Abstract
Previous experience alters the rate of transcriptional induction of many genes in yeast, and this phenomenon persists through several cell division cycles. This phenomenon is called epigenetic transcriptional memory. For the yeast gene INO1, transcriptional memory requires a physical interaction with the nuclear pore complex (NPC) and changes in the chromatin structure of the promoter. These changes lead to binding of a preinitiation form of RNA Polymerase II (RNAPII) to the INO1 promoter, bypassing the need to recruit RNAPII to the promoter during reactivation. In our recent study, we found that in human cells, hundreds of interferon-γ responsive genes exhibit a mechanistically similar form of transcriptional memory. Transcriptional memory requires a homologous nuclear pore protein in yeast and humans, which interacts with the promoters of genes that exhibit transcriptional memory and promotes both alteration of chromatin structure and binding of RNAPII. Whereas the interaction of yeast genes with nuclear pore proteins occurs at the NPC, the interaction of human genes with nuclear pore proteins occurs in the nucleoplasm. Thus, the interaction of nuclear pore proteins with genes plays an important and conserved role in affecting long-term epigenetic changes in transcriptional regulation.
Keywords: nuclear pore complex, nuclear porins, chromatin, epigenetics, transcriptional memory
Introduction
The nuclear pore complex (NPC) is a large (~60 megadalton), universally conserved 8-fold symmetric complex made up of ~30 different nuclear pore proteins (Nups).1-3 This structure plays an essential role in the transit of RNA and protein between the nucleus and the cytoplasm and has been implicated in many diverse processes aside from transport.4-11 Despite early evidence in yeast indicating that the NPC interacts with silenced or repressed portions of the genome,12-14 more recent work has indicated that many active genes in yeast interact with the NPC.15,16 Furthermore, several yeast genes remain associated with the NPC after being repressed and, in the case of the model gene INO1, this interaction leads to an altered chromatin structure and binding of a poised RNA Polymerase II (RNAPII) to the recently repressed promoter.1,17,18 This phenomenon is called transcriptional memory and can be inherited through several generations, indicating that it is epigenetic. The GAL genes and a class of stress-induced genes exhibit a similar behavior.19 The mechanistic details for these genes are similar but not identical to INO1.19 Furthermore, although there is a clear role for the interaction of Nups in promoting transcription in flies and humans,20-23 it was unclear if metazoan Nups were also involved in transcriptional memory. A study published in 2010 suggested that the induction of the human HLA-DRA gene in response to interferon gamma (IFN-γ) was faster in cells that had previously been exposed to IFN-γ.24 This led us to ask if IFN-γ induced genes are regulated by a mechanistically similar form of epigenetic transcriptional memory. In our recent paper, we showed that hundreds of such genes exhibit transcriptional memory and that this form of memory is mechanistically very similar to INO1 memory in yeast.25 Here we discuss these findings in a broader context.
Transcriptional Memory Regulates RNAPII Recruitment
We previously discovered a persistent interaction of a poised, unphosphorylated RNAPII at the INO1 promoter for generations after repression.1 Yeast strains that lack the histone variant H2A.Z or the nuclear pore protein Nup100 do not exhibit transcriptional memory; INO1 relocalizes to the nucleoplasm after repression, RNAPII binding to the promoter is lost and the rate of reactivation of INO1 is significantly slowed (while the rate of the initial activation is unaffected).1 This suggests that transcriptional memory promotes faster reactivation by bypassing the rate-limiting step of RNAPII recruitment. To better understand this, we compared the binding of general transcription factors (GTFs) to the promoter of active and recently repressed INO1 by chromatin immunoprecipitation (ChIP). Every GTF tested (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, TFIIK, TFIIS, and Mediator) was bound to the active INO1 promoter. Most of the GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) were bound to the recently repressed INO1 promoter, but others (TFIIK, TFIIS, and Mediator) were not. This result was consistent with our observation that the RNAPII that was bound to the recently repressed INO1 promoter was not phosphorylated on Ser2 and Ser5 of the carboxyl terminal domain of RNAPII, indicating that this is a preinitiation form of RNAPII.26 The mechanism of INO1 transcriptional memory suggests that transcription can be regulated at the level of RNAPII recruitment, at the level of initiation and at the level of elongation.27,28 Consistent with this conclusion, in both stationary phase yeast cells and G0 lymphocytes, thousands of genes are poised for future expression through binding of preinitiation complex to their promoters.29,30
INO1 transcriptional memory requires an 11 base pair cis acting promoter element called the memory recruitment sequence (MRS). The promoters of stress-induced yeast genes that display transcriptional memory are enriched for a very similar element.19 Furthermore, the MRS is sufficient to recapitulate the chromatin structure associated with transcriptional memory when inserted at ectopic loci.1 However, the MRS is not sufficient to promote PIC association with an ectopic locus, suggesting that PIC association is a context-dependent (and presumably promoter-specific) output of transcriptional memory. Therefore, peripheral localization and chromatin structure probably function to regulate PIC formation and can be uncoupled from these downstream events.
The human gene HLA-DRA is induced by IFN-γ and has much faster induction kinetics if cells have previously been treated with IFN-γ.31,32 This effect persists for up to four cell divisions, suggesting that it is epigenetically inherited through mitosis.24,25 We found that several hundred human genes displayed significantly faster induction kinetics in cells that had been exposed to IFN-γ compared with their initial activation. Transcriptional memory does not require full transcription of HLA-DRA; treatment of cells with IFN-γ for 2 h, which was not sufficient to lead to measurable increase in HLA-DRA, was sufficient to lead to transcriptional memory. After removing IFN-γ, a preinitiation form of RNAPII remained bound to the promoters of genes that exhibited transcriptional memory, suggesting that human genes exhibit a form of transcriptional memory that is mechanistically similar to INO1 memory in yeast.
Non-NPC Bound Roles for Nups
Nups interact with a large number of genes in eukaryotic genomes, leading to both positive and negative impacts on gene expression.15,16,33,34 INO1 transcriptional memory requires interactions with several yeast Nups (Fig. 1).1 The active INO1 promoter interacts with different Nups than the recently repressed INO1 promoter.1 Based on ChIP, active INO1 interacts with Nup2, one of several Nups having Phe-X-Phe-Gly repeats, but does not interact with Nup100, a Gly-Leu-Phe-Gly repeat protein. In contrast, recently repressed INO1 interacts weakly with Nup2 and strongly with Nup100.1 Using antibodies against Phe-X-Phe-Gly or against Nup98, a human homolog of yeast Nup100, we performed ChIP and measured the interaction of each of these antigens with the promoters of human genes that exhibit transcriptional memory. In humans, Phe-X-Phe-Gly Nups include Nup62, Nup153, Nup214, Nup358/RanBP2, and Pom121. Similar to yeast INO1, we observed an interaction of Phe-X-Phe-Gly proteins but not Nup98, with the active promoters and a weaker interaction with Phe-X-Phe-Gly proteins but a strong interaction of Nup98 with previously expressed promoters for up to 96h after removing IFN-γ. Therefore, different human Nups bind to active and recently expressed genes that exhibit transcriptional memory.
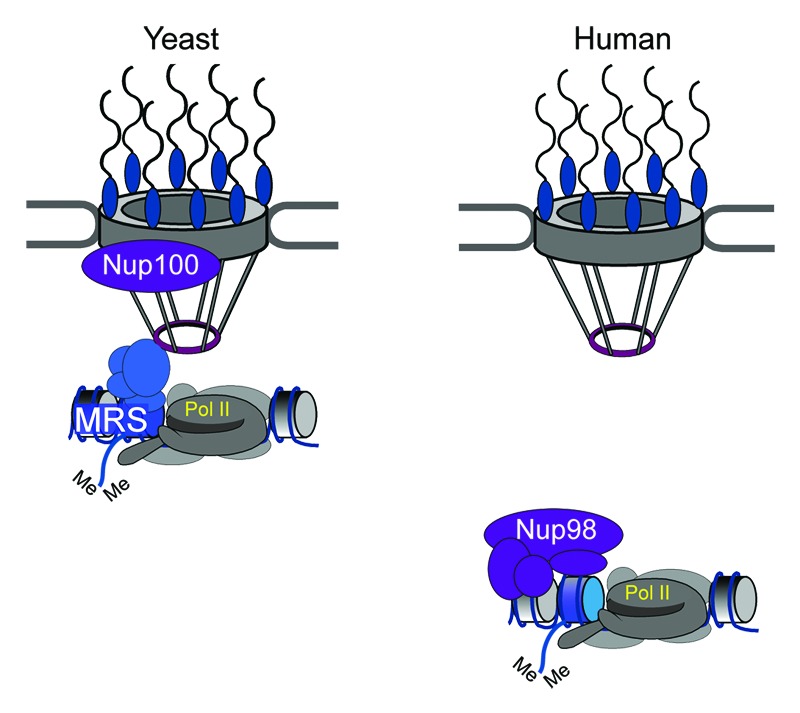
Figure 1. Nup-dependent transcriptional memory in yeast and humans. Left, INO1 transcriptional memory in yeast. After repression, the INO1 gene remains bound to the NPC. RNAPII clears the coding region but remains associated with the promoter in a poised, preinitiation form. Promoter nucleosomes are altered by incorporation of H2A.Z and dimethylation of H3K4 (blue nucleosome). Transcriptional memory persists at the NPC for approximately four generations. Right, IFN-γ transcriptional memory in humans. After the removal of IFN-γ, a poised preinitiation form of RNAPII remains associated with the promoter. Promoter nucleosomes are marked with H3K4me2 (blue) and bind to nucleoplasmic Nups such as Nup98. This epigenetic state is inherited for at least four generations.
Although all of the interactions between yeast genes and Nups occur at the NPC, many Nup-gene interactions in metazoan cells occur in the nucleoplasm, away from the NPC. We localized the HLA-DRA gene with respect to the nuclear periphery using DNA FISH before IFN-γ treatment, during IFN-γ treatment and 48 h after IFN-γtreatment. Consistent with a role for non-NPC-bound Nups in transcriptional memory, HLA-DRA did not co-localize with the nuclear periphery under any condition. This suggests that in HeLa cells, nucleoplasmic Nups interact with HLA-DRA.21,23 Although these interactions occur at a different subnuclear location in yeast and humans, it seems likely that they represent a similar, highly conserved, biochemical process.
Chromatin Structure and Transcriptional Memory
Changes in the chromatin state of gene promoters have been tied to transcriptional memory in both yeast and humans.1,24 In yeast, the histone variant H2A.Z was required for INO1 transcriptional memory and the INO1 MRS was both necessary and sufficient to promote the deposition of H2A.Z.1,17 HLA-DRA transcriptional memory is associated with persistent dimethylation of histone H3 lysine 4 at the promoter (H3K4me2).24 This led us to ask if this chromatin mark was also associated with yeast INO1 transcriptional memory. Indeed, H3K4me2 persists at the recently repressed INO1 promoter. Mutation of the MRS led to loss of H3K4me2 after repression. The MRS was also sufficient to induce dimethylation of H3K4 at an ectopic locus. Moreover, mutants lacking the enzymes responsible for H3K4 methylation blocked INO1 transcriptional memory, suggesting that H3K4 methylation is required for memory. Finally, H3K4me2 deposition occurred in the absence of H2A.Z, suggesting that H3K4me2 incorporation occurred upstream, or in parallel to, H2A.Z deposition.
hNup98 is Required for Transcriptional Memory
Yeast INO1 transcriptional memory specifically requires Nup100.1 Using siRNAs to deplete Nup98 in HeLa cells, we found that loss of Nup98 led to loss of all measures of transcriptional memory: RNAPII and H3K4me2 were lost from the promoters of genes that exhibit IFN-γ memory and the rate of reactivation of these genes was much slower. This effect was specific to Nup98 depletion; Nup107 depletion did not affect transcriptional memory. We also found that mutants lacking yeast Nup100 failed to retain H3K4me2 at the recently repressed INO1 promoter, further supporting the conservation of this mechanism.
Conclusions
Some Nups can interact with promoters to modulate chromatin structure and gene expression in both yeast and humans, suggesting that Nups have functions independent of their role in intranuclear transport. Our recent work has further defined the molecular mechanism of yeast transcriptional memory and found strong evidence that facets of this mechanism are conserved from yeast to humans. Based on this, we predict that many genes are regulated by transcriptional memory in different signaling contexts. Consistent with this notion, the promoters of stress-responsive yeast genes that exhibit a form of epigenetic transcriptional memory are enriched for an element that is very similar to the MRS element.19 Also, these genes require Nup42 for this behavior.19 If these genes are regulated by a mechanism similar to that used by INO1 and HLA-DRA, their promoters may be marked with H3K4me2 and bound to RNAPII following the primary stress.
Transcriptional memory reflects both the cell’s current physiology and previous experiences. In the context of the body, the response of cells and tissues to the same stimulus might differ dramatically based on recent (or not so recent) experience. A complete understanding of gene expression patterns must also consider how these patterns are related to a cell’s or a tissue’s history. Transcriptional memory may allow qualitatively different transcriptional responses to a particular stimulus, by integrating signals with recent experience. If so, this mechanism might provide better responses to infection. However, it is also possible that this mechanism might contribute to pathological outcomes such as chronic inflammation. If so, transcriptional memory may represent a novel pathway for therapeutic intervention for the treatment of inflammatory disorders, which often involve mis-regulation of IFN-γ responsive genes.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/26209
References
- 1.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–25. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell. 2003;4:775–89. doi: 10.1016/S1534-5807(03)00162-X. [DOI] [PubMed] [Google Scholar]
- 3.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 4.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–62. doi: 10.1016/S0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 5.Fischer T, Strässer K, Rácz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–52. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–75. doi: 10.1016/S0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 7.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–23. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–54. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–82. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–78. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–65. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–12. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 13.Hediger F, Dubrana K, Gasser SM. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol. 2002;140:79–91. doi: 10.1016/S1047-8477(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 14.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–89. doi: 10.1016/S0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 15.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–98. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–39. doi: 10.1016/S0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 17.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan Q, Haroon S, Bravo DG, Will JL, Gasch AP. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics. 2012;192:495–505. doi: 10.1534/genetics.112.143016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faria AM, Levay A, Wang Y, Kamphorst AO, Rosa ML, Nussenzveig DR, Balkan W, Chook YM, Levy DE, Fontoura BM. The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity. 2006;24:295–304. doi: 10.1016/j.immuni.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–71. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–39. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–83. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol. 2010;30:2046–56. doi: 10.1128/MCB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–68. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 29.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–83. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–99. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 32.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–95. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 33.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell. 2012;23:1367–75. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–8. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]


