Abstract
Genetic factors have been estimated to account for about 25% of the variation in an adult's life span. The complement component C4 with the isotypes C4A and C4B is an effector protein of the immune system, and differences in the overall C4 copy number or gene size (long C4L; short C4S) may influence the strength of the immune response and disease susceptibilities. Previously, an association between C4B copy number and life span was reported for Hungarians and Icelanders, where the C4B*Q0 genotype, which is defined by C4B gene deficiency, showed a decrease in frequency with age. Additionally, one of the studies indicated that a low C4B copy number might be a genetic trait that is manifested only in the presence of the environmental risk factor “smoking”. These observations prompted us to investigate the role of the C4 alleles in our large German longevity sample (∼700 cases; 94–110 years and ∼900 younger controls). No significant differences in the number of C4A, C4B and C4S were detected. Besides, the C4B*Q0 carrier state did not decrease with age, irrespective of smoking as an interacting variable. However, for C4L*Q0 a significantly different carrier frequency was observed in the cases compared with controls (cases: 5.08%; controls: 9.12%; p = 0.003). In a replication sample of 714 German cases (91–108 years) and 890 controls this result was not replicated (p = 0.14) although a similar trend of decreased C4L*Q0 carrier frequency in cases was visible (cases: 7.84%; controls: 10.00%).
Introduction
Human longevity is considered a multi-factorial phenotype, and genetic factors have been estimated to account for about 25% of the variation in adult life span [1], [2], [3], [4], [5]. Nonagenarians and centenarians have outlived the vast majority of their peers by many decades. Most of them have spent their life in good health and often markedly delay or even escape major age-related diseases [6], [7]. It has been postulated that these long-lived individuals (LLI) carry a reduced number of risk alleles for age-related diseases [8], [9]. A case in point is the apolipoprotein E*ε4 allele (risk factor for Alzheimer's and coronary heart disease) that is significantly depleted in LLI [1].
Complement C4 is a central component of the mannose-binding lectin activation pathways of the complement system that are main effectors of the adaptive and innate immune responses [10], [11], [12]. The C4 protein exists as two isotypes, the acidic C4A and basic C4B form. They are encoded by the C4A and C4B genes located in the major histocompatibility complex (MHC) class III region. C4A and C4B differ from each other only at five single nucleotide polymorphisms (SNPs) [13], [14]. Although the resulting isotypes are antigenetically quite similar (>99% amino acid sequence identity), they exhibit functional differences, for instance in chemical reactivities to substrates [15], [16], [17], [18], [19]. Furthermore, it is thought that the C4A protein is more important in immunoclearance, whereas C4B is more relevant in the defense against microbes [17].
Both C4A and C4B genes are found with varying copy numbers (CNVs), as indicated by the Database of Genomic Variants (http://projects.tcag.ca/variation) [15]. The total number of C4A and C4B genes can range between two and eight (considering homologous chromosomes). While most healthy subjects carry two C4A and two C4B genes each [15], [16], [17], heterozygous deficiency of C4A or C4B has been described in up to 30% of populations of European descent [20]. C4B gene deficiency (zero or one C4B gene in the diploid genome) is traditionally called C4B*Q0 and the corresponding situation for C4A is denoted as C4A*Q0 [12], [15]. In addition, both C4 genes differ in size; they can be short (C4S) or long (C4L; due to the insertion of 6.4 kb from the endogenous retrovirus HERV-K) [21]. In Caucasians about 76% of the C4 genes belong to the long form and 24% to the short form [18], [20]. C4L/C4S gene deficiency is called C4S*Q0 or C4L*Q0, respectively. The copy number and the size of C4 genes strongly determine the plasma C4 protein concentrations and linear correlations between total C4, C4A and C4B gene copy number with their corresponding plasma protein concentration were observed [22]. For instance, individuals containing one or more short C4 genes have consistently higher serum total C4 concentrations than those with long C4 genes only, suggesting a negative epistatic effect of HERV-K retrovirus on the expression of C4 proteins [18].
The frequent variations in C4 size and numbers render the gene an interesting marker for major histocompatibility complex disease associations [23], [24], [25], [26]. Expression and overall gene copy numbers of total C4, C4A, C4B or the C4 gene size (C4S or C4L) may influence the strengths of innate or adaptive immunity and disease susceptibilities [15], [16], [18], [27], [28], [29]. A low C4B copy number has been shown to be a risk factor for cardiovascular diseases [15], [30], [31], [32], [33], [34], [35]. Furthermore, a study by Kramer et al. revealed that a low C4B allele frequency is associated with a shortened life span. The authors observed a significant decrease in C4B*Q0 alleles among healthy old Hungarians (>62 years) compared to a younger (<53 years) control group (from 18% allele frequency in the young, which equals 36% carrier frequency; to 5% allele frequency in the elderly, corresponding to 10% carrier frequency) [36]. In a follow-up study in 1991, Kramer et al. supported their previous findings. They detected a marked decrease in the C4B*Q0 allele frequency (p<0.0001) in a second sample of old Hungarian study participants (>60 years: 5%; equals 10% carrier frequency) compared to younger individuals (<45 years: 16%; equals 32% carrier frequency). To test if this observation was population-specific for the two investigated Hungarian samples, a second collaborative experiment in 381 healthy Icelandic people, including 73 healthy elderly individuals (>59 years) was performed. In this independent replication sample, a significant decrease in the C4B*Q0 carrier frequency was found in the elderly group from 24% to 5% [37]. In a subsequent Icelandic follow-up study, however, no statistically significant age-associated decrease was detected for the C4B*Q0 carrier frequency [30]. Because smoking severely affects health and survival on a global scale [38], [39], the authors tested whether the increased risk of early morbidity observed in carriers of C4B*Q0 could be associated with smoking as an interacting variable. After dividing the whole Icelandic sample into smokers and non-smokers, a significant decrease in C4B*Q0 carrier frequency with age (17–39 years: 19%, 40–49 years: 20%, 50–59 years: 4%, 60–93 years: 0%; χ2 test for trend; p = 0.0079) was seen only among smokers, indicating that C4B low copy number might be a genetic trait that is manifested only in the presence of the environmental risk factor “smoking” [30]. All data on C4B copy number variation in Hungarians and Icelanders are summarized in Szilagyi et al. 2008 [15]. In an Italian study sample of 77 centenarians (100–107 years), 89 elderly subjects (70–89 years) and 235 young subjects (18–49 years) the negative influence of C4B*Q0 alleles on life span was not confirmed [40].
Here, we employed our large study sample of German individuals to i) investigate the role of C4 copy number variation in human longevity, to ii) potentially validate the association of C4B*Q0 carrier state with life span and to iii) test the hypothesis of “smoking” as an effecting variable. About 700 LLI between 94 and 110 years of age and approximately 900 younger individuals (19–75 years) have been analyzed for the C4A, C4B, C4S and C4L allele distribution. To evaluate whether differences in C4B*Q0 carrier frequency could be detected in smokers, we additionally performed a stratified analysis for smokers and non-smokers.
Materials and Methods
Study participants
In this study, we investigated ∼700 LLI (male/female ratio approximately 1/1; age-range 94–110 years) and ∼900 younger controls (male/female ratio approximately 1/1; age-range 19–75 years) drawn from German population-based collections [41], [42] for C4 copy number variation. The proportion of recent smokers was a little above 3% for LLI and a little below 40% for controls. A detailed description of the samples and the recruitment procedure is given elsewhere [43] and in Supplementary Table S1a and S1b in File S1.
The sample used to investigate the C4L alleles in the replication experiment comprised 714 LLI (male/female ratio approximately 1/10; age-range 91–108 years) and 890 controls (male/female ratio approximately 1/10; age-range 18–79), for details see Supplementary Table S1a and S1b in File S1.
All participants gave written informed consent to participate within the study. Approval for the project was obtained by the Ethics Committee of the Medical Faculty of the Christian-Albrechts-University, Kiel.
Genotyping
Genotyping of C4 copy numbers (determination of C4A, C4B, C4L, and C4S dosage) was performed using the TaqMan chemistry-based real-time PCR technique (C4A, Hs07226349; C4B, Hs07226350; C4L, Hs07226352; C4S, Hs07226351) (Life Technologies Corporation, Foster City, CA) [16]. For each sample four replicates were genotyped and samples with data for fewer than three replicates were excluded from further analyses. All copy number data are listed in Data S1.
Statistical analyses
The copy number calculation was performed using the CopyCallerTM Software (Life Technologies Corporation, Foster City, CA). The copy number assignment was cross-checked internally for each individual as the sum of C4A copies plus C4B copies should equal the sum of C4L copies plus C4S copies. If the equality was violated, the corresponding individual was excluded from the analyses. Further quality control for each CNV calling included: a) removal of all samples with analysed replicate number 0, 1 or 2; b) removal of all samples with confidence <0.95 and |CNpredicted-CNcalculated| >0.3.; c) removal of all samples with z score ≥2.65 and d) removal of all samples with 2.65>z score ≥1.75 and |CNpredicted-CNcalculated| >0.3. (criteria for quality control were adopted from the CopyCaller Software User Guide (PN 4400042B)). Sample sizes before and after quality control are given in Supplementary Table S1a and S1b in File S1.
Allele and carrier frequencies were compared between long-lived cases and younger controls by Fisher's exact test. For the comparison of the different age subgroups, the Armitage trend test was applied. To evaluate whether differences in C4B*Q0 carrier frequency were detected in smokers, we performed a stratified analysis for smokers and non-smokers. For C4A, C4B, C4L, and C4S*Q0 carrier frequencies, we additionally applied a sex-stratified analysis.
Primary endpoint of the initial study was the comparison of the C4B*Q0 carrier frequency between LLI and controls, possibly after stratifying for smoking. Secondary endpoints were the C4A, C4L and C4S*Q0 carrier frequency differences between cases and controls and the comparison along three to five age subgroup by a trend analysis. For the replication sample, the C4L*Q0 carrier frequency comparison between LLI and controls served as primary endpoint while the trend analysis over four age subgroups again was the secondary endpoint.
The data of the initial and replication sample for the analysis of C4L*Q0 carrier frequency were combined by mega analysis: We performed a logistic regression with case-control status as outcome and C4L*Q0 carrier status and study (initial or replication) as influential variables.
Sample size and power calculations were performed with the statistics program ‘BiAS. für Windows’ version 8.03 (http://www.bias-online.de/). For the other statistical calculations the statistics program R was used [44].
Registration of smoking habits
Smoking behavior was registered at study entry. Due to missing or indistinct information of smoking habits altogether 490 study participants needed to be excluded from the stratified analysis for smoking. A reported smoking status was available for 672 individuals of the LLI and for 503 controls. Current smokers and recent ex-smokers (quit <3 years ago) were combined as ‘smokers’ and compared to ‘non-smokers’ (never smokers and quit ≥3 years ago). Only cigarette smokers were considered in the analysis. More information about smoking is given in Supplementary Table S1a in File S1.
Results
The power of our study for replicating the difference in C4B*Q0 carrier frequency between the elderly and younger controls, as seen in Hungarians and Icelandic people [36], [37], [45], was calculated to be 100% for the effects in each of the three studies.
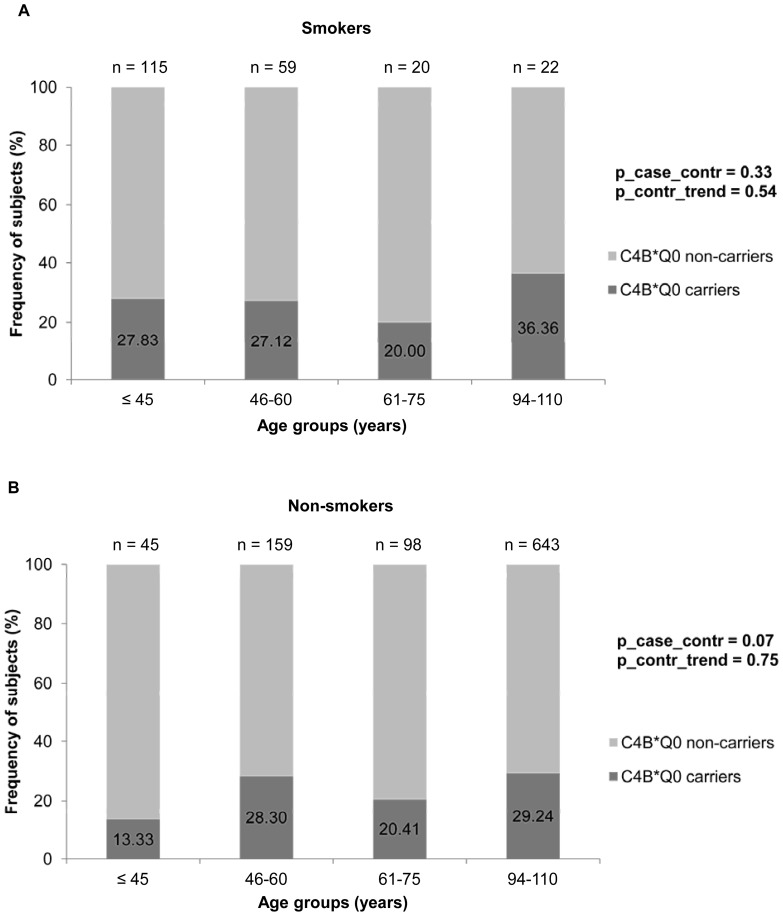
The frequencies for C4B copy numbers are given in Table 1 (for C4A, C4S and C4L see Supplementary Table S2 in File S1). They were similar to those previously described for populations of European descent [17], [37]. No difference in frequency distribution was found between long-lived cases and younger controls (p = 0.32). For the primary endpoint C4B*Q0 carrier state (zero or one C4B gene in the diploid genome), no difference was found between LLI and the whole group of younger controls (carrier frequency LLI: 28.57%, carrier frequency controls: 25.43%, p = 0.16; Table 2). Due to the wide age-range from 19 to 110 years, the study sample was additionally divided into five different subgroups (secondary endpoint analysis) (≤45 years, 46–60 years, 61–75 years, 94–100 years and >100 years). However, no increasing or decreasing trend in C4B*Q0 carrier frequency was observed for the five age subgroups (p value for trend in the control subgroups: 0.85, p value for the comparison of the two long-lived subgroups: 0.93, p value for trend over all five age subgroups: 0.17) (Table 2). The frequencies varied from 23.08% to 28.81% between different age groups without a trend for age (Figure 1). Since genetic variation may potentially influence the longevity phenotype in men and women differently [46], [47], we also performed a sex-stratified analysis. Again, no significant differences between age groups were detected (data not shown). To evaluate whether differences in C4B*Q0 carrier frequencies would be evident in smokers (as previously described in the study of Szilagyi et al. 2008), we performed a stratified analysis for smokers and non-smokers, but no significant differences were observed (Figure 2). Furthermore, we changed our age grouping such that it was concordant with that of Kramer et al. 1989 [36] and Arason et al. 2003 [37] (Supplementary Table S3 in File S1). Still, no significant difference was detected.
Table 1. Frequencies of C4B copy numbers (absolute numbers are in parentheses).
| number of C4B genes | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | p value | |
| German sample | cases (LLI); n = 728 | 2.34% (17) | 26.24% (191) | 65.38% (476) | 5.49% (40) | 0.27% (2) | 0.14% (1) | 0% (0) | 0.14% (1) | |
| controls; n = 920 | 2.28% (21) | 23.15% (213) | 70.00% (644) | 4.35% (40) | 0.22% (2) | 0% (0) | 0% (0) | 0% (0) | 0.32 | |
| European Americans* | females; n = 385 | 2.3% (9) | 25.7% (99) | 64.9% (250) | 6.8% (26) | 0.3% (1)** | ||||
| males; n = 128 | 3.9% (5) | 30.5% (39) | 58.6% (75) | 7.0% (9) | 0% (0)** |
p value: p value for the comparison of allele frequencies between long-lived cases and younger controls (Fisher's exact test).
adapted from [17]; European American female and male controls.
number of C4B genes ≥4.
Table 2. Results of the comparison of Q0 carrier state for the C4B gene (absolute numbers are in parentheses).
| age groups | ≤45 y; n = 299 | 46–60 y; n = 361 | 61–75 y; n = 260 | 94–100 y; n = 275 | >100 y; n = 453 | p value |
| C4B*Q0 carriers | 23.08% (69) | 28.81% (104) | 23.46% (61) | 28.36 (78) | 28.70% (130) | p_contr_trend = 0.85; p_case = 0.93; p_trend = 0.17 |
| 25.43% (234; ≤45–75 y) | 28.57% (208; 94–>100 y) | p_case_contr = 0.16 | ||||
p_contr_trend: p value for trend test of C4B*Q0 frequencies in the three control subgroups (Armitage trend test; secondary endpoint).
p_case: p value for the comparison of C4B*Q0 frequencies in the two case subgroups (Fisher's exact test; secondary endpoint).
p_trend: p value for trend test of C4B*Q0 frequencies in all five age groups (Armitage trend test; secondary endpoint).
p_case_contr: p value for the comparison of C4B*Q0 frequencies between long-lived cases (whole sample) and all younger controls (Fisher's exact test; primary endpoint).
n = number; y = years.
Figure 1. Frequencies of C4B*Q0 carriers (with zero or one C4B gene in the diploid genome) in healthy German individuals of different age-groups.
p_case_contr: p value for the comparison of C4B*Q0 carrier frequencies between long-lived cases and younger controls (Fisher's exact test; primary endpoint). p_contr_trend: p value for trend test of C4B*Q0 carrier frequencies in the three control subgroups (Armitage trend test; secondary endpoint). p_case: p value for the comparison of C4B*Q0 frequencies in the two case subgroups (Fisher's exact test; secondary endpoint). p_trend: p value for trend test of C4B*Q0 frequencies in all five age groups (Armitage trend test; secondary endpoint).
Figure 2. Frequencies of C4B*Q0 carriers in healthy German individuals of different age-groups stratified for (a) smokers (current smokers and quitters <3 years) and (b) non-smokers (never smokers and quitters for ≥3 years).
For abbreviations see legend to Figure 1. Due to the small number of centenarians in the replication sample we did not subdivide the case sample into a nonagenarian and centenarian subgroup.
The investigation of C4A and C4S copy numbers did not yield a significant difference between age groups (data not shown). In the C4L analysis, however, C4L*Q0 demonstrated a different carrier frequency between long-lived cases and younger controls (cases 5.08%, controls 9.12%; p = 0.003, p value for trend in the control subgroups: 0.52, p value for the comparison of the two case subgroups: 0.85, p value for trend over all five age subgroups: 0.004) (Table 3 and Supplementary Table S4 in File S1). The difference between LLI and controls did not depend on gender or smoking status (data not shown). We investigated an additional sample of 714 German LLI and 890 younger controls, which had a power of 86% to replicate our significant results for C4L*Q0. Our primary endpoint analysis revealed a decrease in the C4L*Q0 carrier frequency with age (cases 7.84%, controls 10.00%). Those differences were not statistically significant (p = 0.14), but a significant trend was detected in our secondary endpoint analysis, when the sample was divided into four different age subgroups (≤45 years, 46–60 years, 61–79 years and >91 years, p = 0.03) (Table 4 and Supplementary Table S5 in File S1). When the case-control groups were defined with the same age ranges as the initial screening sample (94–108 and 61–75 years), only marginally different results were obtained (see Supplementary Table S6 in File S1). An additional analysis for males and females separately revealed similar results (data not shown). When the data of the initial and replication sample were combined as a mega analysis (a meta analysis using the original data sets), no significant influence of study (initial or replication) or interaction with C4L*Q0 carrier status was found. However, C4L*Q0 carrier frequency was significantly different between the combined cases and controls (p = 0.002).
Table 3. Results of the comparison of Q0 carrier state for the C4L gene (absolute numbers are in parentheses).
| age groups | ≤45 y; n = 294 | 46–60 y; n = 351 | 61–75 y; n = 254 | 94–100 y; n = 233 | >100 y; n = 416 | p value |
| C4L*Q0 carriers | 9.86% (29) | 9.12% (32) | 8.27% (21) | 4.72 (11) | 5.29% (22) | p_contr_trend = 0.52; p_case = 0.85; p_trend = 0.004 |
| 9.12% (82; ≤45–75 y) | 5.08% (33; 94–>100 y) | p_case_contr = 0.003 | ||||
For abbreviations see legend to Table 2.
Table 4. Replication study in additional German sample: Results of the comparison of Q0 carrier state for the C4L gene (absolute numbers are in parentheses).
| age groups | ≤45 y; n = 344 | 46–60 y; n = 248 | 61–79 y; n = 298 | 91–108 y; n = 714* | p value |
| C4L * Q0 carriers | 11.92% (41) | 10.08% (25) | 7.72% (23) | 7.84% (56) | p_contr_trend = 0.08; p_trend = 0.03 |
| 10.00% (89; ≤45–79 y) | p_case_contr = .14 | ||||
For abbreviations see legend to Table 2.
p_trend: p value for trend test of C4B*Q0 frequencies in all four age groups (Armitage trend test; secondary endpoint).
Due to the small number of centenarians in the replication sample we did not subdivide the case sample into a nonagenarian and centenarian subgroup.
Discussion
The C4 gene size (C4S or C4L) or the copy numbers of total C4, C4A, C4B genes may influence disease susceptibilities or the immune response [15], [16], [18], [27], [28]. As carriers of unfavorable genetic variants are affected by higher mortality, they are expected to decrease in frequency in population strata of increasing age [8], [9]. Previously, an association between C4B copy number and life span was reported for Hungarians [36], [45] showing a decrease of the C4B*Q0 carrier frequency with age from 32% to 10% [45]. This observation indicated that individuals with a low C4B copy number could be selected out from the population due to their increased disease mortality [15]. A replication experiment in an independent Icelandic sample also showed a low C4B*Q0 carrier frequency of 5% in the elderly [37]. However, in an Icelandic follow-up study [30], a significant decrease with age was detected for smokers only. In other samples from Italy, no statistically significant differences in C4B*Q0 allele frequencies were observed [40]. The last four studies had a power above 99% to replicate the original C4B*Q0 association [36], irrespective of any interacting effect of smoking. It has to be taken into account, however, that overestimation of effect sizes is a common phenomenon in discovery samples, which may cause subsequent studies to be underpowered [48].
The previous inconsistent association observations prompted us to investigate the role of C4 copy number variation in our large German longevity sample, taking into consideration the smoking status. No significant frequency differences for C4A, C4B and C4S alleles and no influence of smoking status or gender were detected. In the German sample, the C4B*Q0 carrier frequency for smokers and non-smokers varied from 13.33% to 36.36% for the different age groups without any correlation with age (Figure 2).
For a better comparison with the previous studies, we also subdivided our cases and controls into five age subgroups. Again, no consistent trend for increasing age and no threshold for lower or higher C4B*Q0 carrier frequencies was seen. In particular, the frequencies between nonagenarians and centenarians were very similar.
Considering that our study had a power of 100% to replicate the previous findings, our results suggest that in Germans, the C4B*Q0 copy number plays no role in the ability to reach old age and that smoking does not influence the C4B*Q0 status across age groups as an interacting variable. However, one has to consider that smoking behaviour in prevalence and quality is known to have changed over time during the 20th century [49], [50] and can also vary between different populations. This may result in “smoking” meaning different things in different age groups and populations leading to inconsistent findings. It has also to be taken into account that life expectancy slightly differs between the three investigated populations (Hungary 71.4 years, Germany 77.4 years, Iceland 79.4 years, life expectancies averaged over both sexes according to the U.S. Census Bureau's International Data Base; www.census.gov/). Besides, the negative findings in Germans may be due to population-specific effects, since longevity in different populations is likely to be influenced by varying sets of interacting genetic and environmental factors [51]. With respect to the current investigation, all populations analyzed so far have been of European ancestry and are similar in their C4 gene variation [20], [22].
Moreover, our German sample was more extensive and the 700 German LLI much older (94–110 years) compared to the Hungarian (60–90 years) and Icelandic (60–93 years) old-age groups. The elderly Hungarians and Icelanders, which showed a significant decrease in the C4B*Q0 allele frequency, just comprised 482, 131, 58 and 25 individuals, respectively [30], [36], [37], [45]. These groups might therefore be prone to a large sampling variance or could represent biased samples that do not yield adequately precise estimates for the intended purposes [52]. This assumption is supported by the observation that deviations in C4B*Q0 frequencies – but without a decreasing trend with age – were seen for those age groups of German smokers and non-smokers that included only very few individuals (smokers: 61–75 years, n number of centenarians in the 20, 20.00%; 94–110 years, n number of centenarians in the 22, 36.36%; Figure 2a), (non-smokers: ≤45 years, n number of centenarians in the 45, 13.33%; 61–75 years, n number of centenarians in the 98, 20.41%; Figure 2b). Overall, our data confirm the negative association result of the Italian centenarian study [40] and suggest that C4B*Q0 copy number does not influence survival into old age.
For C4L, independent of gender or smoking status, a significantly lower C4L*Q0 carrier frequency was observed for the long-lived cases compared to younger controls, showing only marginal frequency differences between the three control subgroups on the one hand and between nonagenarians and centenarians on the other. However, our significant case-control association result for the C4L*Q0 allele as primary endpoint could not be replicated in a second German longevity sample, although the same trend towards a decrease in the frequency of C4L*Q0 carriers with age was observed together with a significant trend over four different age groups. Interestingly, the age groups ≤45 years and between 46 and 60 years show a similar C4L*Q0 carrier frequency of around 10 to 12% in contrast to a frequency of around 7 to 8% in the age groups 61 to 79 years and above 91 years (due to the small number of centenarians in the replication sample we did not divide the case sample into a nonagenarian and centenarian subgroup). For the C4L*Q0 replication experiment, a very large analysis population was investigated (1604 individuals), which had a power of 86% to replicate our initial finding. However, it should be mentioned that very often the detected effect size in the first pilot study is considerably overestimated and therefore needs to be adjusted in subsequent replications [48]. Regarding the observed frequency difference for C4L*Q0 carriers in the replication sample (elderly 7.84%; young 10.00%), a sample size of 2825 old-aged cases (with a case-control ratio of 1) would be required for 80% power. Hence, for validation it may be necessary to enlarge sample sizes in future studies or to perform meta-analyses across different longevity populations [53].
To the best of our knowledge, little is known about the association of C4L or C4L*Q0 with disease or other phenotypes. The C4L variant was shown to be associated with less effective gene transcription compared to the C4S variant, raising the possibility of a negative epistatic effect on the expression of C4 proteins [18], [22], [54]. Consequently, the function of the classical complement pathway may affect, via a changed equilibrium or reduction of C4 gene products, complement-dependent pathological processes and hence might influence immunity and lifespan [45]. As the presence of long C4 genes correlates with lower plasma C4 protein concentrations [18], [22], we hypothesize that C4L deficiency in our LLI sample could result in a higher and more balanced C4 protein expression leading to improved health and prolonged life span.
With altogether more than 3000 individuals investigated our study presents the largest C4 copy number investigation in human longevity research to-date. Thus, our analysis results offer a valuable reference point for further genetic studies and help provide a balanced view of the obtained research evidences.
Supporting Information
Combined file of supporting tables. Supplementary Table S1a: Characteristics of samples. Supplementary Table S1b: Quality control of C4A, C4B, C4S and C4L. Supplementary Table S2: Frequencies of C4A, C4S and C4L copy numbers. Supplementary Table S3: Results of the comparison of Q0 carrier state for the C4B gene with age groups as in Kramer et al. 1989 and Arason et al. 2003. Supplementary Table S4: Results of the comparison of Q0 carrier state for the C4L gene (carrier and non-carrier information). Supplementary Table S5: Results Replication study in additional German sample: Results of the comparison of Q0 carrier state for the C4L gene (carrier and non-carrier information). Supplementary Table S6: Replication study in additional German sample: Results of the comparison of Q0 carrier state for the C4L gene with age groups as in the initial study.
(PDF)
Data of copy number calculations performed using the CopyCallerTM Software (Applied Biosystems, Foster City, California, USA) for C4A, C4B, C4S, C4L and C4L Replication.
(XLSX)
Acknowledgments
We thank all the study participants for their cooperation and the laboratory personnel of the Institute of Clinical Molecular Biology and the members of the Popgen Biobank for excellent technical assistance.
Funding Statement
This study was supported by the RESOLVE project (FP7-HEALTH-F4-2008-202047), the Excellence Cluster ‘Inflammation at Interfaces’ and the INTERREG 4 A programme Syddanmark-Schleswig-K.E.R.N (with EU funds from the European Regional Development Fund). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Christensen K, Johnson TE, Vaupel JW (2006) The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet 7: 436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, et al. (1996) The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet 97: 319–323. [DOI] [PubMed] [Google Scholar]
- 3. Ljungquist B, Berg S, Lanke J, McClearn GE, Pedersen NL (1998) The effect of genetic factors for longevity: a comparison of identical and fraternal twins in the Swedish Twin Registry. J Gerontol A Biol Sci Med Sci 53: M441–446. [DOI] [PubMed] [Google Scholar]
- 4. Skytthe A, Pedersen NL, Kaprio J, Stazi MA, Hjelmborg JV, et al. (2003) Longevity studies in GenomEUtwin. Twin Res 6: 448–454. [DOI] [PubMed] [Google Scholar]
- 5. Hjelmborg JvB, Iachine I, Skytthe A, Vaupel JW, McGue M, et al. (2006) Genetic influence on human lifespan and longevity. Hum Genet 119: 312–321. [DOI] [PubMed] [Google Scholar]
- 6. Evert J, Lawler E, Bogan H, Perls T (2003) Morbidity profiles of centenarians: survivors, delayers, and escapers. J Gerontol A Biol Sci Med Sci 58: 232–237. [DOI] [PubMed] [Google Scholar]
- 7. Hitt R, Young-Xu Y, Silver M, Perls T (1999) Centenarians: the older you get, the healthier you have been. Lancet 354: 652. [DOI] [PubMed] [Google Scholar]
- 8. Perls T, Kunkel LM, Puca AA (2002) The genetics of exceptional human longevity. J Am Geriatr Soc 50: 359–368. [DOI] [PubMed] [Google Scholar]
- 9. Flachsbart F, Caliebe A, Nothnagel M, Kleindorp R, Nikolaus S, et al. (2009) Depletion of potential A2M risk haplotype for Alzheimer's disease in long-lived individuals. Eur J Hum Genet 18: 59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carroll MC (1998) The role of complement and complement receptors in induction and regulation of immunity. Annu Rev Immunol 16: 545–568. [DOI] [PubMed] [Google Scholar]
- 11. Walport MJ (2001) Complement. First of two parts. N Engl J Med 344: 1058–1066. [DOI] [PubMed] [Google Scholar]
- 12. Yu CY, Chung EK, Yang Y, Blanchong CA, Jacobsen N, et al. (2003) Dancing with complement C4 and the RP-C4-CYP21-TNX (RCCX) modules of the major histocompatibility complex. Prog Nucleic Acid Res Mol Biol 75: 217–292. [DOI] [PubMed] [Google Scholar]
- 13. Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, et al. (2001) Genetic, structural and functional diversities of human complement components C4A and C4B and their mouse homologues, Slp and C4. Int Immunopharmacol 1: 365–392. [DOI] [PubMed] [Google Scholar]
- 14. Hui J, Oka A, Tomizawa M, Tay GK, Kulski JK, et al. (2004) Identification of two new C4 alleles by DNA sequencing and evidence for a historical recombination of serologically defined C4A and C4B alleles. Tissue Antigens 63: 263–269. [DOI] [PubMed] [Google Scholar]
- 15. Szilagyi A, Fust G (2008) Diseases associated with the low copy number of the C4B gene encoding C4, the fourth component of complement. Cytogenet Genome Res 123: 118–130. [DOI] [PubMed] [Google Scholar]
- 16. Wu YL, Savelli SL, Yang Y, Zhou B, Rovin BH, et al. (2007) Sensitive and specific real-time polymerase chain reaction assays to accurately determine copy number variations (CNVs) of human complement C4A, C4B, C4-long, C4-short, and RCCX modules: elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J Immunol 179: 3012–3025. [DOI] [PubMed] [Google Scholar]
- 17. Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, et al. (2007) Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 80: 1037–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Y, Chung EK, Zhou B, Blanchong CA, Yu CY, et al. (2003) Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J Immunol 171: 2734–2745. [DOI] [PubMed] [Google Scholar]
- 19. Yu CY, Campbell RD (1987) Definitive RFLPs to distinguish between the human complement C4A/C4B isotypes and the major Rodgers/Chido determinants: application to the study of C4 null alleles. Immunogenetics 25: 383–390. [DOI] [PubMed] [Google Scholar]
- 20. Yu CY, Whitacre CC (2004) Sex, MHC and complement C4 in autoimmune diseases. Trends Immunol 25: 694–699. [DOI] [PubMed] [Google Scholar]
- 21. Mack M, Bender K, Schneider PM (2004) Detection of retroviral antisense transcripts and promoter activity of the HERV-K(C4) insertion in the MHC class III region. Immunogenetics 56: 321–332. [DOI] [PubMed] [Google Scholar]
- 22. Saxena K, Kitzmiller KJ, Wu YL, Zhou B, Esack N, et al. (2009) Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: A comparison of Asian-Indian and European American populations. Mol Immunol 46: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chung EK, Yang Y, Rennebohm RM, Lokki ML, Higgins GC, et al. (2002) Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am J Hum Genet 71: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, et al. (1994) The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics 40: 425–436. [DOI] [PubMed] [Google Scholar]
- 25. Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY (1999) Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem 274: 12147–12156. [DOI] [PubMed] [Google Scholar]
- 26. Yu CY (1991) The complete exon-intron structure of a human complement component C4A gene. DNA sequences, polymorphism, and linkage to the 21-hydroxylase gene. J Immunol 146: 1057–1066. [PubMed] [Google Scholar]
- 27. Bishof NA, Welch TR, Beischel LS (1990) C4B deficiency: a risk factor for bacteremia with encapsulated organisms. J Infect Dis 162: 248–250. [DOI] [PubMed] [Google Scholar]
- 28. Fielder AH, Walport MJ, Batchelor JR, Rynes RI, Black CM, et al. (1983) Family study of the major histocompatibility complex in patients with systemic lupus erythematosus: importance of null alleles of C4A and C4B in determining disease susceptibility. Br Med J (Clin Res Ed) 286: 425–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang X, Sun J, Gao Y, Tan A, Zhang H, et al. (2012) Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects. PLoS Genet 8: e1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arason GJ, Kramer J, Blasko B, Kolka R, Thorbjornsdottir P, et al. (2007) Smoking and a complement gene polymorphism interact in promoting cardiovascular disease morbidity and mortality. Clin Exp Immunol 149: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blasko B, Kolka R, Thorbjornsdottir P, Sigurdarson ST, Sigurdsson G, et al. (2008) Low complement C4B gene copy number predicts short-term mortality after acute myocardial infarction. Int Immunol 20: 31–37. [DOI] [PubMed] [Google Scholar]
- 32. Kramer J, Harcos P, Prohaszka Z, Horvath L, Karadi I, et al. (2000) Frequencies of certain complement protein alleles and serum levels of anti-heat-shock protein antibodies in cerebrovascular diseases. Stroke 31: 2648–2652. [DOI] [PubMed] [Google Scholar]
- 33. Kramer J, Rajczy K, Hegyi L, Fulop T, Mohacsi A, et al. (1994) C4B*Q0 allotype as risk factor for myocardial infarction. BMJ 309: 313–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizuno H, Sato H, Sakata Y, Ohnishi Y, Hishida E, et al. (2006) Impact of atherosclerosis-related gene polymorphisms on mortality and recurrent events after myocardial infarction. Atherosclerosis 185: 400–405. [DOI] [PubMed] [Google Scholar]
- 35. Szalai C, Fust G, Duba J, Kramer J, Romics L, et al. (2002) Association of polymorphisms and allelic combinations in the tumour necrosis factor-alpha-complement MHC region with coronary artery disease. J Med Genet 39: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kramer J, Rajczy K, Fust G (1989) Low incidence of null alleles of the fourth component of complement (C4) in elderly people. Immunol Lett 20: 83–85. [DOI] [PubMed] [Google Scholar]
- 37. Arason GJ, Bodvarsson S, Sigurdarson ST, Sigurdsson G, Thorgeirsson G, et al. (2003) An age-associated decrease in the frequency of C4B*Q0 indicates that null alleles of complement may affect health or survival. Ann N Y Acad Sci 1010: 496–99. [DOI] [PubMed] [Google Scholar]
- 38. Peto R, Darby S, Deo H, Silcocks P, Whitley E, et al. (2000) Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ 321: 323–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zaher C, Halbert R, Dubois R, George D, Nonikov D (2004) Smoking-related diseases: the importance of COPD. International Journal of Tuberculosis and Lung Disease 8: 1423–1428. [PubMed] [Google Scholar]
- 40. Bellavia D, Frada G, Di Franco P, Feo S, Franceschi C, et al. (1999) C4, BF, C3 allele distribution and complement activity in healthy aged people and centenarians. J Gerontol A Biol Sci Med Sci 54: B150–153. [DOI] [PubMed] [Google Scholar]
- 41. Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, et al. (2006) PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet 9: 55–61. [DOI] [PubMed] [Google Scholar]
- 42. Steffens M, Lamina C, Illig T, Bettecken T, Vogler R, et al. (2006) SNP-based analysis of genetic substructure in the German population. Hum Hered 62: 20–29. [DOI] [PubMed] [Google Scholar]
- 43. Nebel A, Croucher PJ, Stiegeler R, Nikolaus S, Krawczak M, et al. (2005) No association between microsomal triglyceride transfer protein (MTP) haplotype and longevity in humans. Proc Natl Acad Sci U S A 102: 7906–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Development Core Team 2008. A language and environment for statistical computing. RFoundation for Statistical Computing, Vienna, Austria. ISBN 3–900051–07–0, URL http://www.R-project.org.
- 45. Kramer J, Fulop T, Rajczy K, Nguyen AT, Fust G (1991) A marked drop in the incidence of the null allele of the B gene of the fourth component of complement (C4B*Q0) in elderly subjects: C4B*Q0 as a probable negative selection factor for survival. Hum Genet 86: 595–598. [DOI] [PubMed] [Google Scholar]
- 46. Candore G, Balistreri CR, Listi F, Grimaldi MP, Vasto S, et al. (2006) Immunogenetics, gender, and longevity. Ann N Y Acad Sci 1089: 516–537. [DOI] [PubMed] [Google Scholar]
- 47. Franceschi C, Motta L, Valensin S, Rapisarda R, Franzone A, et al. (2000) Do men and women follow different trajectories to reach extreme longevity? Italian Multicenter Study on Centenarians (IMUSCE). Aging (Milano) 12: 77–84. [DOI] [PubMed] [Google Scholar]
- 48. Ioannidis JPA, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG (2001) Replication validity of genetic association studies. Nat Genet 29: 306–309. [DOI] [PubMed] [Google Scholar]
- 49. Benowitz NL, Jacob P, Bernert JT, Wilson M, Wang LG, et al. (2005) Carcinogen exposure during short-term switching from regular to “Light” cigarettes. Cancer Epidemiol Biomarkers Prev 14: 1376–1383. [DOI] [PubMed] [Google Scholar]
- 50. Franceschi S, Bidoli E (1999) The epidemiology of lung cancer. Annals of Oncology 10: 3–6. [DOI] [PubMed] [Google Scholar]
- 51. Caliebe A, Kleindorp R, Blanche H, Christiansen L, Puca AA, et al. (2010) No or only population-specific effect of PON1 on human longevity: a comprehensive meta-analysis. Ageing Res Rev 9: 238–244. [DOI] [PubMed] [Google Scholar]
- 52. Zaslavsky AM (2001) Statistical issues in reporting quality data: small samples and casemix variation. Int J Qual Health Care 13: 481–488. [DOI] [PubMed] [Google Scholar]
- 53. Flachsbart F, Franke A, Kleindorp R, Caliebe A, Blanche H, et al. (2010) Investigation of genetic susceptibility factors for human longevity – a targeted nonsynonymous SNP study. Mutat Res 694: 13–19. [DOI] [PubMed] [Google Scholar]
- 54. Wahrmann M, Dohler B, Ruhenstroth A, Haslacher H, Perkmann T, et al. (2011) Genotypic Diversity of Complement Component C4 Does Not Predict Kidney Transplant Outcome. J Am Soc of Nephrol 22: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Combined file of supporting tables. Supplementary Table S1a: Characteristics of samples. Supplementary Table S1b: Quality control of C4A, C4B, C4S and C4L. Supplementary Table S2: Frequencies of C4A, C4S and C4L copy numbers. Supplementary Table S3: Results of the comparison of Q0 carrier state for the C4B gene with age groups as in Kramer et al. 1989 and Arason et al. 2003. Supplementary Table S4: Results of the comparison of Q0 carrier state for the C4L gene (carrier and non-carrier information). Supplementary Table S5: Results Replication study in additional German sample: Results of the comparison of Q0 carrier state for the C4L gene (carrier and non-carrier information). Supplementary Table S6: Replication study in additional German sample: Results of the comparison of Q0 carrier state for the C4L gene with age groups as in the initial study.
(PDF)
Data of copy number calculations performed using the CopyCallerTM Software (Applied Biosystems, Foster City, California, USA) for C4A, C4B, C4S, C4L and C4L Replication.
(XLSX)