Abstract
Serial analysis of gene expression (SAGE) is a widely used technique for large-scale transcriptome analysis in mammalian systems. Recently, a modified version called LongSAGE (S. Saha, A.B. Sparks, C. Rago, V. Akmaev, C.J. Wang, B. Vogelstein, K.W. Kinzler [2002] Nat Biotechnol 20: 508-512) was reported by increasing tag length up to 21 bp. Although the procedures for these two methods are similar, a detailed protocol for LongSAGE library construction has not been reported yet, and several technical difficulties associated with concatemer cloning and purification have not been solved. In this study, we report a substantially improved LongSAGE method called Robust-LongSAGE, which has four major improvements when compared with the previously reported protocols. First, a small amount of mRNA (50 ng) was enough for a library construction. Second, enhancement of cDNA adapter and ditag formation was achieved through an extended ligation period (overnight). Third, only 20 ditag polymerase chain reactions were needed to obtain a complete library (up to 90% reduction compared with the original protocols). Fourth, concatemers were partially digested with NlaIII before cloning into vector (pZEro-1), greatly improving cloning efficiency. The significant contribution of Robust-LongSAGE is that it solved the major technical difficulties, such as low cloning efficiency and small insert sizes associated with existing SAGE and LongSAGE protocols. Using this protocol, one can generate two to three libraries, each containing over 4.5 million tags, within a month. We recently have constructed five libraries from rice (Oryza sativa), one from maize (Zea mays), and one from the rice blast fungus (Magnaporthe grisea).
Genome sequencing is becoming an emerging technology for large-scale gene discovery, and many prokaryotic and eukaryotic genomes have been completely sequenced in the last few years. Two model plant species have been sequenced recently: Arabidopsis for dicots (Arabidopsis Genome Initiative, 2000) and rice (Oryza sativa) for monocots (Goff et al., 2002; Yu et al., 2002). Although many genes have been discovered in these two genomes, accurate annotation of the whole genome and identification of all expressed genes continue to be significant challenges because approximately one-half of the predicted genes are unsubstantiated by experimental evidence (Cho and Walbot, 2001; Yuan et al., 2001).
Exhaustive sequencing of expressed sequence tags (ESTs) was the first method used for rapid identification of expressed genes and gene expression profiling (Adams et al., 1995). This method involves the large-scale, single-pass, and partial sequencing of cDNA clones (approximately 500 bp), usually from a large number of libraries representing diverse tissues. ESTs are relatively slow and costly to generate, making it difficult to achieve saturation of a library or to produce quantitative estimates of tissue-specific expression from these data. DNA microarray technology is a new gene profiling technique that has produced a revolution in expression analysis. These “chips” provide a rapid and relatively inexpensive way to monitor in parallel the expression of thousands of transcripts. However, microarrays are subject to inherent limitations, such as background intensities that may rival signals for weakly expressed transcripts, the difficulty of distinguishing between closely related sequences (Duggan et al., 1999), and inability to obtain the transcript variants (Patankar et al., 2001; Jones et al., 2002; Lorenz and Dean, 2002; Gibbings et al., 2003).
Compared with microarrays, serial analysis of gene expression (SAGE) allows both qualitative and quantitative evaluation of thousands of genes without any prior information (Velculescu et al., 1995). It is an extremely powerful, efficient, and global approach for analyzing gene expression profiles, novel gene discovery, revealing novel pathways, and metabolic circuits. SAGE is based on three main principles: (a) short sequences (14-15 bp) are isolated from transcripts, providing sufficient information to provide a defined 3′ position within a transcript; (b) ditags (two ligated individual tags) are concatenated, with as many as 70 to 100 tags per concatemer, and the concatemers are cloned and sequenced; and (c) data output reflects the actual gene expression pattern in a particular condition or stage of an organism and allows visualization of transcript complexity such as transcripts variants, antisense transcripts, etc. (Patankar et al., 2001; Jones et al., 2002; Lorenz and Dean, 2002; Gibbings et al., 2003). One of the major advantages of the SAGE method is that the output information developed is a digital format so that data can be directly compared with data generated by other researchers and laboratories. Virtual tag data can be stored forever, allowing it to be reinterpreted if needed (Aldaz, 2003). Since the first publication of the SAGE technology in 1995 by Velculescu and his colleagues, it has been applied primarily in cancer research, and over 5 million conventional SAGE tags from various cancerous cell lines have been cataloged (Zhang et al., 1997; Boon et al., 2002; Aldaz, 2003; http://cgap.nci.nih.gov/SAGE).
LongSAGE, a modified version of the conventional SAGE, was developed recently for both gene expression and genome annotation studies (Saha et al., 2002). The LongSAGE method uses a different type IIS enzyme, MmeI, which releases 21 bp from each transcript. Saha et al. (2002) and Chen et al. (2002) have shown that longer tags were much more efficient for the identification of novel genes in the complex genomes in comparison with conventional SAGE tags (14-15 bp). Probably because of technical difficulties, such as lower cloning efficiency of concatemers and small insert sizes (approximately 300 bp) associated with library construction, only one Long-SAGE paper has been reported so far. The present study has made several major improvements in the LongSAGE library construction. The new procedure is called Robust-LongSAGE (RL-SAGE) because the cloning efficiency and insert size of LongSAGE clones have been greatly increased. The RL-SAGE procedure is schematically represented in Figure 1. RL-SAGE can generate over 4.5 million tags from a small amount (50 ng) of mRNA using just 20 ditag PCR products. Using this method, we have constructed five libraries from rice, one from maize (Zea mays), and one from blast fungus (Magnaporthe grisea).
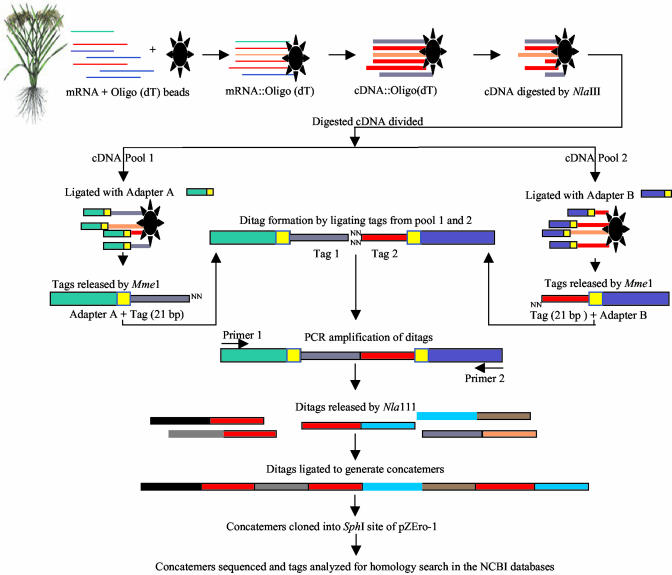
Figure 1.
Schematic representation of LongSAGE technology based on Saha et al. (2002). Total RNA was isolated from rice leaves, and mRNAs was purified and covalently linked to oligo(dT)25 magnetic beads. Single- and double-stranded cDNA was synthesized and digested with NlaIII. The digested cDNA was equally divided into two parts, pool 1 and 2, and ligated with linkers A and B, respectively. These linkers consist of an asymmetric recognition motif for a type IIS enzyme, MmeI, and specific sequences for PCR primer-binding sites. The ligated cassettes (linker::cDNA) were treated with MmeI to release tags from cDNA. The MmeI tags (LongSAGE tags) from pools 1 and 2 were mixed together (sticky end ligation) to form ditag cassettes. These cassettes were PCR amplified using primers specific to each linkers. Then linker sequences were removed from the cassettes by NlaIII digestion. Ditags were ligated together to generate longer molecules called concatemers. Over 0.5-kb concatemers were cloned into SphI site of pZEro-1 and sequenced using M13 primers. Ditags and tags were extracted from a high quality sequence data and homology search was conducted using the National Center for Biotechnology Information (NCBI) databases.
RESULTS
Improvement in Initial mRNA Quantity and Ditag Formation
The major modifications of the RL-SAGE protocol are presented in Table I and briefly described as follows. The RL-SAGE libraries were constructed using a small amount of mRNA (50 ng), compared with 2 to 5 μg mRNA used in conventional SAGE and LongSAGE protocols. The synthesized cDNA was digested with NlaIII for 2.5 h at 37°C as compared with 1 h in conventional SAGE and LongSAGE. We used PCR primers specific for the rice actin (Act1) gene (McElroy et al., 1990) to check cDNA synthesis. Glyceraldehyde-3-phosphate dehydrogenase- and elongation factor-specific PCR primers for human (Homo sapiens) and mouse (Mus musculus), recommended by the I-SAGE kit (Invitrogen, Carlsbad, CA), were not appropriate in our case because the primers are not suitable for PCR amplification in plants. After NlaIII digestion, the supernatant was precipitated and resolved on a 2.5% (w/v) agarose gel to check cDNA synthesis and NlaIII digestion processes (data not shown). Initially, the 3′ ends of cDNAs were ligated with adapters for 2 h as recommended in LongSAGE and conventional SAGE protocols, but we did not obtain any ditag PCR amplifications. When the cDNA adapter ligation was extended overnight, desired ditag PCR products (136 bp) were obtained (Fig. 2A), probably because a 2-hour ligation was insufficient for a complete ligation of adapters to all 3′ cDNA ends. cDNAs were digested with MmeI for 2.5 h as compared with 2.5 h in LongSAGE (Saha et al., 2002). Tags generated from pools 1 and 2 were ligated for 3 h as recommended in LongSAGE (Fig. 1); again, no ditag PCR products were obtained. The next improvement was made by extending the ditag ligation to overnight and obtaining desired PCR products from a 1:100 (v/v) dilution of ditag ligation mixture. This is because in the RL-SAGE protocol, ditags were formed because of sticky-end ligation of two tags with 2-bp overhangs at 3′ cDNA ends generated by MmeI (Fig. 1). Blunt end ligation in conventional SAGE is much less efficient than sticky end ligation, which may affect the formation and subsequent amplification of certain ditags in conventional SAGE.
Table I.
RL-SAGE modifications compared with LongSAGE and conventional SAGE methods
| Library Features | RL-SAGE | LongSAGE (Saha et al., 2002) | Conventional SAGE (Velculescu et al., 1995) | |
|---|---|---|---|---|
| 1. | mRNA quantity | 50 ng | 2 μg | 5 μg |
| 2. | Ligation of cDNAs to adapters | Overnight | Not reported | 2 h |
| 3. | cDNA digestion with N/aIII | 2.5 h | 1 h | 1 h |
| 4. | Ditag ligation | Overnight | 2.5 h | overnight |
| 5. | Number of PCRs | 20 PCRs | Not reported | 300 PCRs |
| 6. | Ditags (136 bp) band purification | Individual PCR resolved | Not reported | PCR products pooled, precipitated, and resolved |
| 7. | Purification of ditags (40 bp) | 16% (w/v) Polyacrylamide gel | Not reported | 12% (w/v) Polyacrylamide gel |
| 8. | Linkers removed | M-280 streptavidin Dynabeads | Not reported | Not reported |
| 9. | Digestion of concatemers | 10 units of N/aIII (37°C, 1 min) | Not reported | Not reported |
| 10. | Purification of concatemers | 6% (w/v) Polyacrylamide gel | Not reported | 8% (w/v) Polyacrylamide gel |
| 11. | Clones screened | Randomly picked | Not reported | Colony PCR screened |
| 12. | Average insert size | 1.00 kb | Not reported | 300—500 bp |
| 13. | Total tags per concatemer | 50 tags (21 bp) | Not reported | 22 tags (14 bp) |
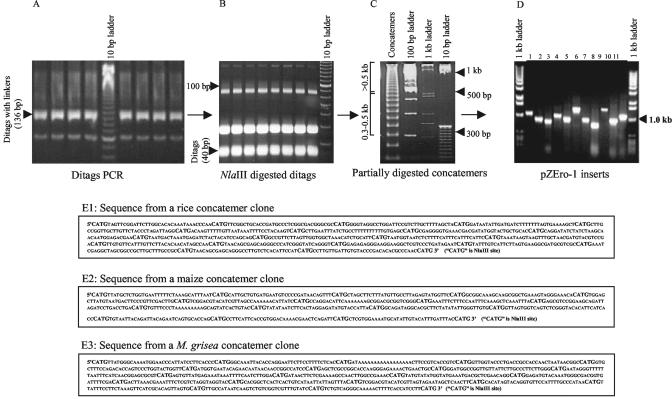
Figure 2.
Experimental procedure for major steps involved in RL-SAGE library construction. A, Ditag PCR products (136 bp) were resolved on a 12% (w/v) polyacrylamide gel. B, PCR products (136 bp) were digested with NlaIII, and the ditag's (40 bp) band was purified from a 16% (w/v) polyacrylamide gel. C, Concatemers from self-ligated ditags were partially digested and resolved on a 6% (w/v) polyacrylamide gel. D, PCR inserts of RL-SAGE clones were separated on a 1.5% (w/v) agarose gel. E1-3, RL-SAGE concatemer clones were sequenced from rice, maize, and blast fungus libraries, and NlaIII sites separating two ditags are shown in bold.
Improvement in Ditag Amplification and Gel Purification
During the initial optimization stage, over 300 ditag PCR amplifications were performed, pooled, and precipitated according to instructions in the I-SAGE kit (Invitrogen). The precipitated ditag PCR products were electrophoresed on a 12% (w/v) polyacrylamide gel as reported in conventional SAGE papers and suggested in the I-SAGE kit protocol. Unexpectedly, it was found that the ditag (136 bp) and linker (100 bp) bands were not separated clearly for gel excision of ditags (data not shown). However, when ditag PCR products from each reaction were loaded directly on a 12% (w/v) polyacrylamide gel without pooling and precipitation, both ditag and linker bands were separated clearly (Fig. 2A). The increased concentration of acrylamide also helped in easy excision of ditag band. Complete digestion of ditags with NlaIII for 3 h was performed as compared with 1.5 h in conventional SAGE. Digested ditags were initially resolved on a 12% (w/v) polyacrylamide gel as recommended by the I-SAGE kit and conventional SAGE protocols. Again, the linker and ditag bands were not separated well for purification. Increased amounts of acrylamide to 16% (w/v) yielded clear separation of both ditag and linker bands (Fig. 2B). This higher acrylamide concentration also might have contributed to decrease the linker contamination and increase the cloning efficiency of concatemers in the subsequent ligation step. Purified ditags (40 bp) were further purified using half of the amount of streptavidin beads by vigorous mixing for 30 min without performing any other steps to remove contaminated linkers from the ditags, as recommended by Powell (1998).
Improvement in Formation of Concatemers and Cloning
In RL-SAGE procedures, ditags (40 bp) with the NlaIII CATG overhangs were purified and selfligated for 3 h to produce longer molecules called “concatemers.” Initially, we performed 300 ditag PCR amplifications and only obtained a library of 100 to 200 clones with an average insert size of 300 to 400 bp. The concatemer ligation mixture was then heated for 15 min at 65°C and quickly chilled on ice for 10 min, as recommended by Kenzelmann and Muhlemann (1999). We also treated concatemers with T4 DNA polymerase for blunt end cloning, which did not result in any clones from the ligation. We repeated this experiment five times and obtained similar results.
We suspected that most of the concatemers became circular during the concatenation process and, thus, were not clonable in the pZEro-1 vector. To release the circularized concatemers, we partially digested them with NlaIII enzyme (37°C, 1 min), purified these concatemers on a 6% (w/v) polyacrylamide gel (Fig. 2C), and cloned into the SphI site of pZEro-1. Interestingly, the partial digestion significantly improved both ligation and transformation efficiency. Subsequently, we scaled down to 20 ditag PCR reactions, as compared with the over 300 ditag PCR reactions indicated in most of the SAGE publications. We obtained an average of 1.0 kb (approximately 50 tags) inserts from the >0.5-kb concatemer fraction (Fig. 2D) and 400 bp from the 0.3- to 0.5-kb concatemer fraction (data not shown). In total, we obtained 2.5 million tags (50,000 clones) from the >0.5-kb fraction and 2 million tags (100,000 clones) from the 0.3- to 0.5-kb fraction. Therefore, about 4.5 million tags could be captured in the library if all clones have been sequenced. We usually sequenced 5,000 to 7,000 individual clones per library because of high cost of sequencing. Using RL-SAGE protocol, one can construct two to three libraries simultaneously within a month from only 20 ditag PCR reactions, in comparison with 2 to 3 months required to construct just one conventional SAGE or LongSAGE library from over 300 ditag PCR reactions.
Sequence Analysis of Three RL-SAGE Clones from the Rice, Maize, and Blast Fungus Libraries
Three randomly selected RL-SAGE clones from the rice, maize, and blast fungus libraries (Fig. 2, E1-E3) were sequenced and analyzed. From the high-quality sequence of each clone, 40, 32, and 38 unique tags were extracted from the rice, maize, and blast fungus clone, respectively (Supplemental Table I). Except for one tag (5′-CATGTAACAGCGAGCAGGGCC-3′, matched to Ramy1, accession no. AY072712) from the rice clone and one tag (5′-CATGGGATGGCCGGTTGTTAT-3′, matched to EST accession no. CA408239) from the blast fungus clone had two identical copies, all other tags were unique. BLAST search in the GenBank showed that most of the tags had matches to either ESTs or genomic sequences or both (Supplemental Table I). About 34 of 40 and 29 of 38 of the tags from rice and blast fungus matched the ESTs or genomic sequences in the GenBank, respectively. In contrast, only 19 of 32 of tags derived from the maize library matched sequences in the NCBI database because fewer genomic and EST sequences from maize are available in the database. About 26 of 40, 18 of 32, and 16 of 38 of rice, maize, and blast fungus tags matched corresponding ESTs in the GenBank, respectively (Table II; Supplemental Table I), suggesting that at least 35% to 55% of the RL-SAGE tags from these libraries could be novel genes that have not been identified in the existing EST collections.
Table II.
Summary of database match of tags isolated from three rice, maize, and blast fungus clones
| Database Match | Rice | Maize | Blast Fungus |
|---|---|---|---|
| Match to EST | 10 (25%) | 15 (46.9%) | 15 (39.5%) |
| Match to gDNAa | 8 (20%) | 1 (3.1%) | 13 (34.2%) |
| Match to both EST and gDNA | 16 (40%) | 3 (9.4%) | 1 (2.6%) |
| Match to either EST or gDNA | 34 (85%) | 19 (59.4%) | 29 (76.3%) |
| No match | 6 (15%) | 13 (40.6%) | 9 (23.7%) |
| Total | 40 | 32 | 38 |
Genomic DNA
DISCUSSION
The SAGE transcript profiling method has enhanced the depth of transcriptome analysis 25- to 50-fold and reduced sequencing costs tremendously in comparison with the EST approach. In the last several years, it has been used widely in the biomedical community but underutilized in the plant community. There have been only few published conventional SAGE reports available for plants to date (Matsumura et al., 1999; Lorenz and Dean, 2002; Gibbings et al., 2003; Jung et al., 2003). Although several laboratories have tried to use this technique for expression profiling, few have been successful because of long and complicated cloning procedures. The RL-SAGE protocol reported in this study will assure high cloning efficiency, large concatemer insert sizes, and deep transcriptome analysis, which are not possible using conventional SAGE or LongSAGE protocols. We critically improved the concatemer cloning step, which avoided colony PCR screening to remove the empty clones and dramatically reduced the time required for RL-SAGE library construction. Because of partial digestion of concatemers, only 20 ditag PCR reactions were sufficient to generate over 4.5 million tags per library, and two to three libraries could be made within a month.
The first change we made was to reduce the initial amount of mRNA required for cDNA synthesis. To overcome the high input requirement for initial RNA, several groups reported an alternative way to solve this problem such as SAGE-Lite (Peters et al., 1999), MicroSAGE (Datson et al., 1999) and SADE (Virlon et al., 1999), PCR-SAGE (Neilson et al., 2000), and SAR-SAGE (Vilain et al., 2003). All these modifications used small amounts of total RNA, ranging from 50 to 1,000 ng, by adopting additional PCR amplifications (PCR amplification of cDNA before ditag formation or re-amplification of ditags by two-step PCR). The additional PCR steps may introduce bias and influence the quantitative flux of gene expression data. Moreover, the modified protocols mentioned above only produced a small number of tags (1,000-3,500 tags) per library and, so far, not many laboratories have successfully adopted these modified protocols in different systems. In addition, over 300 PCR reactions were being performed in these protocols to get sufficient ditags for library construction, which may introduce bias in gene expression patterns. In RL-SAGE, we used only 50 ng of mRNA for cDNA synthesis and conducted only 20 ditag PCR reactions without any prior amplification of cDNAs or re-amplification of ditags. Therefore, RL-SAGE is an ideal protocol for experiments with limited sample quantities (tissue/cells or RNA), when deep transcriptome analysis is required.
We found that a large quantity of mRNA used for cDNA synthesis may lead to an incomplete digestion of cDNA with both NlaIII and MmeI, which can generate multiple tags from the same transcript. If this occurs, it is difficult to distinguish these false tags generated by incomplete digestion from transcript variants such as splicing, antisense, etc. (Patankar et al., 2001; Jones et al., 2002; Lorenz and Dean, 2002; Gibbings et al., 2003). It is ideal to use mRNA on the order of nanograms rather than micrograms to overcome these anomalies. Using our RL-SAGE protocol, it is also possible to generate a complete library from 20 ditag PCR amplifications derived from as low as 5 to 10 ng of mRNA using 1:10 to 1:20 (v/v) dilutions of ditag ligation mixture.
We discovered that concatemers became circular during concatenation process, which has not been reported or addressed previously. The present study resolved this problem by partial digestion of concatemers with NlaIII. The incubation period and amount of NlaIII were critical factors in the partial digestion. From the partial digestion, we obtained RL-SAGE libraries with average insert (concatemers) sizes of 1.0 kb (approximately 50 tags), which is equivalent to 70 tags per concatemer in conventional SAGE. Most conventional SAGE publications reported an average of 22 tags per concatemer (Powell, 1998; Kenzelmann and Muhlemann, 1999). The partial digestion of concatemers reduced 90% of ditag PCR reactions in comparison with conventional SAGE or LongSAGE protocols.
Another major problem in SAGE library construction is the high percentage of clones with small inserts (<200 bp) or empty clones. In many conventional SAGE publications, a tedious method of colony PCR screening of clones was followed to remove undesirable clones for sequencing. For example, Fujii and Amrein (2002) screened 8,640 and 10,848 clones by colony PCR and sequenced 2,236 and 2,496 clones, respectively, for two SAGE libraries of fruitfly (Drosophila melanogaster) head. We estimate that it could have taken at least 2 to 3 months to complete these two libraries because of time-consuming PCR screening of individual clones. In contrast, there were almost no empty clones (<0.5%) in our RL-SAGE libraries constructed so far. A library of over 150,000 clones (4.5 million tags) can be easily generated within 2 weeks without any colony PCR screening because most of the clones have inserts ranging from 400 bp to 2 kb.
The RL-SAGE strategy (Figs. 1 and 2) is not only superior to conventional and LongSAGE, but also has some advantages over a novel transcriptome profiling method called massive parallel signature sequencing (MPSS; Brenner et al., 2000a, 2000b). MPSS can generate maximum 2 million reliable tags from at least 500 ng of mRNA, but RL-SAGE can create over 4.5 million tags from 50 ng of mRNA if all the clones are sequenced. More importantly, MPSS is a complex technique and only available from Lynx Therapeutics, Inc. (http://www.lynxgen.com). For proprietary reasons, this technology may not be easily accessible to certain plant species, whereas RL-SAGE is quite simple and can be used for any species. Theoretically, over 99% of 21-bp RL-SAGE tags can be matched uniquely in the complex genomic sequences, as estimated by Saha et al. (2002). In addition, RL-SAGE tags can be used directly in reverse transcriptase-PCR, RACE or for hybridizing to cDNA libraries to obtain a complete cDNA from a source of interest.
At present, RL-SAGE has two significant limitations. One is the high cost of sequencing of RL-SAGE clones, which prevents large-scale sequencing of an entire library. For example, a library of 20,000 clones (about 1 million tags) will cost at least $120,000 (assuming $6 per clone). This limitation could be solved in the near future with the improvement of DNA sequencing technology. Hopefully, novel technologies like sequencing by hybridization (Halperin et al., 2003) could be adopted to sequence over 4.5 million tags from each RL-SAGE library with low costs in the near future. The other limitation is that RL-SAGE uses NlaIII (CATG occurs once on average in every 256 bp) to cleave cDNA, and some genes (approximately 5%) may be missed in the library because they lack an NlaIII site (Velculescu et al., 1995; Saha et al., 2002). To identify all transcripts, an additional library can be constructed by adopting a different enzyme such as Sau3AI or DpnII. This problem also exists with the MPSS technology, which uses a single 4-bp cutter restriction enzyme DpnII.
In summary, we have made several useful modifications that improved the efficiency of PCR amplification, ditag formation, and concatemer cloning. These modifications have greatly accelerated the RL-SAGE library construction. Twenty PCR reactions (50 μL) are sufficient to generate 4.5 million transcript tags from each RL-SAGE library. The partial digestion of concatemers has reduced number of ditag PCR reactions by over 90% as compared with the original protocol. Using this protocol, we generated five libraries from rice, one from maize, and one from blast fungus. Preliminary sequence analysis of three randomly selected clones from these libraries indicated that at least 35% to 55% of SAGE tags are novel. We believe that our RL-SAGE protocol will facilitate plant transcriptome analysis and will also accelerate discovery of novel genes and annotation of sequenced plant genomes such as rice and Arabidopsis.
MATERIALS AND METHODS
Tissue and RNA Isolation
The rice (Oryza sativa) cv Nipponbare, whose genome has been sequenced, was used for RL-SAGE library construction (Goff et al., 2002). Rice and maize (Zea mays; B73) plants were grown in a Conviron growth chamber under 12 h of light (500 μmol photons m-2 s-1), 20°C at night, 26°C in the day, and 85% relative humidity. Leaf tissue from 3-week old rice and 4-week-old maize plants was harvested for total RNA isolation. Maize inbred line B73 was selected because it is being used for genome sequencing (Tomkins et al., 2002). Rice blast fungus (Magnaporthe grisea) strain 70-15 (Mitchell et al., 2003) was chosen for transcriptome profiling because its draft sequence is available to the public (http://www-genome.wi.mit.edu/annotation/fungi/magnaporthe/). The fungus was grown on a minimum medium (0.2% [w/v] yeast extract and 1% [w/v] Suc) for 3 d (28°C at 200 rpm) and mycelium was harvested for RNA isolation. Two grams of tissue from rice and maize and 2 grams of mycelium from the fungus, respectively, were used for total RNA isolation using the TRIzol solution (Invitrogen). Poly(A+) mRNA was purified using the Oligotex mRNA midi kit (Qiagen Inc., Valencia, CA), according to the manufacturer's instructions.
RL-SAGE Libraries Construction
Because no detailed protocol has been published for LongSAGE library construction, we adopted procedures from conventional SAGE (Velculescu et al., 1995), I-SAGE kit (Invitrogen), and LongSAGE (Saha et al., 2002) methods with several major modifications as described below. About 50 ng of poly(A+) mRNA was bound to magnetic beads with oligo(dT)25, and cDNA was synthesized directly on the oligo(dT) beads. cDNA was digested with NlaIII and divided equally into two parts, pools A and B. These pools were ligated overnight (16°C) with specific linkers. The linker oligonucleotides, i.e. linker 1 (linker 1A [5′-TTTGGATTTGCTGGTGCAGTACAACT AGGCTTAATATCCGACATG-3′] and linker 1B [5′-TCGGATATTAAGCCTAGTTGT ACTGCACCAGCAAATCC-C7 amino-modified-3′]) and linker 2 (linker 2A [5′-TTTCT GCTCGAATTCAAGCTTCTAACGATGTACGTCCGACATG-3′ and linker 2B 5′-TC GGACGTACATCG TTAGAAGCTTGAATTCGAGCAG-C7 amino-modified-3′]) were synthesized and purified on a polyacrylamide gel (Integrated DNA Technologies Inc., Coralville, IA) as reported by Saha et al. (2002). After the beads were washed thoroughly, pools A and B were treated with 20 units of MmeI (37°C, 3 h) (New England Biolabs, Inc., Beverly, MA). The resulting tags from pools A and B were ligated overnight (16°C) in a 10-μL mixture to form ditag cassettes. The ligated ditag mixture was diluted (1:100 [v/v]), and 1 μL was used in a 50-μL PCR mixture. A total of 20 ditag PCR amplifications were performed for 27 cycles using the following primers: forward primer, 5′-biotin GTGCTCGTGGGATTTGCTGGTGCAGTACA-3′; and reverse primer, 5′-biotin GAGCTCGTGCTGCTCGAATTCAAGCTTCT-3′). Individual ditag PCR products (136 bp) were purified on a 12% (w/v) polyacrylamide gel. Linkers (50 bp) were removed from the ditags by NlaIII digestion, and ditag (40 bp) band was purified on a 16% (w/v) polyacrylamide gel with a very short period of UV exposure. Ditags were further purified using half of the amount (100 μL) of M 280 streptavidin beads (Dynal Biotech Inc., Lake Success, NY) as compared with a previous report (Powell, 1998).
Purified ditag cassettes were ligated together (16°C, 3 h) to generate longer molecules (concatemers). In addition, concatemers were partially digested with 10 units of NlaIII (37°C, 1 min), followed by immediate inactivation of the enzyme (75°C, 20 min). Digested concatemers were resolved on a 6% (w/v) polyacrylamide gel, and concatemer fractions ranging from 0.3 to 0.5 kb and over 0.5 kb were purified separately. To avoid DNA damage by UV, marker lanes were cut out from the gel and stained separately with ethidium bromide. The marker lanes were then UV photographed and aligned to their original positions for checking the size of concatemers in the unstained lane. The purified concatemers were cloned into the SphI site of the pZEro-1 plasmid (Invitrogen). The ligated mixture was transformed into TOP10F′ electrocompetent cells (Invitrogen). Positive transformants were selected by plating on low-salt Luria-Bertani plates supplemented with Zeocin (50 μg mL-1; overnight, 37°C). The average concatemer's size was detected by PCR using M13 forward and reverse primers.
Sequencing and Data Analysis
Each RL-SAGE library quality was checked by sequencing randomly selected clones at the Plant and Microbe Genome Facility (Ohio State University, Columbus). The sequence chromatographs were processed with Sequencher 4.1 (Gene Codes, Ann Arbor, MI) software. Ditags (40 bp) were extracted from a high-quality concatemer's sequence. Tag sizes (21 bp) were isolated manually from ditags, and a database homology search was performed using NCBI EST and genomic DNA sequences.
Supplementary Material
Acknowledgments
We thank John J. Dunn (Biology Department, Brookhaven National Laboratory, Upton, NY) for his valuable suggestions during the cloning of concatemers. We also thank Kenneth W. Kinzler and Victor E. Velculescu (Howard Hughes Medical Institute and the Sidney Kimmel Comprehensive Cancer Center, Baltimore) for valuable discussions during construction of RL-SAGE libraries. We are thankful to all members of our laboratory for their valuable help and discussion during this work. Critical reading of the manuscript by Rebecca Nelson (Cornell University, Ithaca, NY) is highly appreciated.
This work was supported by the National Science Foundation-Plant Genome Research Program (grant no. 115642).
The online version of this article contains Web-only data.
References
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O et al. (1995) Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3-174 [PubMed] [Google Scholar]
- Aldaz CM (2003) Serial analysis of gene expression (SAGE) in cancer research. In M Ladanyi, WL Gerald, eds, Expression Profiling of Human Tumors: Diagnostic and Research Applications. Humana Press, Totowa, NJ, pp 47-60
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ et al. (2002) An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA 99: 11287-11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M et al. (2000a) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630-634 [DOI] [PubMed] [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, Mao JI, Luo S, Kirchner JJ, Eletr S et al. (2000b) In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci USA 97: 1665-1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Lee S, Zhou G, Rowley JD, Wang SM (2002) Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc Natl Acad Sci USA 99: 12257-12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Walbot V (2001) Computational methods for gene annotation: the Arabidopsis genome. Curr Opin Biotechnol 12: 126-130 [DOI] [PubMed] [Google Scholar]
- Datson NA, van der Perk-de Jong J, van den Berg MP, de Kloet ER, Vreugdenhil E (1999) MicroSAGE: a modified procedure for serial analysis of gene expression in limited amounts of tissue. Nucleic Acids Res 27: 1300-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM (1999) Expression profiling using cDNA microarrays. Nat Genet 21: 10-14 [DOI] [PubMed] [Google Scholar]
- Fujii S, Amrein H (2002) Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J 21: 5353-5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings JG, Cook BP, Dufault MR, Madden SL, Khuri S, Turnbull CJ, Dunwell JM (2003) Global transcript analysis of rice leaf and seed using SAGE technology. Plant Biotechnol J 1: 271-285 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100 [DOI] [PubMed] [Google Scholar]
- Halperin E, Halperin S, Hartman T, Shamir R (2003) Handling long targets and errors in sequencing by hybridization. J Comput Biol 10: 483-497 [DOI] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA (2002) Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res 11: 1346-1352 [DOI] [PubMed] [Google Scholar]
- Jung SH, Lee JY, Lee DH (2003) Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Mol Biol 52: 553-567 [DOI] [PubMed] [Google Scholar]
- Kenzelmann M, Muhlemann K (1999) Substantially enhanced cloning efficiency of SAGE (serial analysis of gene expression) by adding a heating step to the original protocol. Nucleic Acids Res 27: 917-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, Dean JF (2002) SAGE Profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiol 22: 301-310 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R (1999) Technical advance: transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J 20: 719-726 [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TK, Thon MR, Jeong J-S, Brown D, Deng J, Dean RA (2003) The rice blast pathosystem as a case study for the development of new tools and raw materials for genome analysis of fungal plant pathogens. New Phytol 159: 53-61 [DOI] [PubMed] [Google Scholar]
- Neilson L, Andalibi A, Kang D, Coutifaris C, Strauss JF 3rd, Stanton JA, Green DP (2000) Molecular phenotype of the human oocyte by PCR-SAGE. Genomics 63: 13-24 [DOI] [PubMed] [Google Scholar]
- Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF (2001) Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol Biol Cell 12: 3114-3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DG, Kassam AB, Yonas H, O'Hare EH, Ferrell RE, Brufsky AM (1999) Comprehensive transcript analysis in small quantities of mRNA by SAGE-Lite. Nucleic Acids Res 27: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J (1998) Enhanced concatemer cloning: a modification to the SAGE (serial analysis of gene expression) technique. Nucleic Acids Res 26: 3445-3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW (2002) Using the transcriptome to annotate the genome. Nat Biotechnol 20: 508-512 [DOI] [PubMed] [Google Scholar]
- Tomkins JP, Davis G, Main D, Yim Y, Duru N, Musket T, Goicoechea JL, Frisch DA, Coe EH Jr, Wing RA (2002) Construction and characterization of a deep-coverage bacterial artificial chromosome library for maize. Crop Sci 42: 928-933 [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484-487 [DOI] [PubMed] [Google Scholar]
- Vilain C, Libert F, Venet D, Costagliola S, Vassart GR (2003) Small amplified RNA-SAGE: an alternative approach to study transcriptome from limiting amount of mRNA. Nucleic Acids Res 31: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlon B, Cheval L, Buhler JM, Billon E, Doucet A, Elalouf JM (1999) Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci USA 96: 15286-15291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79-92 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Quackenbush J, Sultana R, Pertea M, Salzberg SL, Buell CR (2001) Rice bioinformatics: analysis of rice sequence data and leveraging the data to other plant species. Plant Physiol 125: 1166-1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW (1997) Gene expression profiles in normal and cancer cells. Science 276: 1268-1272 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.