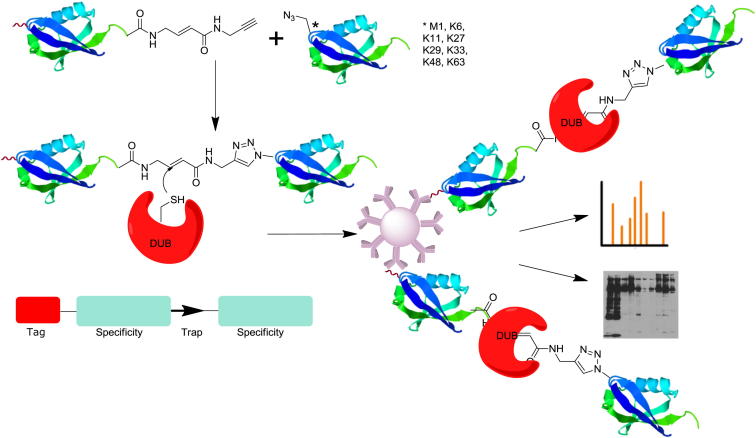
Di-Ub Probe-Labeling Profile and Proteome Analysis in Cell Lysates
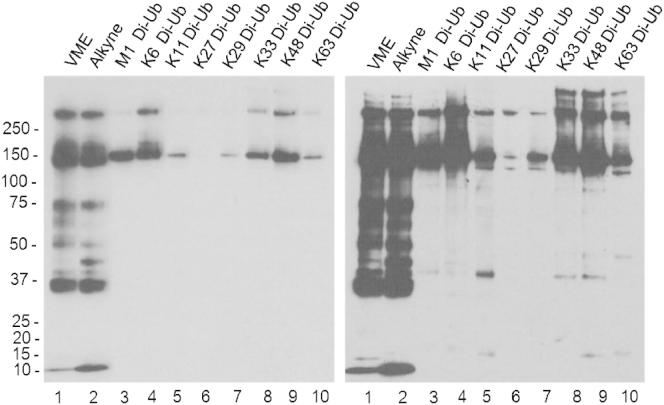
In order to compare differences in the cellular labeling pattern of the eight Di-Ub probes, the HA-Ub-VME and HA-Ub-alkyne probes, the respective probe and HEK293T cell lysate were incubated and analyzed by SDS-PAGE and anti-HA immunoblotting (Figure 6). Interestingly, Di-Ub probe labeling profiles are distinct from one another and notably so from the Mono-Ub probes, suggesting a higher DUB selectivity. To investigate this in more detail, cell lysate probe labeling followed by immunoprecipitation (IP) with anti-HA antibody agarose beads was carried out for all ten Ub probes. The elution fractions of all IPs were analyzed by tandem mass spectrometry to identify ubiquitin processing enzymes trapped by the probes.
Figure 6.
Lysate Labeling with Mono- and Di-Ub Probes Representing All Ub-Linkages
Lysate labeled with a panel of mono- and Di-Ub probes. Reactions were separated by SDS-PAGE and analyzed by anti-HA western blotting. For better visualization, two exposure times of the anti-HA western blot (left panel, low exposure; right panel, long exposure) are shown.
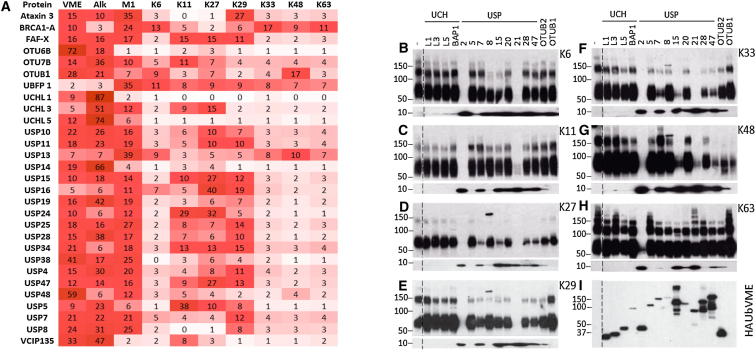
In total, 29 DUBs were identified by LC-MS/MS in all ten probe IPs combined including 18 USPs, four UCHs, five OTUs, one MJD, and one sumo deconjugase (Figure 7A). The mass spectrometric intensities of peptide signals for detected DUBs in all experiments were subjected to relative quantitation in order to compare DUB selectivity for the various Ub linkages. In separate experiments, poly-Ub chains of defined linkage were incubated in vitro with various recombinant members of the UCH, USP and OTU families to compare and further explore DUB preferences for different Ub linkages. Samples were analyzed by SDS-PAGE and anti-Ub immunoblotting to visualize disassembly of the respective Ub chains (Figures 7B–7I). As a positive control all recombinant DUBs were labeled with HA-Ub-VME, demonstrating that all enzymes used in this panel were active (Figure 7J).
Figure 7.
Cellular DUB Specificity for Ub-Linkage Topology
(A) DUBs detected by mass spectrometry analysis of eluted material from immunoprecipitation of cell lysates labeled with Di-Ub probes are displayed in a heat map representing their relative abundance. MS-based relative quantitation of DUBs per IP is shown ranging from low (white) to high abundance (bright red). The numbers express the percentage of the total amount of the observed DUB across all Di-Ub probe pull-downs (set to 100). For additional information, see DNA Sequences for Wild-Type and Mutant Ubiquitin and Table S2.
(B–I) Poly-Ub cleavage specificity of a panel of recombinant DUBs. Recombinant human enzymes UCH-L1/L3/L5, BAP1, USP2, USP5, USP7, USP8, USP15, USP20, USP21, USP28, USP47, and OTUB1/2 were incubated with the poly-Ub chains containing K6, K11, K27, K29, K33, K48, and K63 linkages and analyzed by SDS-PAGE and anti-Ub immunoblotting. –, no enzyme added.
(J) Activity-based profiling of a panel of recombinant DUBs using HA-Ub-VME, followed by SDS-PAGE separation and anti-HA immunoblotting, demonstrating the activity of the recombinant DUBs used.
Probing DUB Ub-Linkage Specificity
The monomeric HA-Ub-VME and alkyne probes showed generally the lowest labeling specificity across the panel of cellular DUBs detected (Figure 6, lanes 1 and 2), whereas interestingly, the M1 Di-Ub probe was the one with the most promiscuous specificity among the Di-Ub probes (Figure 7, lane 3). It should be noted that although the Di-Ub probes display the electrophilic trap in the appropriate position to mimic the native isopeptide bond (Figures 1 and S1), the M1 probe is linked via the amino acid side chain (M1 position) rather than the protein N terminus. The different relative position of the distal ubiquitin and the greater degree of flexibility afforded by this side chain linkage in comparison to the native peptide bond apparently cause decreased specificity of this probe (Komander et al., 2009). Although many DUBs showed little reactivity toward the different Di-Ub linkage types in comparison to the Mono-Ub probes, several were selective for particular Di-Ub probes tested.
We detected all four cysteine protease members of the UCH family in our probe IP experiments. Consistent with the observation that UCH enzymes are inefficient at ubiquitin chain cleavage (Kulathu and Komander, 2012, Zhou et al., 2012), we observed low general reactivity toward the Di-Ub probes in comparison to the monomeric probes in our IP samples (Figure 7A). We also detected little evidence of cleavage in our recombinant enzyme assays (Figure 7B).
Five OTU-containing enzymes were detected in the IP material. With OTU6B, we observe little reactivity toward the Di-Ub probes in comparison to the monomeric probes. OTU7B, previously reported to be K11-specific (Bremm et al., 2010), does indeed reflect this specificity in our assay showing the highest reactivity among the Di-Ub probes, although the specificity we observe is somewhat more modest than that seen within the previously reported in vitro assays with recombinant enzymes (Mevissen et al., 2013). OTUB1 displayed K48 linkage-specificity among the Di-Ub probes (Figure 7A), in line with both literature observations (Edelmann et al., 2009, Virdee et al., 2010) and results of our poly-Ub cleavage assay (Figure 7G). VCIP interestingly displayed a preference for the K11 linkage among the Di-Ub probes in line with in vitro assays using truncated VCIP (Mevissen et al., 2013), although little activity was seen with the K48 Di-Ub linked probe as would be expected from the in vitro assays using the truncated form (Mevissen et al., 2013).
For USPs, it has previously been noted that these enzymes are generally specific to ubiquitination substrates rather than the linkage of the ubiquitin chain, with a low degree of selectivity observed between the various ubiquitin linkages (Faesen et al., 2011). Our poly-Ub cleavage assays and probe labeling experiments confirm this observation, although we do see some examples of apparent specificity in the reactivity toward different Di-Ub probes.
For instance, USP13, USP25, and USP40 showed high relative affinity for the M1 probe. Interestingly, previous in vitro assays suggested little reactivity with linear Ub chains for these DUBs (Faesen et al., 2011, Kulathu and Komander, 2012). This discrepancy could be due to either the artificial nature of the in vitro assay or the greater flexibility of the linkage present in our M1 Di-Ub probe as compared to the native peptide bond (Komander et al., 2009). Interestingly, OTULIN, a DUB exclusively specific for cleaving linear poly-Ub chains (Keusekotten et al., 2013), was not detected with the M1 Di-Ub probe, possibly due to similar reasons.
USP15 in our IP results showed a degree of selectivity for K27-linked probe (Figure 7A). The corresponding poly-Ub cleavage assay showed that USP15 in its recombinant form was indeed active in cleaving K27 chains, but also other linkages (Figures 7B–7I).
USP16 showed a high relative reactivity toward the K27 and to a lesser extent the K29 probe in our assay. Literature precedence suggests that USP16 is active against these chain types although a less striking selectivity was observed previously (Faesen et al., 2011).
Di-Ub probe labeling data also suggested K11 selectivity by USP5 (Figure 7A), whereas our poly-Ub cleavage data suggested a very low degree of reactivity generally (Figures 7B–7I). Previous data has, however, shown USP5 activity against K11 chains (Bremm et al., 2010).
USP47 was found to have a high degree of specificity toward K27 linkages based on reactivity toward the Di-Ub probes and the recombinant DUB assays, and USP24 was highlighted to have a high degree of specificity toward the K27 and K11 probes.
We also detected Ataxin 3, principally in the M1 and K29 probe IPs despite previous suggestions that it has low activity toward chains of four Ubs or less (Mao et al., 2005).
Ub fusion protein 1 was also detected, and the highest relative abundance was observed with the M1 Di-Ub probe. BRCA1-A showed little selectivity among the different probes.
Taken together, our engineered probes representing all naturally occurring Di-Ub linkages provide clues on topology preferences of cellular DUBs for different poly-Ub chains, some of which were validated in vitro. Interestingly, many DUBs appear also to recognize noncanonical Ub-linkages other than K48/63 scaffold, suggesting the processing of a wider range of ubiquitinated substrates with different Ub linkage topologies.
Quantitative mass spectrometry analysis of Di-Ub probe IP material also revealed components of the Ub conjugation machinery were enriched by the different Di-Ub probes, as has been previously observed with mono-Ub probes (Love et al., 2009). A panel of conjugating enzymes was detected including those from HECT and RBR E3 families along with RING E3 ligases and E2 family members (Table S2).
Significance
The application of engineered activity probes based on Di-Ub scaffolds allows us to address DUB Ub-linkage specificity in a cellular context, which generally appears less biased toward canonical Ub-linkages than previously anticipated. Certain DUBs, in particular OTUs, retain selectivity for distinct poly-Ub chains as predicted using recombinant enzyme assays. The approach described provides insights into the selectivity of DUBs within the cognate environment of substrates as part of multiprotein complexes, which is often problematic to reconstitute with in vitro experiments. The detection of many enzymes of the ubiquitin conjugation machinery is noteworthy and may also provide us with a useful tool to study such enzymes in a cellular environment. These advances provide a framework for further understanding the role of DUBs and Ub conjugating enzymes in cellular functions under normal physiological conditions as well as in disease. A greater understanding of DUB Ub-linkage specificity may also offer inroads to assess cellular effects and specificity of small molecule inhibitors for future pharmacological intervention strategies.
Experimental Procedures
Reagents were purchased from Aldrich and used as supplied. Deionized water (minimum resistivity 18.2 MΩ) was used for preparation of buffers and chemical and biochemical reactions. Precision plus protein standard all blue gel marker (BioRad) was used for SDS-PAGE. HA-Ub-alkyne, HA-Ub-VME probes, and azidohomoalanine were synthesized in house according to literature procedures (Borodovsky et al., 2002, Link et al., 2007, McGouran et al., 2012) using a HA-Ub(1-75)-intein-chitin binding domain construct. HEK293T cells were cultured in DMEM (Dulbecco) medium supplemented with 10% FCS (Sigma) and 1% penicillin/streptomycin at 37°C with 5% CO2.
Preparation of Ubiquitin Mutants
Following overnight culture bacteria were harvested and the wild-type plasmid DNA containing the Ubiquitin sequence cloned into the pET15b (Addgene) vector was isolated, using Wizard Plus SV Minipreps DNA Purification System (Promega) according to the manufacturer’s protocol. DNA concentration was analyzed by measuring absorption at 260 nm wavelength using a NanoDrop spectrophotometer. Point mutants of Ubiquitin were created using the QuickChange II site directed mutagenesis kit by Stratagene. Mutant I2A was used as template for all following substitutions: K6M, K11M, K27M, K29M, K33M, K48M, and K63M. The primers used for the design of the Ubiquitin mutants are listed in Table S1. Mutant constructs were verified by DNA sequencing (Source Bioscience, Department of Biochemistry, University of Oxford). Sequencing results can be found in the DNA Sequences for Wild-Type and Mutant Ubiquitin.
Expression of Ubiquitin Mutants, Azidohomoalanine Incorporation, and Protein Purification
For the expression of WT and mutant Ub, methionine auxotroph B834(DE3) competent cells were used (Novagen) according to manufacturers protocol. Glycerol stocks were prepared by picking a single colony from an agar plate and incubating it in LB medium with shaking (160 rpm) at 37°C for 18 hr. A 100 μl cell suspension was mixed with 900 μl of 60% glycerol in water and stored at −80°C.
Overnight culture (10 ml) was inoculated from the respective glycerol stock. Bacteria were incubated in SelenoMet medium (Molecular Dimensions) containing 40 μg/ml methionine at 37°C for 18 hr. After transferring the bacteria to up to 500 ml SelenoMet medium containing and methionine, bacteria were grown with shaking (180 rpm) at 37°C until they reached an OD600 of 0.9. After harvesting, the cells were washed two times with methionine-free SelenoMet medium before resuspension in SelenoMet medium containing 40 μg/ml azidohomoalanine (Aha) and 0.4 mM IPTG. Protein expression was accomplished at 37°C for 4 hr with continuous shaking (180 rpm). Subsequently the harvested bacteria were resuspended in lysis buffer (100 mM Tris pH 7.5, protease inhibitors [Roche], 0.02% NP40, 0.4 mg/ml lysozyme) before 1.2 mg/ml MgSO4 and 12 μg/ml DNaseI were added. Vigorously vortexed cell lysate was cleared by centrifugation (16,000 × g, 30 min, 4°C) and treated with 0.5% perchloric acid on ice. Remaining Ub in the supernatant was dialyzed in 50 mM NaOAc pH 4.5 for 18 hr at 4°C.
The Ub mutants were resolved on an 18% Bis-Tris gel and visualized by silver staining. Linear mode MALDI-TOF MS analysis (Bruker) was also performed (Figure 2). A further aliquot of each mutant was subjected to digestion by trypsin and the resulting peptides analyzed by LC-MS/MS (Mackeen et al., 2010) to confirm Aha incorporation. In brief, proteins were resuspended in 6 M Urea, reduced with dithiothreitol and alkylated with iodoacetamide before proteolysis with trypsin. Desalted samples were separated on a 75 μm × 250 mm reversed phase Nano-Acquity-UPLC (Waters) and analyzed by a coupled Q-TOF Premier mass spectrometer (Waters). MS/MS spectra were searched against a human database containing the Ub mutant sequences (86,755 sequences) in Mascot (Matrix Science v2.4) allowing one missed cleavage and 50 ppm/0.1 Da mass deviation in MS and MS/MS, respectively. Oxidation of methionine, methionine to azidohomoalanine exchange, and deamidation of asparagine and glutamine were used as variable modifications. MS/MS spectra showing incorporation of Aha in the positions M1, K6, K11, K27, K29, K33, K48, and K63 are shown in Figures S2A–S2H.
Di-Ub Probe Synthesis by 1,4-Triazole Formation
HPLC-purified HA-Ub-alkyne (77 μl, 2 mg/ml) was mixed with the appropriate azidohomoalanine incorporated Ub mutant (805 μl, 0.56 mg/ml) in 50 mM Na2HPO4/NaH2PO4 pH 8 buffer. Tris[(1-ethoxycarbonylmethyl-1H-1,2,3-triazol-4-yl)-methyl]amine (90 μg in 15 μl CH3CN) and Cu(I)Br (120 μg in 15 μl CH3CN) were added before shaking at room temperature. After 30 min, a second portion of Cu(I)Br (120 μg in 15 μl CH3CN) was added, prior to a further 30 min incubation with shaking. The Di-Ub probe was buffer exchanged into 50 mM NaOAc pH 4.5 using gel filtration (PD-10, GE Healthcare). The resulting probes were HPLC (Agilent 1100) purified using a SCX biomonolith column (5.2 × 4.95 mm, Agilent) with a linear gradient from 0% to 100% buffer B, 50 mM NaOAc (pH 4.5, buffer A), 50 mM NaOAc (pH 4.5), and 1 M NaCl (buffer B) at a flow rate of 0.5 ml/min. Product-containing fractions were identified based on SDS-PAGE separation and anti-HA immunoblotting (Figure 3) and concentrated. The correct linkage was confirmed by LC-MS/MS analysis for seven out of the eight Di-Ub probes (Figures 5A and S3A–S3F). A minority of purifications, most notably for K33 Di-Ub, contained Tri-Ub species possibly resulting from insufficient cleavage of the N-terminal azidohomoalanine leading to double ligation. These by-products were removed.
In Vitro DUB Labeling
One hundred nanograms of recombinant DUBs OTUB1, OTUB2, UCH-L1, UCH-L3, UCH-L5, BAP1, USP2, USP5, USP7, USP8, USP15, USP20, USP21, USP28, and USP47 (all obtained from Progenra) or 20 μg HEK293T cell lysate, prepared as described (McGouran et al., 2012), were incubated with the relevant Ub probe at varying concentrations. Labeling was conducted in presence or absence of 10 mM N-ethylmaleimide (NEM) to confirm specificity. Volumes were adjusted to 15 μl with 50 mM NaH2PO4/Na2HPO4 pH 8.0 buffer. All reactions were incubated at 37°C for 3 hr followed by addition of 5 μl of 3× reducing sample buffer and heating to 95°C for 5 min. Samples were resolved on a NuPAGE 4%–12% Bis-Tris gradient gel and visualized by silver staining or western blotting methods using a directly coupled anti HA-HRP antibody (1:10,000 dilution, Sigma Aldrich H6533).
An inactivated linear Di-Ub probe was also generated to discount the possibility of conjugation through the Di-Ub C terminus. Two micrograms of linear Di-Ub probe was incubated in 10 μl inactivation buffer (50 mM MeSNa, 50 mM NaH2PO4/Na2HPO4 pH 8.0) at 37°C for 24 hr. A total of 1.5 μl of 50 mM MeSNa was added along with 10 μl 50 mM NaH2PO4/Na2HPO4 pH 8.0 and Di-Ub probe was incubated for a further 40 hr at room temperature (RT). The buffer was exchanged to 50 mM NaH2PO4/Na2HPO4 pH 8.0 by centrifugal filter devices.
Alternatively, 1 μM recombinant DUBs were incubated with 100 ng of poly-linked Ub chains (K6/K11/K27/K29/K33/K48, or K63, BIOMOL) for 3 hr at 37°C in 50 mM tris (pH 8.0) and 1 mM DTT, separated by NuPAGE 4%–12% Bis-Tris gradient gel electrophoresis, and analyzed by anti-Ub immunoblotting (1:3,000 dilution, BD Pharmingen).
Surface Plasmon Resonance
Surface plasmon resonance experiments were performed on a BIACORE 2000 instrument equipped with research grade CM5 sensor chips (BIAcore AB). All surfaces were prepared by an amine-coupling method (Johnsson et al., 1991) using reagents available from BIAcore AB (N-ethyl-N′-dimethylaminopropyl-carbodiimide [EDC], N-hydroxysuccinimide [NHS], and 1 M ethanolamine-HCl). Flow cells were activated for 15 min at 5 μl/min with a solution containing 50 mM NHS and 0.2 mM EDC. K48 Di-Ub WT and K48 Di-Ub probe were diluted to 20 μg/ml in 10 mM sodium acetate (pH 4.5) and injected until immobilization levels of 12,000–14,000 RU were achieved. The remaining reactive groups were blocked with a 12 min injection of 1 M ethanolamine-HCl at pH 8.5. A nonderivatized flow cell served as a reference surface. OTUB1 (C91S mutant) binding data were collected at 20°C at a flow rate of 5 μl/min in a solution of running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, and 0.005% P20) for both reference and reaction surfaces. OTUB1 C91S was dissolved directly in the running buffer at a stock concentration of 100 μM. Serial dilutions were made in running buffer across the concentration series from 10 nM to 3 μM. Samples were injected for 300 s followed by re-equilibration with running buffer. The raw response data were zeroed on both the response and time axes prior to the start of the injection. To correct for bulk refractive index changes, responses from a reference surface without protein were subtracted from the reaction surface data. To extract equilibrium dissociation constants, we plotted binding responses at equilibrium against ligand concentration and measured concentration values at 50% site occupation (Figure 5B).
IP and Analysis by Mass Spectrometry
Preparation of cell lysate and labeling of endogenous active DUBs using the HA-Ub-alkyne, HA-Ub-VME, and HA-Di-Ub probes was performed according to literature procedures (Borodovsky et al., 2002). For the quantitative mass spectrometry experiments, 1 mg of HEK293T cell lysate was incubated with 20 μg of Di-Ub probe (or 10 μg of the Mono-Ub probe) in NET buffer (50 mM Tris pH 7.5, 5 mM EDTA, 150 mM NaCl, 0.5% NP-40) in a total volume of 180 μl for 3 hr at 37°C. SDS was then added to 0.5% final concentration and the protein mixture was incubated at 4°C for 1 hr. The SDS concentration was reduced to 0.05% by adding NET buffer before the mixture was added to monoclonal anti-HA agarose beads (Sigma Aldrich). Immunoprecipitation was carried out at 4°C for 18 hr followed by three washing steps with high salt NET-buffer (50 mM Tris pH 7.5, 5 mM EDTA, 300 mM NaCl, 0.5% NP-40). Elution was accomplished by addition of HA peptide to a final concentration of 3 mg/ml.
The immunoprecipitated eluate was desalted using chloroform-methanol extraction, followed by overnight in-solution tryptic digestion and subsequent desalting as described previously (Altun et al., 2011). In brief, proteins were chloroform-methanol precipitated prior to resuspension in 6 M Urea, reduced with dithiothreitol, and alkylated with iodoacetamide before proteolysis with trypsin. Tryptic digests were desalted and subjected to LC-MS/MS (75 μm × 250 mm reversed phase Nano-Acquity-UPLC, Waters) analysis using a Thermo LTQ Orbitrap Velos (30,000 Resolution, Top 20, collision-induced dissociation) workflow and a gradient of 1%–40% acetonitrile in 48 min at a flow rate of 250 nl/min. Samples were analyzed in duplicate. Peptides were detected and quantified with Progenesis LC-MS software (version 3.1.4003.30577) using default settings (no deconvolution/deisotoping, 200 most intense MS/MS peaks). A merged peak list generated by Progenesis LC-MS was searched against the UniProt human database (version.3.80, 20,339 entries) using Mascot v2.4, allowing one missed cleavage and 20 ppm/ 0.5 Da mass deviations in MS and MS/MS. Carbamidomethylation of cysteine was a fixed modification. Oxidation of methionine and deamidation of asparagine and glutamine were used as variable modifications.
For label-free protein quantitation, Mascot results were imported into Progenesis LC-MS. Raw protein abundance values from Progenesis LC-MS were first normalized using quantile normalization and technical replicates aggregated by taking the median normalized value for each protein in each sample. Quantitation of enzyme abundance based on the different probe immunoprecipitations was based on at least two unique peptides, confidence score >50 and ANOVA (p value) <0.05 in order to generate the heat maps for Figure 7A.
Acknowledgments
We thank Dr. Katalin di Gleria, Dr. Nicola Ternette, Dr. Roman Fischer, and Shenliang Yu for expert help and assistance with experiments and aspects of the mass spectrometry analysis. We also thank members of Prof. Chris O’Callaghan’s group for help with the SPR experiments. We are indebted to Dr. Benjamin Nicolson (Progenra) for providing a panel of recombinant DUB enzymes. H.B.K. is supported by the Wellcome Trust OXION initiative. B.M.K. is supported by the John Fell OUP Research Award and the Biomedical Research Centre (NIHR), Oxford, UK. M.A. is supported as a Hållsten Academy fellow by the Swedish Medical Research Council, the StratNeuro, Åke Wibergs, and the Magnus Bergvalls Foundation (Sweden).
Published: November 27, 2013
Footnotes
Supplemental Information includes DNA sequences for wild-type and mutant Ubiquitin, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2013.10.012.
Contributor Information
Holger B. Kramer, Email: holger.kramer@dpag.ox.ac.uk.
Benedikt M. Kessler, Email: benedikt.kessler@ndm.ox.ac.uk.
Supplemental Information
References
- Altun M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J., Kumar K.G.S., Konietzny R., Fischer R., Kogan E. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Kessler B.M., Casagrande R., Overkleeft H.S., Wilkinson K.D., Ploegh H.L. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Bremm A., Freund S.M., Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda C., Liu J., Chaturvedi A., Nowicka U., Cropp T.A., Fushman D. Nonenzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J. Am. Chem. Soc. 2011;133:17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A., Merkx R., Berlin I., Rodenko B., Wijdeven R.H., El Atmioui D., Yalçin Z., Robson C.N., Neefjes J.J., Ovaa H. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. ChemBioChem. 2012;13:2251–2258. doi: 10.1002/cbic.201200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T., Weikart N.D., Mootz H.D., Waller M.P. Naturally and synthetically linked lys48 diubiquitin: a QM/MM study. RSC Advances. 2013;3:16122–16129. [Google Scholar]
- Edelmann M.J., Iphöfer A., Akutsu M., Altun M., di Gleria K., Kramer H.B., Fiebiger E., Dhe-Paganon S., Kessler B.M. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- Eger S., Scheffner M., Marx A., Rubini M. Synthesis of defined ubiquitin dimers. J. Am. Chem. Soc. 2010;132:16337–16339. doi: 10.1021/ja1072838. [DOI] [PubMed] [Google Scholar]
- El Oualid F., Merkx R., Ekkebus R., Hameed D.S., Smit J.J., de Jong A., Hilkmann H., Sixma T.K., Ovaa H. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew. Chem. Int. Ed. Engl. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen A.C., Luna-Vargas M.P., Geurink P.P., Clerici M., Merkx R., van Dijk W.J., Hameed D.S., El Oualid F., Ovaa H., Sixma T.K. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Iphöfer A., Kummer A., Nimtz M., Ritter A., Arnold T., Frank R., van den Heuvel J., Kessler B.M., Jänsch L., Franke R. Profiling ubiquitin linkage specificities of deubiquitinating enzymes with branched ubiquitin isopeptide probes. ChemBioChem. 2012;13:1416–1420. doi: 10.1002/cbic.201200261. [DOI] [PubMed] [Google Scholar]
- Johnsson B., Löfås S., Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Keusekotten K., Elliott P.R., Glockner L., Fiil B.K., Damgaard R.B., Kulathu Y., Wauer T., Hospenthal M.K., Gyrd-Hansen M., Krappmann D. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiick K.L., Saxon E., Tirrell D.A., Bertozzi C.R. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Komander D., Reyes-Turcu F., Licchesi J.D., Odenwaelder P., Wilkinson K.D., Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y., Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- Kumar K.S.A., Spasser L., Erlich L.A., Bavikar S.N., Brik A. Total chemical synthesis of di-ubiquitin chains. Angew. Chem. Int. Ed. Engl. 2010;49:9126–9131. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- Link A.J., Vink M.K., Tirrell D.A. Preparation of the functionalizable methionine surrogate azidohomoalanine via copper-catalyzed diazo transfer. Nat. Protoc. 2007;2:1879–1883. doi: 10.1038/nprot.2007.268. [DOI] [PubMed] [Google Scholar]
- Love K.R., Pandya R.K., Spooner E., Ploegh H.L. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem. Biol. 2009;4:275–287. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackeen M.M., Kramer H.B., Chang K.H., Coleman M.L., Hopkinson R.J., Schofield C.J., Kessler B.M. Small-molecule-based inhibition of histone demethylation in cells assessed by quantitative mass spectrometry. J. Proteome Res. 2010;9:4082–4092. doi: 10.1021/pr100269b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Senic-Matuglia F., Di Fiore P.P., Polo S., Hodsdon M.E., De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc. Natl. Acad. Sci. USA. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGouran J.F., Kramer H.B., Mackeen M.M., di Gleria K., Altun M., Kessler B.M. Fluorescence-based active site probes for profiling deubiquitinating enzymes. Org. Biomol. Chem. 2012;10:3379–3383. doi: 10.1039/c2ob25258a. [DOI] [PubMed] [Google Scholar]
- Mevissen T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Pickart C.M., Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- Piotrowski J., Beal R., Hoffman L., Wilkinson K.D., Cohen R.E., Pickart C.M. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev V.V., Green L.G., Fokin V.V., Sharpless K.B. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Tsou W.L., Sheedlo M.J., Morrow M.E., Blount J.R., McGregor K.M., Das C., Todi S.V. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS ONE. 2012;7:e43112. doi: 10.1371/journal.pone.0043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdee S., Ye Y., Nguyen D.P., Komander D., Chin J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- Virdee S., Kapadnis P.B., Elliott T., Lang K., Madrzak J., Nguyen D.P., Riechmann L., Chin J.W. Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Winblade Nairn N., Johnson R.S., Tirrell D.A., Grabstein K. Processing of N-terminal unnatural amino acids in recombinant human interferon-beta in Escherichia coli. ChemBioChem. 2008;9:324–330. doi: 10.1002/cbic.200700379. [DOI] [PubMed] [Google Scholar]
- Weikart N.D., Mootz H.D. Generation of site-specific and enzymatically stable conjugates of recombinant proteins with ubiquitin-like modifiers by the Cu(I)-catalyzed azide-alkyne cycloaddition. ChemBioChem. 2010;11:774–777. doi: 10.1002/cbic.200900738. [DOI] [PubMed] [Google Scholar]
- Weikart N.D., Sommer S., Mootz H.D. Click synthesis of ubiquitin dimer analogs to interrogate linkage-specific UBA domain binding. Chem. Commun. (Camb.) 2012;48:296–298. doi: 10.1039/c1cc15834a. [DOI] [PubMed] [Google Scholar]
- Wiener R., DiBello A.T., Lombardi P.M., Guzzo C.M., Zhang X., Matunis M.J., Wolberger C. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 2013;20:1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Pasunooti K.K., Li F., Liu X.W., Liu C.F. Synthesis of K48-linked diubiquitin using dual native chemical ligation at lysine. Chem. Commun. (Camb.) 2010;46:7199–7201. doi: 10.1039/c0cc01382j. [DOI] [PubMed] [Google Scholar]
- Ye Y., Blaser G., Horrocks M.H., Ruedas-Rama M.J., Ibrahim S., Zhukov A.A., Orte A., Klenerman D., Jackson S.E., Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.R., Zhang Y.H., Liu S., Song A.X., Hu H.Y. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.