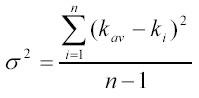Abstract
Light scattering and standard stopped-flow techniques were used to monitor rapid association of ribosomal subunits during initiation of eubacterial protein synthesis. The effects of the initiation factors IF1, IF2, IF3 and buffer conditions on subunit association were studied along with the role of GTP in this process. The part of light scattering theory that is essential for kinetic measurements is high-lighted in the main text and a more general treatment of Rayleigh scattering from macromolecules is given in an appendix.
Keywords: Ribosomes; Prokaryotic Initiation Factor-2; Scattering, Radiation
Introduction
In eubacteria, association of ribosomal subunits and initiation of protein synthesis require the three initiation factors IF1, IF2 and IF3 (1-3). In eukaryotes, subunit association and initiation of translation are more complex and require at least twelve initiation factors (2). All three prokaryotic initiation factors have their corresponding functional homologues in eukaryotes. Initiation factors IF1 and IF2 are close sequence and functional homologues of the eukaryotic initiation factors eIF1A (4) and eIF5B (3), respectively. Initiation factor IF3 has no sequence homology with any of the eukaryotic initiation factors (2). It has, however, several functions in common with eukaryotic eIF3 as well as with eukaryotic eIF1. The latter factor associates with eIF3 in mammals and is one of the subunits of eIF3 in yeast (5).
Termination of protein synthesis in eubacteria is carried out by either one of the class-1 peptide release factors RF1 or RF2 in a stop codon dependent way (6). After peptide release, rapid dissociation of the class-1 release factor is accomplished by the GTP-dependent action of the class-2 release factor RF3 (7-9). Subsequently, the ribosome is split by the combined activities of RRF, EF-G and IF3 (10), making the ribosomal 30S and 50S subunits ready for a new round of initiation of protein synthesis. Here, the 30S subunit, in complex with IF3, binds a messenger RNA, IF1, IF2:GTP and initiator tRNA (fMet-tRNAfMet) in a 30S pre-initiation complex (1), which rapidly recruits the 50S subunit in the formation of a 70S initiation complex. After GTP hydrolysis, IF2 rapidly dissociates from the 70S initiation complex, thereby making the ribosome ready to form the first peptide bond in a nascent protein (11). Subunit joining is an essential step in initiation of protein synthesis, but has in the past received comparatively little attention.
Subunit association or dissociation can be directly monitored by light scattering (12, 13) or ultracentrifugation (14) methods. More recently, Rayleigh light scattering, in combination with stopped-flow techniques, was used to study rapid formation of translation competent ribosomes from different pre-initiation 30S complexes and the 50S subunit (11).
In contrast to the rapid and non-invasive light scattering techniques, ultra centrifugation methods provide little kinetic information on subunit association or dissociation, but have been used to monitor the extent of eukaryotic 80S assembly (15). Ultracentrifugation methods are of non-equilibrium type and subunits, originally in ribosome complexes, may become separated during a centrifugation run. This potential problem is aggravated by the high pressure that develops in the rotor during a centrifuge run which further promotes subunit dissociation (16).
In this work, we describe the principles of Rayleigh light scattering and explain how this method can be combined with stopped-flow techniques to monitor the kinetics of formation or disruption of macromolecular complexes.
We apply the method, using a standard stopped-flow instrument (11), to initiation of protein synthesis in eubacteria, and present novel experiments that high-light the roles of IF3 and GTP on IF2 for selective and rapid 70S initiation complex formation.
The method of light scattering is general and can also be used to study the formation or disruption of macromolecular complexes other than the ribosome.
Materials and Methods
Chemicals and buffers
Nucleoside triphosphates (ATP, UTP, and GTP), radioactive amino acids and unlabelled nucleotides were from Amersham (USA). Non-hydrolysable GTP analogue GDPNP (GMPPNP), CTP, phosphoenolpyruvate (PEP), myokinase (MK), pyruvate kinase (PK), putrescine, spermidine, puromycin dihydrochloride, and non-radioactive amino acids were from Sigma (USA). All other chemicals were of analytical grade from Merck (Germany). Before use in binding and exchange assays, the guanine nucleotides GTP and GDP were further purified as described (8). All experiments were carried out in polymix buffer (17) which has the following final composition: [95 mM KCl, 5 mM NH4Cl, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate (KP) (pH 7.5) and 1 mM DTE]. One ml of this buffer is prepared by adding 0.1 ml of 10 times polymix, 0.05 ml of 20 times KP and 0.02 ml of 50 mM DTE to 0.83 ml of water. Preparation of 10 times polymix buffer is described in the protocol section. It contains the components of the polymix buffer at 10 times concentration but does not contain KP and DTE to avoid precipitation of calcium phosphate.
Components of the translation system
Synthetic mMFTI mRNA, encoding the tetra-peptide Met-Phe-Thr-Ile, was prepared according to (18). 70S ribosomes, 50S and 30S subunits were prepared from the E. coli strain MRE 600, using sucrose gradient zonal ultracentrifugation according to (19). Initiation factors were purified from overproducing strains according to (20). [3H]fMet-tRNAfMet and Phe-tRNA synthetase (PheRS) were prepared according to (7). Elongation factors EF-Tu, EF-Ts and tRNAPhe were purified according to (21).
Kinetics of macromolecular complex formation analyzed by stopped-flow and light scattering
In a typical light scattering experiment to monitor a binary complex formation between particles of type A and B, a solution containing particles A is rapidly mixed with a solution containing particles B and the intensity of light scattered perpendicular to the beam of illuminating light is recorded as a function of time. Initially, the mixture contains particles A and B at concentrations a(0) and b(0), respectively, while the concentration, c(0), of complex C is zero. The scattering intensity, I(t), at a time t after the mixing is the sum of the scattering intensities from free A-particles, free B-particles and C-complexes:
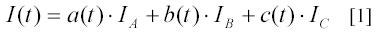
a(t) and b(t) are the concentrations of free particles A and B, c(t) is the concentration of complexes, C. IA , IB and IC are the scattering intensities per unit concentration for the corresponding particles and complexes. Since, for every C complex that is formed, one particle A and one particle B are consumed, a(t) and b(t) are related to the initial concentrations a(0) and b(0) as: a(t) = a(0) – c(t) and b(t) = b(0) – c(t). Introducing these mass relations in expression [1] gives

where ΔIC is the increase in light scattering intensity when the two particles A and B form a complex C. For particles with dimensions much smaller than the wave length of the illuminating light, the scattering intensity of a particle is proportional to the square of its molecular mass and does not depend on particle shape (12) (see also Appendix 1). Since the complex C between particles A and B is just a bigger particle, ΔIC can be estimated as:
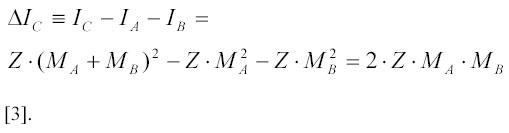
MA and MB are the molecular masses of the A and B particles, respectively, and Z is a proportionality coefficient for the particular experimental set up. Ribosomes and their subunits do not extend more than 30 nm, which is less than one tenth of the wave length (λ= 430 nm) of illuminating light used in most light scattering experiments on these particles (11-13). Therefore, relation [3] holds very well and it follows from this expression that, with x=MA/MB , the scattering intensity increases by a factor of 1+2x/(1+x2) when molecules A and B form a complex. The largest relative increase is by a factor of two, when MA=MB so that x=1.
Relation [2] shows that the increase, I(t)-I(0), in scattering intensity with time is directly proportional to the concentration c(t) of formed complexes. When complex formation has reached equilibrium, the plateau value, Ieq , of the scattered intensity is given by

Combining the experimentally measured parameters I(0), I(t) and Ieq , and using the relations [2] and [4] one gets:
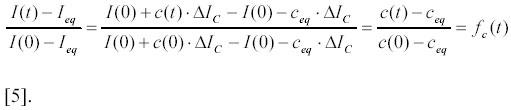
The time evolution of the function fc(t), which is one at time zero and zero at infinite time, contains all kinetic information about the complex formation. The ratio fc(t) is the difference between the current and the equilibrium concentration of the complex C, normalized to the value of this difference at time zero. Notice that fc(t) can be obtained from the experimentally measured intensities I(0), I(t) and Ieq without knowledge of the absolute value of ΔIC . Therefore, kinetic experiments can be interpreted without knowledge of the coefficient Z in relation [3], which depends on the experimental set up.
To exemplify the kind of kinetic information one can get from light scattering experiments, we consider the irreversible formation of a complex C from A- and B-particles that have the same initial concentration [A]0

The corresponding rate equation is:
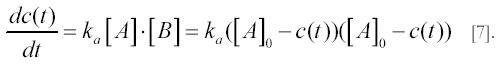
Its analytical solution is
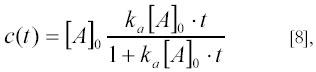
as can be verified by substituting c(t) in [7] with the expression for c(t) in [8]. After a long time all particles A and B will eventually end up in complexes C and an equilibrium concentration of complexes ceq=[A]0 will be reached.
Substituting the above expressions for c(t) and ceq=[A]0 into [5] one obtains a very simple expression for fc(t):
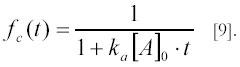
Accordingly, a plot of 1/fc(t) versus time gives a straight line with slope ka[A]0 , from which the association rate constant ka can be obtained from linear regression and knowledge of the initial concentrations [A]0 . In general, however, it is better to use non-linear regression methods to obtain ka along with normalization parameters. For this, the scattered intensity I(t) can be written as:
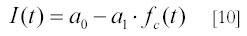
This relation follows from Eq. [5], with a0 equal to Ieq and a1 equal to Ieq - I(0). A best fit of this theoretical expression for I(t) to its experimental counterpart by variation of the parameters ka , a0 and a 1 , e.g. with the Marquardt algorithm (22), gives an estimate of ka along with the expected error (standard deviation) of this estimate. The content of this section is all that is required to apply Rayleigh light scattering to the kinetic analysis of macromolecular complex formation. An extended and more detailed description of light scattering theory and its experimental applications can be found in Appendix 1.
Light scattering experiments
Association of ribosomal subunits was monitored with light scattering after their rapid mixing in an SX-18MV stopped-flow instrument (Bio-sequential SX-18MV, Applied Photophysics, Leatherhead, UK) equipped with Xenon arc light source. To study the role of IF3 in subunit association, two mixtures, A and B, were prepared. Mixture A contained 4µM mMFTI mRNA, 0.5 mM ATP, 0.5 mM of GTP, 2 µM 30S and either (i) no additional components, (ii) 4 µM IF3, (iii) 2 µM IF1, 4µM IF2 or (iv) 4 µM IF3, 2 µM IF1, 4µM IF2 and 4 µM [3H]fMet-tRNAfMet as indicated. Mixture B contained 0.5 mM ATP, 0.5 mM GTP and 2 µM 50S. To remove dust particles, the mixtures were spun for 3 min at 14000 rpm in an Eppendorf centrifuge before they were loaded into the syringes of the stopped-flow instrument and pre-incubated at 37°C for at least 5 minutes. After rapid mixing, light scattering at 436 nm at right angle to the illuminating light was recorded as a function of time. The instrument was in emission detection mode, the photomultiplier voltage set to 520 V, and the time constant of the noise reduction filter to 5 ms. The volume of mixes (around 1 ml each) loaded into the syringes of the stopped-flow instrument was sufficient for at least ten independent time traces. Kinetic parameters were obtained for each individual trace by non-linear regression (see the section “Curve fitting” below) and used to obtain average estimates of rate constants and the standard deviations of these estimates.
To study the effects of GTP and GDP on the kinetics of subunit association in the presence of IF2, two mixtures, A and B, were prepared. Mixture A contained 0.5 mM ATP, 1 mM PEP, 0.5 mM of either GTP, GDPNP or GDP, 4 µM [3H]fMet-tRNAfMet, 2 µM 30S, 4µM mMFTI mRNA, 2 µM IF1, 4 µM IF2 and 4 µM IF3. Mixture B contained 0.5 mM ATP, 1 mM PEP, 2 µM 50S. Both mixtures were centrifuged for 3 min at 14000 rpm, loaded into the syringes of the stopped-flow instrument and pre-incubated at 37°C for at least 5 min before fast mixing.
Effects of buffer composition on the kinetics of 70S initiation complex formation were studied as follows. Two mixtures, A and B, were prepared and loaded into the syringes of the stopped-flow instrument. Mixture A contained 2 µM 70S ribosomes, 4µM mMFTI mRNA, 4 µM IF1, 4 µM IF3, 0.5 mM ATP, 0.5 mM GTP and indicated concentrations of PEP and Mg (OAc)2 in polymix buffer. Mixture B contained 0.5 mM ATP, 0.5 mM GTP, 4 µM [3H]fMet-tRNAfMet, 4 µM IF2 and the same concentrations of PEP and Mg(OAc)2 in polymix buffer as in mixture A. Mixture A was pre-incubated for at least 10 min at 37°C to ensure ribosome dissociation into 30S and 50S subunits. The formation of 70S initiation complexes was then initiated by mixing the mixtures A and B in a stopped-flow instrument as described above.
The kinetics of association of naked ribosomal subunits was studied in the following way. Two mixtures, A and B, were prepared and loaded into the syringes of the stopped-flow instrument. Mixture A contained 2 µM 30S ribosomal subunits, 0.5 mM ATP, 0.5 mM GTP, 1.5 mM PEP and indicated concentrations of Mg(OAc)2 in polymix buffer. Mixture B contained 2 µM 50S ribosomal subunits, 0.5 mM ATP, 0.5 mM GTP and the same concentrations of PEP and Mg(OAc)2 in polymix buffer as in mixture A. The formation of 70S initiation complexes was then initiated by mixing mixtures A and B in a stopped-flow instrument as described above.
Dipeptide-Formation assay
The effect of IF3 on the formation of translation-competent 70S initiation complexes was also studied with a dipeptide formation assay. To this end, two mixtures, A and B, were first prepared. Mixture A contained 0.5 mM ATP, 2 mM PEP, 0.5 mM GTP, 1.5 µM 30S, 2.5 µM mMFTI mRNA, 2.5 µM IF1, 2.5 µM IF2. Mixture B contained 0.5 mM ATP, 2 mM PEP, 2 µM 50S, 3 µM EF-Tu, 5 µM tRNAPhe, 30 µM phenylalanine, 1 µg/ml PK, 0.1 µg/ml MK and 10 U/ml PheRS (1 U of PheRS aminoacylates one pmol of tRNA per second). Then, 2.5 µM [3H]fMet-tRNAfMet was added either to mixture A or to mixture B. After pre-incubation for 10 min at 37°C the mixtures A (0.025 ml) and B (0.025 ml) were loaded into a quench flow instrument (KinTech, USA), mixed and quenched after the indicated times by 50% formic acid. The samples were centrifuged and the amount of formed fMet-Phe-tRNAPhe in the pellet was determined by HPLC as described previously (18).
Curve fitting
The association rate constant (ka) for subunit association in the stopped-flow light scattering experiments was estimated by non-linear regression (22) or by the Origin Program, using the three-parameter relation Eq. [10].
Results
IF3 as anti-association factor
In the absence of initiation factors, the 30S:mRNA complex associated with the 50S subunit with an association rate constant ka=1.2 μM-1 s-1 (Fig. 1A). Fig. 1C shows that the addition of IF1, IF2 and GTP to the 30S:mRNA complex resulted in a faster association of 30S with 50S (ka=4.1 μM-1 s-1). In the presence of only IF3, there was no complex formation between 50S and 30S:mRNA alone (Fig. 1B), or together with IF1, IF2 and GTP (not shown).
Fig. 1. The anti-association activity of IF3.

The extent of 70S initiation complex formation was monitored as a function of time by light scattering after rapid mixing in the stopped-flow instrument of a volume containing 30S subunits, mRNA, GTP and initiation factors as indicated with a volume containing 50S subunits. Time traces obtained with no initiation factors added to the 30S subunits (Panel A), with only IF3 added (Panel B), with IF1 and IF2 added (Panel C) and with IF1, IF2, IF3 and [3H]fMet-tRNAfMet added (Panel D).
The results of these experiments, summarized in Table 1, demonstrate the ability of IF3 to block subunit association when the 30S pre-initiation complex lacks initiator tRNA. When, however, fMet-tRNAfMet was present in the pre-initiation 30S:mRNA complex together with IF1, IF2 and GTP, the block was removed and the ribosomal subunits joined with an association rate constant ka = 12 μM-1s-1 (Fig. 1D).
Table 1.
Association rate constants of 50S subunits with 30S:mRNA in the presence of different combinations of Initiation Factors and fMet-tRNA.
| Factors added | kass(μM-1s-1) | |
| none | 1.2 ± 0.13 | |
| +IF3 | <0.0 | |
| +IF1+IF2:GTP | 4.1 ± 0.3 | |
| +IF1+IF2:GTP+IF3 | < 0.01 | |
| +IF1+IF2:GTP+IF3+fMet-tRNA | 12.2 ± 1.6 | |
| +IF1+IF2:GTP+IF3+fMet-tRNA (*) | 8.6 ± 1.2 | |
| +IF1+IF2:GDP+IF3+fMet-tRNA (*) | 0.13 ± 0.03 | |
| The association rate constant kass was calculated as an average (kav ) over rate constants (ki ) obtained from n (between six and eight) individual time traces of light scattering in a stopped-flow experiment. The error is the standard deviation σ obtained from the variance, estimated as | ||
| ||
| (*) The reaction buffer contained 1 mM phosphoenolpyruvate (PEP). | ||
The vital importance of IF3 for proper initiation of protein synthesis in eubacteria is further illustrated by quench-flow experiments that monitored the rate of dipeptide formation in the absence of IF3 (Fig. 2). In one experiment, pre-initiation 30S:mRNA complex together with fMet-tRNAfMet, IF1 and IF2 was mixed with 50S complex and all factors needed for peptide-bond formation (Fig. 2A). In an otherwise identical parallel experiment, fMet-tRNAfMet was present in the 50S, rather than in the 30S, mixture when the rate of peptide bond formation was followed (Fig. 2B). The rate of peptidyl-transfer was fast in the former (Fig. 2A), but virtually zero in the latter experiment (Fig. 2B). The absence of dipeptide formation in the second experiment reflects the rapid formation of a translationally inactive 70S complex lacking initiator tRNA. This complex is of the type seen with light scattering in Fig. 1C. This inactive ribosome complex was, in other words, formed before initiator tRNA had time to bind to the 30S:mRNA pre-initiation complex.
Fig. 2. Rate of initiation in the absence of IF3 monitored by di-peptide formation.

The extent of dipeptide formation was monitored as a function of time after rapid mixing in a quench flow instrument of a volume containing 30S subunits, mRNA, GTP, IF1 and IF2 with an equal volume containing 50S subunits. Initiator tRNA was present either in the 30S or in the 50S mix. Time curves obtained with [3H]fMet-tRNAfMet added with 30S (Panel A), [3H]fMet-tRNAfMet added with 50S (panel B).
The role of GTP in subunit association
It was recently shown that GTP on IF2 is important for fast association of a 30S pre-initiation complex with the 50S subunit (11). Those experiments were carried out with the functionally active β-form of IF2, lacking part of the N-terminal domain of the α-form of the factor (23, 24). Since differences in the GTP dependency of these factors cannot be excluded, we present here a similar study, but with a his-tagged version of the full-length α-form of IF2. Pre-initiation 30S complexes were formed with IF1, IF2, IF3, mRNA and fMet-tRNA in the presence of GTP, GDP or the GTP analogue GDPNP. Subsequently, these were rapidly mixed with 50S subunits in the stopped-flow instrument and the intensity of the scattered light was recorded. The rate constant for subunit association was around 8.5 μM-1 s-1 with GTP (Fig. 3A) or GDPNP (not shown) and 0.13 μM-1 s-1 with GDP (Fig. 3B). This means that GTP accelerated subunit formation sixty-fold compared to the rate obtained with GDP, in line with our previous results with the β-form of IF2 (11), but in contrast to results obtained by others (25).
Fig. 3. The effects of G-nucleotides on the association of 30S pre-initiation complex with 50S subunits.

The extent of 70S initiation complex formation was monitored as a function of time by light scattering after rapid mixing of pre-initiation 30S complexes with 50S subunits in a stopped-flow instrument. Traces obtained with GTP (Panel A) and GDP (Panel B).
Dependence of the subunit association rate constant on buffer composition
It is well known that the concentration of magnesium ions, ionic strength and composition of the buffer have a profound effect on the rate and accuracy of protein synthesis (21). It has also been demonstrated that the association rate constant of ‘empty’ ribosomal subunits increases by almost an order of magnitude when the Mg2+ concentration in the buffer increases from 4 to 8 mM (13). It was therefore of considerable interest to study the effect of Mg2+ and other components, like phospho-enolpyruvate (PEP), usually included in buffers for in vitro translation on the rate of formation of ‘real’ 70S initiation complexes. In the light scattering experiments described below ribosomes were first dissociated into their subunits in the presence of mRNA and initiation factors IF1 and IF3 in a buffer of indicated composition and then initiation factor IF2:GTP was added together with fMet-tRNA to dissociated 70S ribosomes in the stopped-flow instrument.
Fig. 4 shows that the rate constant of subunit association increased by only 50% from 7 µM-1s-1 to about 10 µM-1s-1 when the free Mg2+ concentration increased from 3 to 7 mM in our standard polymix buffer (compare Fig. 4A and 4B). At the same time, the addition of 10 mM PEP to polymix buffer (Fig. 4C) resulted in a three-fold decrease in the rate constant for formation of the 70S initiation complex to 2.3 µM-1s-1.
Fig. 4. The effects of buffer composition on the rate of 70S initiation complex formation.
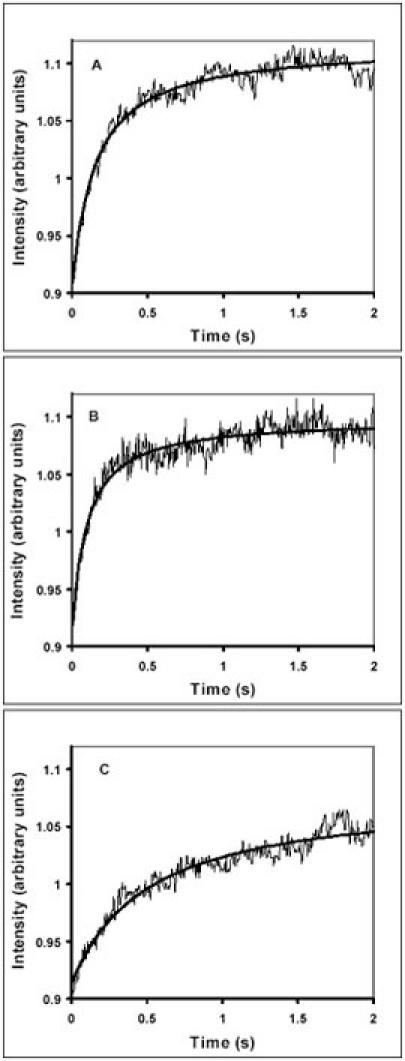
The extent of 70S complex formation was monitored as a function of time by light scattering after rapid mixing of mixture A containing dissociated 70S ribosomes together with IF1, IF3 and mRNA with mixture B containing IF2:GTP together with fMet-tRNA. Complex formation in polymix buffer (PM) with 3 mM of free Mg2+ (Panel A), in PM buffer with 7 mM of free Mg2+ (Panel B) and in PM buffer with 3 mM free Mg2+ plus 10 mM of PEP (Panel C).
An unexpectedly modest effect of Mg2+ on the association rate of 50S subunits with pre-initiated 30S complexes (see Table 2) compared to the large effects seen for association of “naked” 30S and 50S subunits observed by Wishnia et al. (13) may be due to the presence of mRNA, fMet-tRNA or initiation factors in the 30S pre-initiation complex. Alternatively, the difference could be due to the different ionic milieu in the polymix buffer compared to that in the buffer used in (13). The latter work employed a simple TMN buffer containing 10 mM Tris pH 7.5, 50 mM NH4Cl, 7 mM β-mercaptoethanol and different concentrations of MgCl2.
Table 2.
Dependence of association rate constants of 70S initiation complex (IC) or naked 70S ribosomes on buffer conditions. 70S initiation complexes were formed from 30S and 50S subunits in the presence of mRNA, IF1, IF2:GTP, IF3 and fMet-tRNA.
| Free Mg++ | PEP | kass (μM-1 s-1) | 70S type |
| 3 | 0 | 6.9± 0.7 | IC |
| 7 | 0 | 9.6± 1.5 | IC |
| 3 | 10 | 2.3± 0.4 | IC |
| 3 | 1.5 | 10.1± 0.9 | “naked” |
| 7 | 1.5 | 18.3± 1.8 | “naked” |
| The association rate constant kass and errors were calculated as for Table 1. | |||
To discriminate between these possibilities we have measured the association rate of naked 30S and 50S subunits in polymix buffer containing either 3 or 7 mM of free Mg2+. The results shown in Fig. 5 clearly demonstrates that the increase in Mg2+ concentration in polymix buffer from 3 to 7 mM results in about 80% increase in the subunit association rate from approximately 10 µM-1s-1 to 18 µM-1s-1 which is comparable to the 50% increase observed for pre-initiated 30S complexes and 50 subunits (see Table 2). Thus, the different effect of Mg2+ on subunit association in the two buffer systems is probably due to the difference in composition of the buffers and not to the presence or absence of initiation factors, mRNA or fMet-tRNA.
Fig. 5. The effects of the level of magnesium on the rate of naked 70S formation formation from its subunits.

The extent of 70S complex formation was monitored as a function of time by light scattering after rapid mixing of mixture A containing 30S ribosomes with mixture B containing 50S subunits. 70S formation in polymix buffer (PM) with 3 mM of free Mg2+ (Panel A), in PM buffer with 7 mM of free Mg2+ (Panel B).
Discussion
This work demonstrates the power of combining stopped-flow and light scattering techniques for experimental studies of how ribosomal subunits join during initiation of protein synthesis. Light scattering techniques were used in early experiments to determine how the equilibrium constant for subunit association depends on translation initiation factors (12). The kinetics of association of naked 30S and 50S subunits and its dependence on buffer conditions have previously been studied with stopped-flow techniques (13) and the effect of IF3 on the rate of ribosome splitting has been addressed with light scattering and manual mixing (12). However, under near-physiological conditions used in our in vitro experiments (21), subunit association catalyzed by initiation factors (Figs. 1 and 2; Antoun et al. (11)) and ribosome splitting, catalyzed by EF-G, RRF and IF3 (10), are rapid processes and their study therefore requires the combination of stopped-flow techniques and light-scattering. Here, we used stopped-flow with light scattering techniques to demonstrate the anti-association property of IF3 in the absence of initiator tRNA (Fig. 1), and complemented these measurements with quench-flow experiments, performed under similar conditions, to follow the rate of formation of the first peptide bond after initiation of protein synthesis (Fig. 2). The IF3 dependent block in the association of ribosomal subunits can be removed by the presence of initiator tRNA, IF1 and IF2. In cases when the subunits associate in the absence of IF3 and initiator tRNA the formed 70S ribosomes are unable to participate in protein synthesis. This suggests that IF3 plays a fundamental role in preventing premature ribosome formation in the absence of initiator tRNA.
We also demonstrated the fundamental role of GTP for fast subunit association catalyzed by the α-form of IF2 during initiation of eubacterial protein synthesis, in line with previous results obtained with the β-form of IF2 (11). During exponential growth of bacteria, ribosomes load on to the 5’ end of an mRNA each four seconds (26). The distance between ribosomes in a polysome is around 230 nucleotides (26). With a rate of 20 codons/s for protein elongation (27) and the need to clear the occluded ribosome binding site (1) to allow for the binding of the next ribosome, the lower limit for the initiation rate is about 0.3 s-1. This rate includes 30S docking to mRNA, fMet-tRNA and IF2 binding and subunit joining. Taking into account the results in Table 1 and that the concentrations of free ribosomal subunits in the cell are around 1 μM (1) one can conclude the rate of subunit joining catalyzed by IF2:GTP observed here is compatible with the rate of initiation in vivo.
The presented light scattering experiments show also the importance of a proper choice of buffer conditions to study the kinetics of ribosomal reactions. All experiments presented in this paper were performed in polymix buffer that mimics the ionic milieu of the bacterial cell (21). We have found, however, that some additional, supposedly “neutral,” components, often included in standard in vitro translation systems, like phosphoenolpyruvate (PEP) may result in considerable alteration in the rate of 70S complex formation (Table 2). Even an addition of small amounts of PEP (1 mM) to the reaction mixture results in a noticeable decrease of the rate of association of pre-initiated 30S complexes with 50S subunits (Table 1). The reason for this PEP effect is not clear at present. The effect of an increase of the free Mg2+ concentration from 3 to 7 mM in polymix buffer on the rate of 70S initiation complex was however much more modest, i.e. only 50%. This 50% effect is, nevertheless, quite comparable with an 80% increase in the association rate for “naked” ribosomal subunits upon the same increase in free Mg2+ concentration (see Table 2). Comparison with published data (13) on association of “naked” subunits shows, however, that the increase in free Mg2+ concentration from 3 to 7 mM results in a drastic increase in the association rate of ‘naked’ subunits from 0.63 μM-1s-1 to about 20 μM-1s-1. This discrepancy is likely to be due to the absence of organic polyamines such as putrescine and spermidine in the TNM buffer system [10 mM Tris pH 7.5, 50 mM NH4Cl, 2 to 8 mM MgCl2 and 7 mM β-mercaptoethanol] used in the previous work (13). Addition of polyamines corresponds, to a first approximation, to an effective increase of Mg2+ concentration in the buffer since polyamines mimic the most important, electrostatic, contribution of Mg2+ in shielding phosphates of rRNA and reducing the electrostatic repulsion between the subunits (13). The association rate constant of 10 μM-1s-1 at 3 mM free Mg2+ in polymix buffer is similar to the rate constant of 9.2 µM-1s-1 observed by Wishnia et al. (13) at 5.5 mM of free Mg2+. It seems therefore more appropriate to compare our results with those in TNM buffer upon the increase of Mg2+ from 5.5 to 9.5 mM. Published data (13) show that the association rate of naked subunits plateaus around 7.5 mM Mg2+ reaching 22 μM-1s-1. If this value of 22 μM-1s-1 is really a plateau, we will get a very good agreement for the effect of Mg2+ on subunit association in two different buffer systems.
The experiments described here were performed with a standard stopped-flow instrument (SX-18MV, Applied Photophysics, Leatherhead, UK) in fluorescence mode. The required ribosome concentration was in the μM range, and all solutions were centrifuged for 3 min at 14000 rpm to remove dust particles and aggregates before they were loaded into the syringes of the stopped-flow instrument. The stopped-flow measurements successfully covered a broad range of subunit association times, from 10 ms to 30 min.
As described, the scattering intensity is proportional to the molar concentration of particles and to the square of their molecular weight. Therefore, the scattering intensity from a 1 μM solution of 30S subunits (Mw 900 kD) equals the scattering intensity from a 100 μM solution of 90 kD proteins. Accordingly, light scattering methods can be used to monitor the association kinetics also of proteins with considerably smaller molecular weights than the ribosome and its subunits, albeit at higher protein concentrations and with a larger investment in the total amount of protein.
Acknowledgments
We thank Gun Stenberg and Gunnar Johansson for helping us with the light scattering experiments. This work was supported by the Swedish Research Council, the Estonian Science Foundation and the Wenner-Grenska Samfundet Foundation and the Egyptian mission department.
Appendix
Protocols
Only double distilled water was used in all protocols described in this paper. All stock solutions were filtered through 0.45 or 0.22 μm BA85 membrane filters (Schleicher-Schuell, Germany).
Preparation of 10 times Polymix buffer
Preparation of putrescine solution
Put 100 g of putrescine powder (Sigma) in a heat resistance glass, add 600 ml of water and warm to 90°C with eventual stirring until putrescine dissolves completely. Cool the solution to room temperature.
Adjust the pH to 8.0 with concentrated acetic acid (approximately 100 ml is required) and wait until it cools to the room temperature again. Add more acetic acid to lower pH to 7.6 and adjust the solution volume to 1134 ml.
Add one 100 ml cup of activated coal and stir under hood for 30 min. Filter through a paper filter first and then through a 0.45 µm BA85 membrane filter. Rap in foil and store at +4 oC.
Preparation of 10 times Polymix (2 L)
Add 141.66 g KCl; 5.35 g NH4Cl; 21.44 g Mg (OAc)2 and 1.47 g CaCl2 to a big glass beaker with a stirring bar and dissolve salts in approximately 1.5 L water.
Add slowly 160 ml putrescine while stirring relatively fast and then 5.1 g spermidine dissolved in a small volume of water.
Adjust the pH to 7.5 with either concentrated acetic acid or 5M KOH. Adjust the volume to exactly 2000 ml and filter through 0.22 μm BA85 membrane filters. Disperse in several smaller bottles and store at -20°C. The final composition of 10 times Polymix is the following: [950 mM KCl, 50 mM NH4Cl, 50 mM Mg (OAc)2, 5 mM CaCl2, 80 mM putrescine and 10 mM spermidine] (17).
[3H]fMet-tRNAfMet preparation from over expressing cells
Cell preparation and growth
Add 2 µl of pSBtRNAfMet plasmid (28) to 100 µl of the MRE600 competent cells, and keep on ice for 30 min.
Transfer cells into 42°C (heat shock) for 45 sec, then put them on ice for 2 min, add 0.8 ml 2*YT medium (29).
Incubate cells at 37°C for 45 min, then plate 100 µl of transformed cells onto plates with 0.1 g/l ampicillin and incubate overnight at 37°C.
Pick a colony from the plate, suspend it into the 1.5 L of 2*YT medium with 0.1 g/l ampicillin and grow overnight with shaking at 37°C for 20 hours.
Collect cells by centrifugation at 4000 g for 45 min at 4°C, re-suspend them in 10 ml of [10 mM Tris-HCl (pH 7.5) with 100 mM NaCl] in Falcon tube and pellet by centrifugation at 4000 g for 20 min.
Extraction and purification
Re-suspend the cell pellet in 20 ml of the AM buffer containing [30 mM NaOAc (pH5.1), 10 mM MgCl2] and add an equal volume of phenol saturated with the same AM buffer. Vortex cells with phenol for 1 hour at room temperature and collect then the water phase after 40 min centrifugation at 17,000 g (12,000 rpm for SS34 rotor (Sorvall)) at 4°C.
Add 1/10 volume of 3 M NaOAc (pH5.1) to the aqueous phase and precipitate nucleic acids with 2 volumes of ethanol. After 2 hours at -20°C collect the precipitate by 20 min centrifugation at 4,000 g at 4°C.
Dissolve the pellet in water and add 4M NaCl to 1 M final concentration. Keep the solution on ice for 20 min to precipitate high molecular weight DNA and RNA; then clarify the solution by spinning down at 34,500 g (17,000 rpm for SS34 rotor) at 4°C for 30 min. Precipitate tRNA from the supernatant by adding 1/10 volume of 3M NaOAc (pH 5.1) followed by 2 volumes of ethanol.
After 2 hours at -20°C, collect the precipitate by 20 min centrifugation at 4,000 g. Dissolve the pellet in 8 ml of 1 M Tris-HCl (pH 8.1) and incubate at 37°C for 2 hours to de-acylate tRNA.
Dilute tRNA solution to 100 ml with water and apply directly to a 120 ml Sepharose Q-FF column (Pharmacia) equilibrated with 80 mM Tris-HCl (pH 8.1).
Wash the column with TMEDN200 buffer containing [20 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 0.1 mM EDTA, 1 mM DTE and 200 mM NaCl] until OD reaches a base-line and start then a 250 ml plus 250 ml gradient from 380 mM to 700 mM NaCl in TMED buffer containing [20 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 0.1 mM EDTA, 1 mM DTE].
Identify tRNAfMet by charging assay described below. Pool fractions containing tRNAfMet and precipitate them by adding 1/10 volume of 3M NaOAc pH 5.1 followed by 2 volumes of ethanol.
Collect the precipitate by centrifugation at 4000 g for 20 min (4°C) and dissolve the pellet in 2 ml Polymix (PM) buffer. Measure the amount of tRNA by the charging assay.
Charging assay: Add 10 µl of assayed chromatographic fraction to 90 µl of charging mix containing, at final concentration in the reaction mix, [10 mM PEP; 1 mM ATP; 0.05 mg/ml pyruvate kunase (KP), 0.003 mg/ml myokinase (MK); 30 U methionine tRNA synthetases (MetRS) and 0.2 mM [3H] methionine with specific activity of at least 50 cpm/pmol. Incubate for 10 min at 37°C, precipitate charged tRNA by adding 5 ml of ice-cold 5% trichloroacetic acid (TCA), filter through GF/C filters (Millipore), dry filters, add 4 ml Ready Protein scintillation cocktail (Beckman Coulter, USA) and count. 1 U of an aminoacyl-tRNA synthetase enzyme is equal to the amount of the enzyme that charges one picomole of cognate tRNA per second.
Preparation of 10-THF
This procedure was adapted from (30). Dissolve 25 mg of folinic acid Ca2+ salt (Sigma) in 2 ml of 50 mM β-mercaptoethanol. Add 0.22 ml 1 M HCl and dissolve precipitated material by shaking at 37˚C. Incubate at room temperature for at least 3 hours. Adjust the concentration to approximately 15 mM by adding 0.1 M HCl to get approximately 10 ml final volume of 5:10 methenyltetrahydrofolate (5:10 cyclic THF).
Measure the final concentration using a millimolar extinction coefficient 25 at 355 nm. Disperse in Eppendorf tubes and store at -80°C.
Before use, transform 5:10 cyclic THF into its active 10-THF form by neutralizing with 1/10 volume of 1 M KOH and adding 1/50 volume of 1M Tris-HCl (pH 7.5). Incubate 15 min on ice before adding to the Charging/Formylation mixture.
Charging and formylation of tRNAfMet
Assemble the Charging / Formylation mixture in a 50 ml Falcon tube by adding the following components: 4 ml water, 10 µl RNA guard (Amersham), 1 ml 10* TMK buffer containing [10 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2], 1 ml of neutralized 10-THF, 12 μl of 14 M β-mercaptoethanol, 0.1 mg PK, 0.01 mg MK, 0.2 ml of 100 mM PEP and 0.1 ml of 100 mM ATP.
Add then tRNAfMet and [3H] methionine in a two times molar excess over tRNAfMet and adjust the final reaction volume to 10 ml. Finally, add 4000 U of MetRS and 2000 U of Formyl Methionine Transferase (FMT). FMT was purified as described in (31).
Incubate the reaction mixture at 37°C for 20 min, take 10 µl samples both into 3 ml ice cold 5% TCA and into 0.1 ml 20% formic acid at 1, 10, 15 and 20 min to follow the kinetics of charging and formylation as described below. Filter the TCA precipitate through GF/C filters (Millipore), dry, add Ready Protein scintillate, and count. The radioactivity retained on the filter corresponds to the amount of aminoacylated tRNA.
The formic acid precipitate was processed as follows: the samples were centrifuged for 15 min at 14,000 rpm (20, 800 g) in an Eppendorf centrifuge and the amount of formed fMet-Phe-tRNAPhe in the pellet was determined by hydrolyzing the pellet with 200 µl of 0.5M KOH for 10 minutes at room temperature. Then 15 µl of 100% formic acid was added and samples were centrifuged for 15 min at 14,000 rpm in Eppendorf centrifuge. Supernatants containing ([3H]fMet) were then analyzed with the HPLC using 90% water: 10% methanol mobile phase with 0.1% TFA and a C18 reverse phase column (LiChrosphere 100 RP-18 (5 µm ), Merck, Germany). At this concentration of methanol the formylated [3H]fMet separates well from [3H]Met and the purity of [3H]fMet-tRNAfMet can be determined from the areas of [3H]Met and f[3H]Met peaks.
Purification of [3H]fMet-tRNAfMet on BD Sepharose column
Note that this procedure requires BD-Sepharose, which is not available commercially. BD-Sepharose can be prepared by the procedure similar to that described in (32) with some modifications. In short, the BD cellulose drying step before the pyridine addition step in the procedure of Gillam and Tener (32) was replaced with ethanol washing followed by dimethylformamide washing to remove water (J.C. Janson, personal communication). An alternative procedure of fMet-tRNA preparation that relies entirely on commercially available products is described in (33).
Stop the charging / formylation reaction by adding 1.4 ml 4M NaCl and 1 ml 1M NaOAc and load the reaction mix directly onto a 60 ml BD-Sepharose column equilibrated with AEN240 buffer containing [20 mM NaOAc, 1 mM EDTA, 3 mM β-mercaptoethanol and 240 mM NaCl]. Wash the column with AEN550 buffer containing 550 mM NaCl instead of 240 mM until OD reaches the base line; then develop a 250 ml plus 250 ml gradient from 0.55 M to 1.6 M NaCl in AEN buffer [20 mM NaOAc, 1 mM EDTA, 3 mM β-mercaptoethanol].
Identify [3H]fMet-tRNAfMet by taking 10 µl of each fraction on filters and counting as described above. Pool fractions containing purified [3H]fMet-tRNAfMet and precipitate them by adding 1/10 volume of NaOAc (pH 5.1) followed by 2 volumes of ethanol.
Centrifuge at 4000 g for 20 min (4°C) and dissolve [3H]fMet-tRNAfMet in a minimal volume (2 ml) of PM buffer. Dialyze against 500 ml of PM buffer twice, 60 min for each step, and then check the final concentration and store at -80°C.
Purification of His-Tagged IF2
Grow the His-tagged IF2 (IF2_HT) overproducing cells BL21 (DE3) with a pAF2H plasmid (34) at 37°C in 2 L of LB medium supplemented with 0.5% glucose and 50 mg/L kanamycin with moderate shaking until OD600 = 0.7. Induce IF2_HT over expression by adding IPTG to 2 mM final concentration and continue the growth for 4 more hours. Harvest cells by centrifugation at 5,000 rpm for 40 min (GS-3 rotor, Sorvall). Re-suspend cells in 50 mM Tris-HCl (pH7.5) buffer and pellet them again in Falcon tubes by centrifugation at 4000 g for 20 min. Store at -80°C before use.
Prepare the following buffers:
TNM200 buffer [20 mM Tris-HCl (pH 8.1), 200 mM NaCl, 5 mM MgCl2]
TMNDN120 buffer: [40 mM Tris (pH 7.5), 80 mM NaCl ,40 mM NH4Cl, 5 mM MgCl2,1 mM DTE]
TMNDN70 buffer [40 mM Tris (pH 7.5), 30 mM NaCl ,40 mM NH4Cl, 5 mM MgCl2, 1 mM DTE]
Take IF2_HT cells (20 g), thaw them at room temperature and re-suspend in 20 ml of TNM200 buffer. Add 100 mM PMSF to 0.2 mM final concentration and 0.02 ml of 1 mg/ml DNAse I (Sigma). Open cells with French press (Thermo IEC, 6000 psi) and remove the cell debris by centrifugation in SS34 rotor (Sorvall) at 16,000 rpm (30,000 g) for 40 min at 4˚C.
Dilute clear supernatant to 150 ml with TNM200 buffer and load it immediately onto a C16/40 column (Pharmacia) packed filled with 20 ml Ni-CAM matrix (Sigma) and pre-equilibrated with TNM200 buffer.
After sample application wash the column with TNM200 buffer containing 3 mM imidazol until OD280 reaches base line. Elute the protein with 200 mM imidazol in TNM200 buffer. Identify IF2 containing fractions with 10% SDS PAGE and pool those containing most of IF2.
Precipitate the protein with 0.6 g/ml (NH4)2SO4. After 3 h, collect the precipitate by 40 min centrifugation in SS34 rotor (Sorvall) at 18,000 rpm (38,000 g) and dissolve the pellet in 6 ml of TMNN120 buffer. Clarify protein solution by centrifugation and apply it by gravity to a 600 ml (C26/120) Sephacryl S200 (Pharmacia) gel-filtration column equilibrated with TMNDN120 buffer. Eluted protein with the same buffer at 40 ml/h.
Use 10% SDS PAGE to identify IF2 containing fractions. Pool fractions containing most pure IF2 and dilute the solution twice with water. Apply the diluted solution directly to a small 5 ml Sepharose SP-Fast Flow column (Pharmacia) equilibrated with TMNDN70 buffer. Wash the column with 3-5 volumes of TMNN70 buffer and elute IF2 in TMNDN70 buffer with 1 M NaCl.
Precipitate the protein with 0.6 g/ml of (NH4)2SO4. After 3 h collect the precipitate by 40 min centrifugation in SS34 rotor at 18,000 rpm and dissolve the pellet in 100 ml of 20 mM Tris-HCl (pH 7.5). Load the protein solution directly onto a 18 ml Sepharose Q-FF column equilibrated with TMNDN70 and wash extensively with the same buffer. Elute IF2 in TMNDN70 buffer with 1 M NaCl and precipitate it with 60 g/ 100 ml of (NH4)2SO4.
After at least 3 h collect the precipitate by 40 min centrifugation in SS34 rotor at 18,000 rpm and dissolve the pellet in a minimum volume of TMNDN70 buffer. Transfer the solution to a dialysis bag and dialyze against PM buffer. Take the protein from dialysis bag, spin for 15 min in the Eppendorf centrifuge at 14,000 rpm (20,800 g) to get rid of dialysis precipitate and store at -80°C in 0.3 ml aliquots.
A 10% SDS PAGE of the this IF2_HT preparation in Fig. 6 shows only the band of an expected molecular weight of around 100 kD indicating that the protein should be more than 95% pure.
Fig. 6. Purity of His-Tagged IF2 (IF2_HT) as seen on 10% SDS PAGE.

Lanes 1 and 5 contain protein molecular weight marker. Lanes 2, 3, and 4 contain increasing amounts of IF2_HT.
Purification of EF-Tu
Prepare the following buffers:
TMD buffer:[40mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM DTE]
TMDK80 buffer: [40mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM DTE, 80 mM KCl]
Re-suspend 400 g of thawed E. coli MRE 600 cells in 250 ml of TMD buffer containing 1 ml of 100 mM PMSF. Add 0.2 ml of 1 mg/ml DNase I (Sigma) and break cells using the French press (at 5000-7000 psi). Add TMD buffer up to 1 L final volume and stir in the cold rum for 10-20 min. Centrifuge then at 9,000 rpm in GS3 rotor (Sorvall) for 90 min.
Collect carefully the clear supernatant and apply it directly onto a 600 ml DEAE- Sepharose CL-6B column (Amersham) pre-equilibrated with the TMD buffer at 300 ml/h. Wash the column with 1-2 column volumes of TMD and develop a 2.5 L +2.5 L gradient from 20 mM to 400 mM NaCl in TMD buffer at 300 ml/h.
Assay column fractions for PheRS/LeuRS activity. To run the assay, add 5 µl of assayed chromatographic fraction to 95 µl of charging mix containing at final concentration in the reaction mix [10 mM PEP; 1 mM ATP; 0.05 mg/ml pyruvate kunase (KP), 0.003 mg/ml myokinase (MK); 5 µM tRNAPhe, 15 µM tRNALeu, 0.2 mM [3H] Phe and 0.2 mM [14C] Leu, both with specific activity of at least 50 cpm/pmol. The charging mix should be pre-incubated at 37°C for 5 min before adding the assayed fraction. Incubate for 15 s at 37°C and precipitate charged tRNA by adding 5 ml of ice-cold 5% trichloroacetic acid (TCA), filter through GF/C filters (Millipore), dry, add 4 ml Ready Protein scintillation cocktail (Beckman Coulter, USA) and count. Run the fractions with LeuRS activity on 10% SDS PAGE. Use EF-Tu and EF-G standards to identify the corresponding bands on gel. Pool EF-Tu fractions avoiding EF-G contaminations and precipitate with 30 g of (NH4)2SO4 per 100 ml of the protein solution.
After 3 h collect the precipitate by 40 min centrifugation in SS34 rotor (Sorvall) at 18,000 rpm and re-suspend the pellet in a minimal volume of TMDK80 buffer. Transfer the slurry into a dialysis bag and dialyse for 1-2 h until all precipitate dissolves.
Clarify the dialyzed solution by 30 min centrifugation in SS34 rotor at 17,000 rpm and apply the EF-Tu solution to a 700 ml C26/120 AcA44 (Amersham) gel filtration column equilibrated with TMK80 buffer. The sample volume should not exceed 30 ml. Run the column at 30 ml/h and analyze eluted fractions by 12% SDS PAGE.
Pool major EF-Tu containing fractions and apply them directly onto a small 30 ml Q-Sepharose FF column (Amersham) equilibrated with TMK80. Develop a 400 ml +400 ml gradient from 80 mM to 400 mM NaCl in TMD buffer at 200 ml/h. Identify EF-Tu containing fractions by SDS-PAGE and precipitate the almost pure EF-Tu with 60% (NH4)2SO4.
To further purify EF-Tu, collect the precipitate by 40 min centrifugation in SS34 rotor at 18,000 rpm (38,700 g) and re-suspend the pellet in a minimal volume of TMDK80 buffer. Transfer the slurry into a dialysis bag and dialyze for 1-2 h until all precipitate dissolves. Clarify the solution. If its volume exceeds 10 ml reduce it below 10 ml using Amicon 10 kD cut off spin concentrators. Apply EF-Tu to the 700 ml C26/120 AcA44 column equilibrated with TMK80 buffer. Run the column at 30 ml/h and identify fractions containing pure EF-Tu by 12% SDS PAGE.
Pool Tu-fractions and precipitate them with 0.5g/ml of (NH4)2SO4. After at least 3 h collect the precipitate by 40 min centrifugation in SS34 rotor at 18,000 rpm (38,700 g). Dissolve the pellet in a minimal volume of PM buffer and dialyze against polymix with 3 dialyze changes. Disperse in Eppendorf tubes (0.1 ml per tube) and store at -80°C.
Preparation of MFTI-mRNA
DNA template for mRNA
Order two DNA oligoes:
Forward:(5')GGTACCGAAATTAATACGACTCACTATAGGGAATTCGGGCCCTTGTTAACAATTAAGGAGG and
Reverse:(5')TTTTTTTTTTTTTTTTTTTTTCTGCAATTAAATCGTAAACATAGTATACCTCCTTAATTGTTAACAAGGGCCCG
Assemble a PCR reaction by mixing required amount of water, 100 µl 10x PCR buffer (Promega), 20 µl each of dNTP (25 mM stock), 1000 pmol of both Forward and Rev oligoes in 1 ml water. Disperse in several PCR tubes and put in a thermo cycler.
Warm to 96°C for 5 min and anneal oligoes by cooling to 42°C. Add 10 U of Taq DNA polymerase (Promega) to each PCR tube and run 5-8 cycles: 42°C (1 min) =>72°C(1 min) =>93°C (1 min) to fill in the DNA template. Cool down to 4°C.
Extract twice with equal volume of a standard phenol/ chloroform mix and precipitate the water phase by adding 1/10 volume of 3 M NaOAc (pH 5.1) followed by 2 volumes of ethanol. After 30 min at –20°C collect the precipitate by centrifugation at 14,000 rpm in Eppendorf centrifuge for 25 min, wash pellet with 70% ethanol, dry and dissolve in 0.5 ml of water.
mRNA transcription and purification
Assemble the transcription mixture (4 ml final volume) by mixing 2 ml of water with 0.4 ml of 10* TMS buffer containing [400 mM Tris-HCl (pH7.5), 220 mM MgCl2, 10 mM spermidine]. Add then 20 µl of 1% Triton X100, 80 µl of 50 mM DTE, 160 µl of each ATP (100 mM), UTP (100 mM), CTP (100 mM), GTP (100 mM), 100 µl of DNA template. Adjust the volume to 4 ml with water and then add 40 µl T7 RNA polymerase (5 mg/ml) and 5 µl RNA guard (Amersham). Incubate at 37˚C for 3 hours.
Add 0.2 ml of 0.5 M EDTA and 0.6 ml 4 M NaCl to stop the reaction and centrifuge the mixture for 10 min at 14,000 g to pellet Mg2+:pyrophosphate precipitate. Collect supernatant and adjust its volume to 12 ml with TEN500 buffer containing [20 mM Tris-HCl (pH 7.5), 0.5 mM EDTA and 500 mM NaCl].
Apply the supernatant directly onto a small 1-2 ml oligo-dT column (Pharmacia) equilibrated with TEN500 buffer. Wash the column with TEN500 buffer until UV reaches a base line and elute mRNA with TEN0 buffer containing [20 mM Tris-HCl (pH 7.5) and 0.5 mM EDTA].
Pool mRNA fractions and precipitate by adding 1/10 volume of 3 M NaOAc (pH 5.1) followed by 2 volumes of ethanol. After at least 30 min at –20°C, collect the precipitate by centrifugation at 14,000 rpm in Eppendorf centrifuge for 30 min, wash the pellet with 70% ethanol, dry and dissolve in 0.5 ml of water. Store at -80°C before use.
Appendix 1: Theory and Applications of Rayleigh Light Scattering
Textbooks on light scattering seldom consider scattering molecules (particles) explicitly. Instead, they describe how the presence of particles affects the index of refraction of the solution (35, 36). This makes the simple relation between light scattering and molecular properties hard to grasp. In what follows, we will therefore derive the theory of light scattering with explicit reference to the scattering molecules and their properties. Light that traverses a solution will induce oscillating dipoles in the molecules that are present, and these dipoles will be secondary light sources that are responsible for the scattered light. The scattered intensity can be observed by a detector situated at an angle θ from the direction of the incident light at a distance R from the illuminated volume V. The textbook expression for the intensity, I, of scattered light that passes through a 1 cm2 of detector surface is:
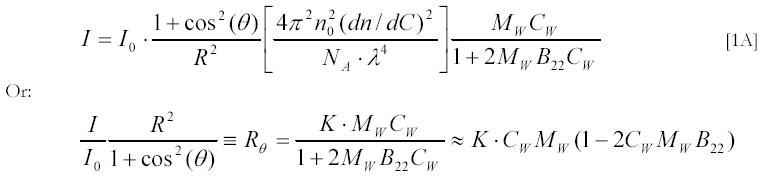
The best known variant of relation (36) is:

Here, Rθ is the so called Rayleigh ratio, n0 is the refraction index of the solvent, dn/dC is its increment by the solute, NA is the Avogadro number and K is the combination of constants in the square brackets in [1A]; MW is the molecular weight of dissolved particles, CW is their weight concentration (in g/l) and B22 is the second virial coefficient for particle-particle interactions. A semi-rigorous derivation of [1A] that relies on thermodynamic theory of solution fluctuations can be found in the second edition of van Holde's 'Physical Biochemistry' (36). The appearance of the cos2(θ) term in [1A] is due to the use of non-polarized light (36). In what follows we will derive expression [1A] starting from particles and particle properties.
Consider first two very small particles in solution at positions r1 and r2 that scatter the incident light. A geometrical consideration shows (36) that the amplitude A2 of scattered light from these two particles on the detector is:
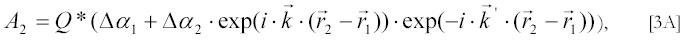
Δα1 is the polarizability of the first particle in excess over the polarizability of the solvent, Δα2 is the excess polarizability of the second particle and the exponential terms reflect the phase differences of the light scattered by the first and second particle when it arrives at the detector. The wave vector k’ of the scattered light makes an angle θ with the wave vector k of the incident light. The proportionality factor Q for the light polarized perpendicularly to both k and k' is :

where E0 is the amplitude of the incident light (36). To simplify the formulae we will in what follows omit the proportionality factor Q and re-introduce it in the final equations.
The relation [3A] can be further simplified by introducing a scattering vector s :

directed along the difference between the wave-vector k of incident and the wave-vector k' of scattered light. Note also that according to [5A] s and θ are uniquely determined by each other for monochromatic light of wave-length λ.
Omitting Q and introducing s one can re-write [3A] as:
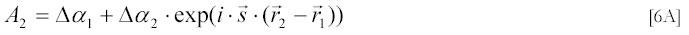
The intensity I2 of scattered light measured by the detector is the square of the module of the amplitude on the detector, i.e.
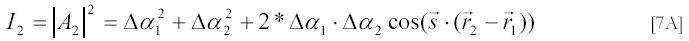
This relation shows that the intensity depends on the polarizabilities of particles 1 and 2 and also on the inter-particle distance. Since particles 1 and 2 move freely in solution, all directions of the vector r12 = r2 - r1 are equally probable and an average intensity <I2 > for a given distance r12 between the particles can be obtained by averaging [7A] for all directions of the vector r12 . This averaging can be done analytically:

This expression leads to the following fundamental relation for the intensity of scattered light:

where r is the distance between two particles in solution. This relation tells us that the intensity I2 depends on the product of s and r:
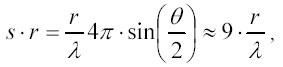
where the last estimate is valid for the scattering angle θ = 90° representative for most experimental set ups. If two particles form a tight complex the distance r would be constant. For particles with dimensions d much smaller than the light wave-length λ the product s*r is much less then 1. Therefore:

Assuming r=10 nm and λ = 400 nm one can estimate that the contribution of s-dependent terms in the Taylor expansion would not exceed 2% and for all practical purposes the scattering from the complex of two particles which are much smaller than the wave-length is:
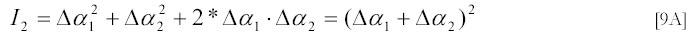
In cases when particles do not form a tight complex the scattering intensity in [8A] should be averaged for all possible separations between particles. This averaged scattering intensity is given by:

where V is the illuminated volume and P(r)/V is the probability density of finding the two particles separated by distance r; Pinf /V is this probability for very large separations. If the interactions between particles can be described by a potential U(r) of mean force (which is negative in case of attraction and vanishes to zero for large separations between particles) one can re-write the above relation as:
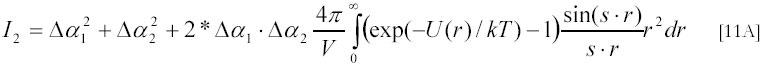
This relation shows that the s-dependence of the intensity of scattered light (or its dependence on the scattering angle θ) contains information about inter-particle interactions. In principle, the potential U(r) of this interaction, can be determined from this s-dependence (37). For solid non-interacting spherical particles of radius R (hard spheres) the above relation simplifies to:
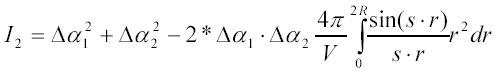
since the potential U(r) goes to + infinity for distances between particle centers less then 2*R and is zero for separations larger than 2*R:

Integrating the above expression for I2 one gets:

Here, v is the particle volume, and since v<<<V the last, "excluded volume," term in the above expression is negligible which leads to the usual expression for the scattering intensity from two non-interacting particles:

The "excluded volume" term may become important for concentrated solutions as discussed below. Comparison of [9A] and [12A] tells us that the gain of scattering intensity due to complex formation is 2*Δα1*Δα2 . Since the polarizabilities Δα of particles are proportional to their molecular weights, the excess scattering ΔIC due to complex formation will also be proportional to the product of their molecular weights.
The relation [6A] for the amplitude of scattered light from two small particles can be extended directly to the case of N such particles. The amplitude at the detector of scattered light from N particles is given by:
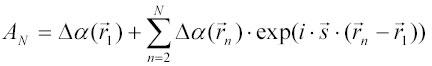
Since the amplitude can be multiplied by exp(ix) (where x may be any real number) without changing the intensity, we can multiply AN by exp(isr1) to get a more symmetrical expression for AN :

The corresponding scattering intensity, IN , becomes:
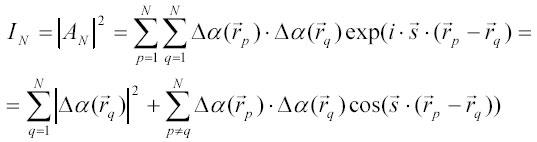
Again, one can average IN first assuming that all directions of the vector rpq = rp -rq are equally probable in solution and get by analogy with [8A]:
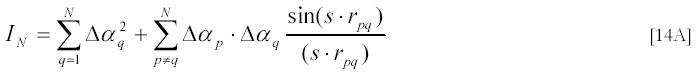
In case when N particles form a tight complex with a maximal dimension much smaller than the wave length of the incident light one can neglect the s-dependence in [14A] and re-write [14A] in the form:
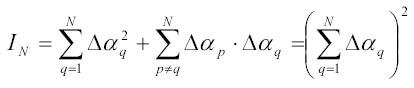
This last relation is a direct generalization of relation [9A] for small multi-component particles. It tells us that the scattering intensity is proportional to the square of particle volume (or mass) but does not depend on particle shape.
In case of solution of N particles the second term in [14A] should be averaged for all separations between particles. For the solution of N identical particles this averaging simplifies to:
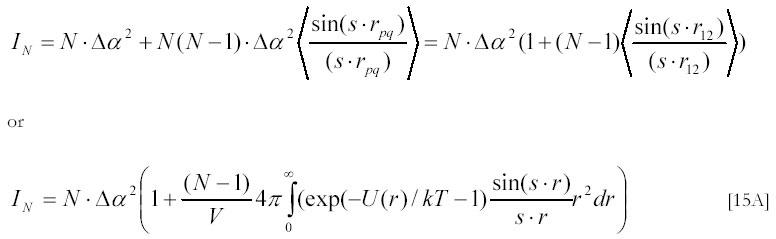
The number of particles, N, in an illuminated volume V can be expressed through their weight concentration CW (in g/l):

Neglecting 1 in comparison with N the expression [15A] can be re-written as:
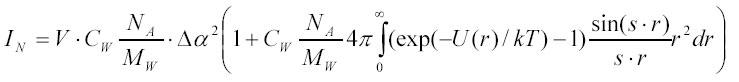
If short range interactions predominate between particles or one measures light scattering at very low angles (s ® 0) one can neglect the s-dependence in the integral and re-write the above expression as:

where the expression in square brackets was denoted as B*22 . It can be shown (37) that the so defined B*22 is proportional to a thermodynamically defined second virial coefficient B*22 (th) for particle-particle interactions in solution:
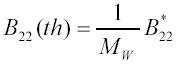
For a solution of non-interacting hard-spheres of radius R, the second virial coefficient B*22 is simply 4*V:

where v is the volume of a particle (in l) and V is the specific volume (in l/g) of the particle. In this case the product 2CW*B*22 is simply the fraction of illuminated volume occupied by particles (multiplied by 8). For a 1 µM ribosomal solution (approximately 3 mg/ml) the value of 2CW*B*22 in hard sphere approximation is less than 0.01 and can be neglected in comparison with 1 in [16A]. Light scattering of a solution of weakly interacting particles at low concentrations is therefore dominated by the first term in relations [14A] and [16A], which is the sum of scattering intensities of individual particles in solution. This justifies the use of expression [1] in the section "Kinetics of macromolecular complex formation analyzed by stopped-flow and light scattering" and the subsequent analysis of complex formation presented in that section. However, analysis of the concentration dependence of light scattering from a solution of interacting particles is one of the most straightforward approaches to determine the second virial coefficient B*22 for this interaction (38). Assuming that the excess polarizability Δα is proportional to the particle MW with the proportionality coefficient γ one can rewrite [16A] in the form:
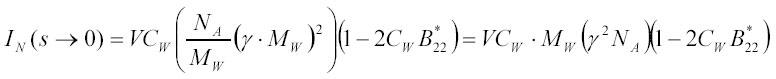
which shows explicitly that the scattering intensity is proportional to the weight concentration and to the molecular weight of a particle. This relation between the scattering and particle molecular weight is the basis for applications of light scattering for routine measurements of molecular weights with commercially available instruments (39).
After introducing the missing proportionality factor Q one can re-write the above relation in a more complete form:
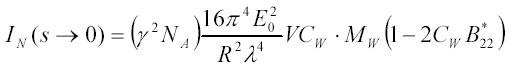
Taking into account that B*22 = MW*B22 (th) and that incident light is non-polarized (36) the above relation can be re-written in the form:
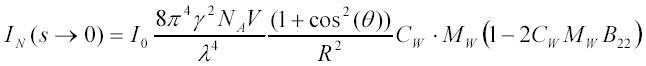
very similar to the classical relation [1A] obtained from thermodynamic considerations in (36).
References
- Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. The structure and function of initiation factors in eukaryotic protein. Cell Mol Life Sci. 2000;57:651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Shin BS, Dever TE, Burley SK. Engaging the ribosome: universal IFs of translation. Trends Biochem Sci. 2001;26:705–709. doi: 10.1016/S0968-0004(01)02024-2. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? Embo J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. Embo J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. Embo J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M, Dessen P, Pantaloni D, Godefroy-Colburn T, Wolfe AD, Dondon J. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J Mol Biol. 1975;94:461–478. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- Wishnia A, Boussert A, Graffe M, Dessen PH, Grunberg-Manago M. Kinetics of the reversible association of ribosomal subunits: stopped-flow studies of the rate law and of the effect of Mg2+. In: J Mol Biol, vol. 93. pp. 499-415; 1975: 499-415. [DOI] [PubMed]
- Blumberg BM, Nakamoto T, Kezdy FJ. Kinetics of initiation of bacterial protein synthesis. Proc Natl Acad Sci USA. 1979;76:251–255. doi: 10.1073/pnas.76.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Noll M, Hapke B, Noll H. Structural dynamics of bacterial ribosomes. II. Preparation and characterization of ribosomes and subunits active in the translation of natural messenger RNA. J Mol Biol. 1973;80:519–529. doi: 10.1016/0022-2836(73)90419-1. [DOI] [PubMed] [Google Scholar]
- Jelenc PC, Kurland CG. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci USA. 1979;76:3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Freistroffer DV, MacDougall J, Buckingham RH, Ehrenberg M. Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. Embo J. 1997;16:4134–4141. doi: 10.1093/emboj/16.13.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci USA. 1995;92:1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffientini A, Lorenzetti R, Gastaldo L, Parlett JH, Spurio R, La Teana a, Islam K. Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr Purif. 1994;5:118–124. doi: 10.1006/prep.1994.1018. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M, Bilgin N, Kurland C. Design and use of a fast and accurate in vitro translation system. In: Ribosomes and Protein Synthesis. A practical Approach. pp. 101-128. Oxford: Oxford University Press; 1990: 101-128.
- Marquardt DW. An algorithm for least squares estimation of nonlinear parameters. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- Cenatiempo Y, Deville F, Dondon J, Grunberg-Manago M, Sacerdot C, Hershey JW, Hansen HF, Petersen HU, Clark BF, Kjeldgaard M et al. The protein synthesis initiation factor 2 G-domain. Study of a functionally active C-terminal 65-kilodalton fragment of IF2 from Escherichia coli. Biochemistry. 1987;26:5070–5076. doi: 10.1021/bi00390a028. [DOI] [PubMed] [Google Scholar]
- Sacerdot C, Vachon G, Laalami S, Morel-Deville F, Cenatiempo Y, Grunberg-Manago M. Both forms of translational initiation factor IF2 (alpha and beta) are required for maximal growth of Escherichia coli. Evidence for two translational initiation codons for IF2 beta. J Mol Biol. 1992;225:67–80. doi: 10.1016/0022-2836(92)91026-l. [DOI] [PubMed] [Google Scholar]
- Tomsic J, Vitali LA, Daviter T, Savelsbergh A, Spurio R, Striebeck P, Wintermeyer W, Rodnina MV, Gualerzi CO. Late events of translation initiation in bacteria: a kinetic analysis. Embo J. 2000;19:2127–2136. doi: 10.1093/emboj/19.9.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham JL, Maaloe O, Neidhardt FC. Growth of the Bacterial Cell. Sunderland, MA 01375, USA: Sinauer Associates Inc.; 1983.
- Farewell A, Neidhardt FC. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Blanquet S. Maturation of pre-tRNA(fMet) by Escherichia coli RNase P is specified by a guanosine of the 5'-flanking sequence. J Biol Chem. 1995;270:15908–15914. doi: 10.1074/jbc.270.26.15908. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Preparation and transformation of competent Ecoli using callcium chloride. In: Molecular Cloning (A Laboratory Manual) Edited by Spedding G. pp. 116-118. New York: Cold Spring Harbor Laboratory Press; 2001: 116-118.
- Dubnoff JS, Maitra U. Isolation and properties of polypeptide chain initiation factor FII from Escherichia coli: evidence for a dual function. Proc Natl Acad Sci USA. 1971;68:318–323. doi: 10.1073/pnas.68.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh V, Gite S, Li Y, RajBhandary UL. Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: role of a 16-amino acid insertion module in initiator tRNA recognition. Proc Natl Acad Sci USA. 1997;94:13524–13529. doi: 10.1073/pnas.94.25.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam IC, Tener GM. The Use of BD-Cellulose in Separating Transfer RNAs. Methods in Enzymology. 1971;20:55–71. [Google Scholar]
- Rodnina MV, Semenkov YP, Wintermeyer W. Purification of fMet-tRNA(fMet) by fast protein liquid chromatography. Anal Biochem. 1994;219:380–381. doi: 10.1006/abio.1994.1282. [DOI] [PubMed] [Google Scholar]
- Forster AC, Weissbach H, Blacklow SC. A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Anal Biochem. 2001;297:60–70. doi: 10.1006/abio.2001.5329. [DOI] [PubMed] [Google Scholar]
- Cantor CR, Schimmel PR. Light Scattering. In: Biophysical Chemistry. pp. 838-842. New York: W.H. Freeman and Company; 1980: 838-842.
- van Holde KE. Scattering. In: Physical Biochemistry. pp. 209-224. Englewwood Cliffd, NJ 0732: Prentice-Hall; 1985: 209-224.
- Belloni L. Interacting monodisperse and polydisperse spheres. In: Neutron, X-Ray and Light Scattering Edited by Linder P, Zemb T. pp. 135-155. Amsterdam: Elsevier Science; 1991: 135-155.
- Velev OD, Kaler EW, Lenhoff AM. Protein interactions in solution characterized by light and neutron scattering: comparison of lysozyme and chymotrypsinogen. Biophys J. 1998;75:2682–2697. doi: 10.1016/S0006-3495(98)77713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]