Abstract
Plant cell walls are composed of a large number of complex polysaccharides, which contain at least 13 different monosaccharides in a multitude of linkages. This structural complexity of cell wall components is paralleled by a large number of predicted glycosyltransferases in plant genomes, which can be grouped into several distinct families based on conserved sequence motifs (B. Henrissat, G.J. Davies [2000] Plant Physiol 124: 1515-1519). Despite the wealth of genomic information in Arabidopsis and several crop plants, the biochemical functions of these coding regions have only been established in a few cases. To lay the foundation for the genetic and biochemical characterization of putative glycosyltransferase genes, we conducted a phylogenetic and expression analysis on 10 predicted coding regions (AtGT11-20) that are closely related to the MUR3 xyloglucan galactosyltransferase of Arabidopsis. All of these proteins contain the conserved sequence motif pfam 03016 that is the hallmark of the β-d-glucuronosyltransferase domain of exostosins, a class of animal enzymes involved in the biosynthesis of the extracellular polysaccharide heparan sulfate. Reverse transcriptase-polymerase chain reaction and promoter:β-glucuronidase studies indicate that all AtGT genes are transcribed. Although six of the 10 AtGT genes were expressed in all major plant organs, the remaining four genes showed more restricted expression patterns that were either confined to specific organs or to highly specialized cell types such as hydathodes or pollen grains. T-DNA insertion mutants in AtGT13 and AtGT18 displayed reductions in the Gal content of total cell wall material, suggesting that the disrupted genes encode galactosyltransferases in plant cell wall synthesis.
The plant cell wall represents a complex extracellular matrix that functions both in the control of expansion growth and the mechanical strength of the plant body (Carpita and Gibeaut, 1993). The primary (i.e. growing) cell wall is composed of three interconnected networks of polysaccharides that are classified as cellulose, hemicelluloses, and pectins (McNeil et al., 1984; Bacic et al., 1988; McCann and Roberts, 1991). Cellulose is a linear (1→4)-β-d-glucan, which is organized into load-bearing microfibrils typically consisting of 36 parallel polysaccharide chains (Delmer, 1999). These microfibrils are coated and cross-linked by hemicelluloses (primarily xyloglucans [XyGs] and glucuronoarabinoxylans) that strongly bind to cellulose via hydrogen bond interactions. The cellulose-hemicellulose network is in turn embedded in a negatively charged matrix of pectic material encompassing homogalacturonans and rhamnogalacturonans (RGs) I and II. In addition to these polysaccharides, plant cell walls contain proteoglycans and structural proteins, many of which are heavily glycosylated (Cassab, 1998; Gaspar et al., 2001). One distinguishing feature of plant cell wall components is the large variety of glycosidic linkages connecting many different neutral and acidic monosaccharides. Unlike most bacterial surface polymers, plant cell wall polysaccharides are not assembled from defined building blocks but are synthesized by a multitude of glycosyltransferases that act on backbone structures formed by polysaccharide synthases. For this reason, the side chains of many cell wall glycans do not have a specific subunit structure but can only be defined by their overall monosaccharide composition and the types of linkages between them.
One of the main challenges in plant cell wall research is the characterization of glycosyltransferases in polysaccharide synthesis to understand their catalytic properties and the functional significance of the glycosidic linkages they establish. Substantial progress has been made in the identification of plant cellulose synthase (CesA) genes using both mutant approaches and sequence similarities to bacterial counterparts (Pear et al., 1996; Richmond and Somerville, 2000; Williamson et al., 2001). Although the Arabidopsis genome contains 10 distinct CesA genes, genetic evidence suggests that there is little functional redundancy within this gene family. This can be ascribed partially to the observation that cellulose in primary walls is synthesized by different enzymes than cellulose in secondary walls (Arioli et al., 1998; Taylor et al., 1999; Fagard et al., 2000; Caño-Delgado et al., 2003). Furthermore, there is substantial evidence that the cellulose-synthesizing complexes in the plasma membrane require more than one CesA isoform to be functional (Taylor et al., 2000; Perrin, 2001).
Biochemical approaches have led recently to the identification of XyG fucosyltransferase genes in Arabidopsis and pea (Pisum sativum; Perrin et al., 1999; Faik et al., 2000) and a galactomannan galactosyltransferase gene in fenugreek (Trigonella foenum-graecum; Edwards et al., 1999). Interestingly, an Arabidopsis homolog of the fenugreek enzyme acts as a cellopentaose xylosyltransferase in vitro, establishing the same type of α-(1→6) linkage found in XyGs. For this reason, the Arabidopsis gene (AtXT1) is presumed to encode a xylosyltransferase in XyG biosynthesis (Faik et al., 2002). Both XyG fucosyltransferase (AtFUT1) and AtXT1 belong to multigene families with 10 and seven members, respectively (Sarria et al., 2001; Faik et al., 2002). Because a mutation in AtFUT1 eliminates XyG fucosylation throughout the plant, this gene does not appear to be functionally redundant with any of the other AtFUT family members (Vanzin et al., 2002; Perrin et al., 2003). Considering that Fuc residues have been found in RG-I, RG-II, and arabinogalactan-proteins (AGPs) from Arabidopsis (Zablackis et al., 1995; van Hengel and Roberts, 2002), it is tempting to speculate that members of the AtFUT gene family encode the respective fucosyltransferases with little if any genetic redundancy. Similar considerations apply to the six AtXT1 homologs in the Arabidopsis genome because the XyG core structure contains three Xyl residues, which may be attached by separate xylosyltransferases.
To identify cell wall-related coding regions by a genetic approach, we isolated Arabidopsis mutants with changes in the monosaccharide composition of total cell wall material leading to the identification of 11 complementation groups (mur1-mur11; Reiter et al., 1993, 1997). The MUR1 and MUR4 genes were shown to encode nucleotide sugar interconversion enzymes in the de novo synthesis of GDP-l-Fuc and UDP-l-Ara, respectively (Bonin et al., 1997; Burget et al., 2003), whereas mur2 is defective in XyG fucosyltransferase AtFUT1 (Vanzin et al., 2002). The mur3 mutation has been shown recently to eliminate galactosylation of the third Xyl residue within the XXXG core structure of XyG, whereas galactosylation of the second Xyl residue is markedly enhanced (Madson et al., 2003). Because of the substrate specificity of XyG fucosyltransferase, mur3 plants lack the entire α-l-fucosyl-(1→2)-β-d-galactosyl-(1→2) side chain that is normally attached to the XXXG core structure of most XyGs. Positional cloning of the MUR3 gene and enzyme assays on the recombinant MUR3 protein revealed that it represents a XyG galactosyltransferase specific for the third Xyl residue within the XXXG repeat unit, a finding that is consistent with the altered XyG structure of mur3 plants (Madson et al., 2003). The MUR3 protein contains the structural motif pfam03016 that represents the glucuronosyltransferase domain of exostosins. This class of animal enzymes catalyzes the formation of the extracellular matrix component heparan sulfate, a glycosaminoglycan that is initially synthesized as a linear chain of alternating GlcA and GlcNAc units, and later modified by epimerization and sulfation events (Esko and Selleck, 2002). As outlined in the results section of this contribution, MUR3 belongs to a multigene family of putative glycosyltransferases encompassing 10 closely related members (AtGT11-20) and 28 more distantly related coding regions. Here, we describe the transcriptional and mutational analysis of selected members of the AtGT gene family, which will lay the foundation for the functional characterization of these putative glycosyltransferases in plant cell wall synthesis.
RESULTS
Identification of a Family of Arabidopsis Genes Homologous to the MUR3 XyG Galactosyltransferase
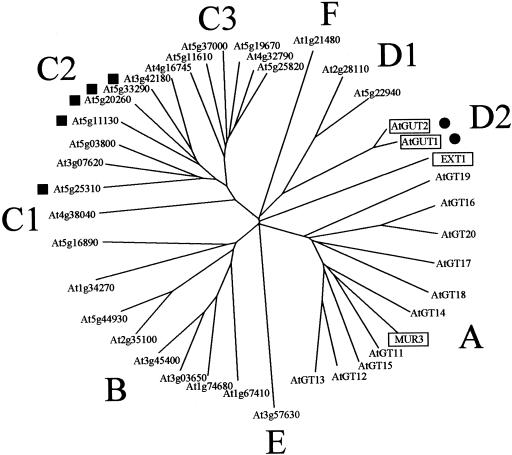
PSI-BLAST searches (Altschul et al., 1997) with the MUR3 protein as the query sequence identified 38 coding regions within the Arabidopsis genome with significant sequence similarity to MUR3. All of these putative proteins belong to glycosyltransferase family 47 as defined by Henrissat and Davies (2000; see http://afmb.cnrs-mrs.fr/CAZY/) and contain the structural motif pfam03016, which represents the glucuronosyltransferase domain of animal exostosins. A phylogenetic tree generated with the amino acid sequences of the GT47 family reveals several subgroups, which were designated A, B, C1, C2, C3, D1, D2, E, and F (Fig. 1). Of these, groups C1, E, and F contain only a single member. Most of the putative proteins are predicted to contain a single transmembrane domain at the N terminus, suggesting that they are targeted to a membrane system such as the endoplasmic reticulum or the Golgi. Gene products At2g31990 (AtGT15) in group A and At3g07620 in group C2 are predicted to contain two and three transmembrane domains, respectively. Five of the GT47 family members (all of them in group C2) are predicted to lack signal peptides or transmembrane domains, and the two proteins within group D2 (AtGUT1 and AtGUT2) are predicted to contain a signal peptide for secretion but to lack a membrane anchor. Both of these proteins are >90% identical to the putative RG-II glucuronosyltransferase from N. plumbaginifolia (Iwai et al., 2002) but contain N-terminal extensions of 74 and 71 amino acids, respectively, when compared with the N. plumbaginifolia enzyme. Gene product At1g21480 (group F) is predicted by SignalP-HMM to contain a signal anchor sequence (72% probability) but received only a 15% probability score for a transmembrane domain by the TMHMM algorithm.
Figure 1.
Phylogenetic tree of the GT47 family members of Arabidopsis. The alignment was generated using ClustalX, and the branching pattern was visualized by TreeView. Squares, Gene products predicted to be cytoplasmic by both the SignalP-HMM and Transmembrane Hidden Markov model (TMHMM) algorithms; circles, gene products predicted to have a cleaved signal peptide. All other putative proteins are likely to contain at least one transmembrane domain. AtGUT1 and AtGUT2 are close homologs to the putative RG-II glucuronosyltransferase from Nicotiana plumbaginifolia (Iwai et al., 2002), and EXT1 denotes the glucuronosyltransferase domain of exostosin 1 from humans (Homo sapiens). Proteins with known (EXT1 and MUR3) or predicted functions (At-GUT1 and AtGUT2) are boxed.
Subgroup A of GT47 contains MUR3 and 10 closely related sequences, which were designated AtGT11 through AtGT20 for Arabidopsis glycosyltransferases 11 through 20. Pair-wise comparisons of the amino acid sequences of the putative catalytic domain of these proteins showed between 31% and 73% identity (Table I; Fig. 2). Although all of these proteins were predicted to contain an N-terminal membrane anchor and a conserved globular domain, substantial length differences were observed in the spacer region that separates these two main structural elements (Fig. 3). We also noticed a substantial variability in the lengths of the N-terminal sequences preceding the transmembrane domain and the carboxy-terminal sequences extending beyond the conserved catalytic domains (Fig. 3). The 10 MUR3 paralogs are dispersed over four chromosomes with the exception of AtGT12/AtGT13, which are arranged in tandem on chromosome II. AtGT12, AtGT13, AtGT14, AtGT16, AtGT18, AtGT19, and AtGT20 are predicted to lack introns, whereas AtGT11, AtGT15, and AtGT17 are predicted to contain one intron. Reverse transcriptase-polymerase chain reaction (RT-PCR) results supported the intron/exon structure of these genes (data not shown). In the case of AtGT11, AtGT12, AtGT14, AtGT15, and AtGT16, we noticed discrepancies in the predicted protein length between the current annotation by the Arabidopsis Genome Initiative (AGI) and the coding regions predicted by the GlimmerM algorithm. In all of these cases, GlimmerM predicted translation initiation at an AUG codon upstream of the N-terminal Met in the AGI annotation. To determine the most likely coding regions, we conducted RT-PCR experiments with primers flanking the longest reading frame for each gene. With the exception of AtGT16, products of the expected lengths were obtained (data not shown), which suggests that the translation products predicted by GlimmerM are produced in vivo even though we cannot exclude the possibility that some AUG codons are bypassed during protein synthesis. RT-PCR data for AtGT16 supported the protein structure of the AGI annotation, which is by 23 amino acids shorter than the longest open reading frame. In case of AtGT14, two small introns are predicted in the AGI annotation, which would remove parts of the catalytic domain. However, an evaluation of this coding region by the GlimmerM algorithm suggested that AtGT14 is an intronless gene. This prediction was supported by a full-length cDNA in the Arabidopsis database (clone no. C103172; accession no. BT006172) and our own RT-PCR results (data not shown).
Table I.
Amino acid identities among Arabidopsis MUR3-like proteins The sequences shown in Figure 2 were used in pair-wise BLAST alignments to obtain the identity values.
| AtGT11 | AtGT12 | AtGT13 | AtGT14 | AtGT15 | AtGT16 | AtGT17 | AtGT18 | AtGT19 | AtGT20 | |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| MUR3 | 59 | 50 | 47 | 54 | 48 | 40 | 44 | 42 | 36 | 40 |
| AtGT11 | - | 52 | 49 | 55 | 51 | 39 | 45 | 40 | 38 | 39 |
| AtGT12 | - | - | 67 | 53 | 52 | 38 | 41 | 39 | 37 | 39 |
| AtGT13 | - | - | - | 50 | 50 | 38 | 37 | 37 | 37 | 38 |
| AtGT14 | - | - | - | - | 54 | 39 | 41 | 42 | 39 | 40 |
| AtGT15 | - | - | - | - | - | 39 | 37 | 39 | 39 | 39 |
| AtGT16 | - | - | - | - | - | - | 37 | 38 | 35 | 73 |
| AtGT17 | - | - | - | - | - | - | - | 41 | 31 | 38 |
| AtGT18 | - | - | - | - | - | - | - | - | 35 | 37 |
| AtGT19 | - | - | - | - | - | - | - | - | - | 34 |
Figure 2.
Amino acid sequence alignment between all members of Arabidopsis GT47 subgroup A corresponding to positions 151 to 538 in MUR3. Identical and similar amino acid residues are shaded in black and gray, respectively. Asterisks below the sequences denote amino acid residues that are completely conserved between the sequences.
Figure 3.
Comparison of structural elements between MUR3 and the AtGT gene products. Red, Predicted transmembrane domains; green, conserved catalytic domains (see Fig. 2). The highly conserved motif FCLQPX16GCIPV within this domain is marked in blue. Yellow, N- and C-terminal extensions and the spacer region between the transmembrane domain and the presumptive catalytic domain.
Expression Profiles of the MUR3-Like Genes
One approach to study the function of the MUR3 paralogs is to determine changes in the cell wall composition of insertion mutants. To be able to select appropriate tissues or organs for cell wall analysis, we determined the expression patterns of MUR3 and all 10 AtGT genes using RT-PCR analysis and promoter:β-glucuronidase (GUS) fusions. Semiquantitative RT-PCR results showed that all 10 MUR3 paralogs were expressed, although at different levels. Amplification products from leaves, stems, flowers, and roots were clearly visible for MUR3, AtGT15, AtGT17, and AtGT19 after 31 cycles, for AtGT11, AtGT13, and AtGT14 after 35 cycles, and for AtGT12, AtGT16, AtGT18, and AtGT20 after 40 cycles (Fig. 4). Only small amounts of RT-PCR products were detected in leaves and roots for AtGT12 and AtGT20, even after 40 cycles.
Figure 4.
Expression of MUR3 and its 10 close homologs in 3-week-old leaves (L) and one-month-old stems (St), flowers (F), and roots (R). Total RNA (1 μg) was used for RT-PCR with gene-specific primers. RT-PCR amplification of the gene for the translation elongation factor EF1αA4 was used as an internal control. RT-PCR products shown for AtGT15, AtGT17, and AtGT19 were amplified for 31 cycles, RT-PCR products shown for AtGT11, AtGT13, and AtGT14 were amplified for 35 cycles, and RT-PCR products shown for AtGT18, AtGT16, AtGT12, and AtGT20 were amplified for 40 cycles.
To obtain gene expression data at a higher resolution, promoter:GUS fusions for all 10 MUR3 paralogs were introduced into Arabidopsis plants. Staining results indicated that the AtGT genes could be divided into three groups in regard to their GUS expression patterns. The first group encompasses AtGT11, AtGT13, AtGT14, AtGT15, AtGT18, and AtGT19, whose transgenic plants showed GUS activities throughout the young seedlings including cotyledons, hypocotyls, true leaves, and roots. These genes were also expressed in the inflorescences including flowers and siliques (Figs. 5 and 6). AtGT13: GUS showed a gradient of activity in the root, with most intense staining in the oldest regions, whereas no GUS activity was observed in the root tips and lateral roots (Fig. 5). In the case of AtGT14 transgenic lines, no GUS activity could be detected in the hypocotyls of 1-week-old seedlings, although high GUS activities were observed in the hypocotyls of seedlings 2 weeks after germination (Fig. 5). Interestingly, the AtGT genes showed distinct expression patterns within flowers. For example, AtGT19:GUS showed highest activity in the stamens, especially the pollen grains (Fig. 6), whereas AtGT14:GUS was expressed strongly in both stamens and carpels (Fig. 5). AtGT11: GUS and AtGT15:GUS were strongly expressed in the sepals, whereas AtGT13:GUS was expressed primarily in petals and carpels (Fig. 5).
Figure 5.
GUS activities within Arabidopsis plants transformed with promoter:GUS fusions for AtGT11, AtGT12, AtGT13, AtGT14, and AtGT15. Two to three-week-old seedlings, 2-week-old roots, and 5-week-old inflorescences were stained and photographed. Insets in the inflorescence panels show flowers and anthers at higher magnifications, and the inset in the AtGT14 seedling panel shows a plantlet at the cotyledon stage.
Figure 6.
GUS activities of Arabidopsis plants transformed with promoter:GUS fusion for AtGT16, AtGT17, AtGT18, AtGT19, and AtGT20. Two to 3-week-old seedlings, 2-week-old roots, and 5-week-old inflorescences were stained and photographed. The inset in the “AtGT19 inflorescence” panel shows intense staining of pollen grains, and the inset in the “AtGT20 seedling” panel shows staining of hydathodes.
The second group of genes encompasses AtGT16 and AtGT17, where GUS activities were observed in the roots and parts of the rosette but not in the inflorescences (Fig. 6). Strong AtGT16:GUS activities were found in young leaves and older parts of the roots, whereas AtGT17:GUS activities were detected in the hypocotyls and vascular tissue of the primary roots.
The third group of genes encompasses AtGT12 and AtGT20, whose GUS expression was restricted to specific tissues: AtGT12 transgenic plants showed GUS activities exclusively in the pollen grains (Fig. 5), whereas AtGT20:GUS was only detected in hydathodes (Fig. 6).
Identification of T-DNA-Tagged Mutants
To obtain lines with gene disruptions in the MUR3 paralogs, we screened collections of T-DNA insertion mutants at the Salk Institute Genome Analysis Laboratory (La Jolla, CA) and the University of Wisconsin knockout facility (Madison). Homozygous lines with insertions in exons were obtained in case of AtGT13 and AtGT18, leading to gene disruptions 172 and 1,120 bp downstream of the predicted ATG initiation codons, respectively (Fig. 7A). Homozygosity of the mutant lines was verified by PCR with gene-specific primers flanking the T-DNA inserts, which resulted in products of 6.9 and 7.9 kb for AtGT13 and AtGT18, respectively, whereas no PCR products corresponding to the wild-type fragments (1.4 and 2.0 kb, respectively) could be identified in the mutant lines (Fig. 7B).
Figure 7.
A, Schematic diagram of locations of T-DNA inserts in the AtGT13 and AtGT18 coding regions; B, Identification of homozygous mutants of atgt13 and atgt18. Primers flanking the T-DNA insertions were used for PCR amplification from genomic DNA.
Cell Wall Composition of the atgt13 and atgt18 Insertion Mutants
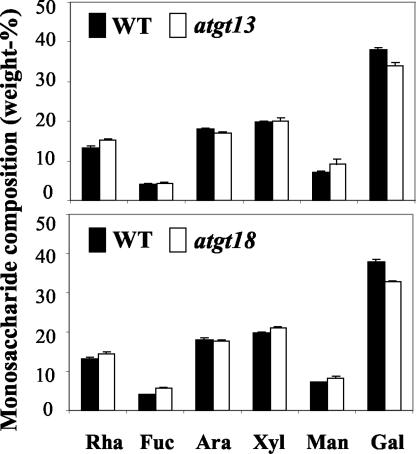
To determine whether the gene disruptions of AtGT13 and AtGT18 caused an alteration of the monosaccharide composition of total cell wall material, the relative amounts of neutral monosaccharides were determined for wild-type plants and both mutant lines by gas-liquid chromatography of alditol acetates. Leaf material was chosen for this analysis because of its availability in large quantities and because both genes are expressed in leaves based both on RT-PCR and promoter:GUS results. Compared with wild-type plants, the atgt13 and atgt18 mutants showed 10.3% and 13.5% reduction in Gal content, respectively, which was counterbalanced by slight increases in all other monosaccharides except Ara (Fig. 8).
Figure 8.
Monosaccharide content in leaf-derived cell wall matrix polysaccharides (pectins and hemicelluloses). Identical amounts of cell wall material were used for the wild type (black bars) and T-DNA insertion mutants (white bars). Bars = mean of 15 samples ± se.
RT-PCR experiments on RNA from atgt13 and atgt18 plants indicated the absence of transcripts from these genes in the mutant background, whereas amplification products were readily detectable in wild-type controls (data not shown). These results support the idea that the T-DNA insertions in these genes are responsible for the reduced Gal content. Nonetheless, we cannot rule out the possibility that the observed changes in cell wall composition are caused by unidentified background mutations. Complementation experiments with the respective wild-type alleles are currently under way to address this point.
DISCUSSION
The cell walls of higher plants are primarily composed of complex polysaccharides, which contain at least 13 different monosaccharides that are attached to each other in a vast array of linkages (Carpita and Gibeaut, 1993). Assuming that the formation of each unique linkage is catalyzed by a separate enzyme, it has been estimated that 53 distinct glycosyltransferases are needed to catalyze the synthesis of pectic cell wall components (Ridley et al., 2001). Additional glycosyltransferases are needed in the biosynthesis of cellulose, callose, and hemicellulosic polysaccharides such as XyG, glucuronoarabinoxylans, and (gluco) mannans. Finally, the apoplastic space contains a variety of heavily glycosylated proteins such as Hyprich glycoproteins and AGPs, which are expected to require a separate set of glycosyltransferases to catalyze the formation of protein-glycosyl junctions, and unique linkages within side chains. For this reason, it comes as no surprise that plant genomes contain several hundred coding regions for putative glycosyltransferases, which can be grouped into 35 distinct families based on conserved sequence motifs (Henrissat and Davies, 2000; Henrissat et al., 2001). Despite the high complexity of cell wall polymers, the number of coding regions is considerably higher than the bare minimum needed to catalyze the formation of all unique linkages. This may be explained by genetic redundancy, cell type-specific expression patterns, the involvement of heterooligomeric proteins in the formation of specific linkages, or the presence of pseudogenes.
As an initial step to characterize an interesting family of glycosyltransferase genes, we conducted an expression analysis and initial mutant characterization on a subgroup of family GT47 that contains the MUR3 XyG galactosyltransferase. All members of GT47 contain conserved protein domain pfam03016, which was originally defined as the signature motif of the β-(1→4)-d-glucuronosyltransferase domain of animal exostosins. Genetic data on the nolac H18 mutant of N. plumbaginifolia suggest that the mutated gene (GT47 member NpGUT1) encodes a β-(1→4)-d-glucuronosyltransferase involved in the synthesis of RG-II (Iwai et al., 2002). This is indicative of a conservation of the donor substrate and the type of linkage between related animal and plant enzymes. On the other hand, the MUR3 gene encodes a β-(1→2)-d-galactosyltransferase, which suggests that GT47 is a heterogeneous family of glycosyltransferases establishing a variety of linkages with several donor and acceptor substrates. The only common denominator between animal exostosins, NpGUT1 and MUR3, is the use of UDP sugars as donor substrates and the inversion of configuration during glycosyl transfer. Because the GT47 members selected for our study form a distinct subgroup within a larger family of enzymes (Fig. 1), we speculate that AtGT11 through AtGT20 encode galactosyltransferases establishing a β-linkage during glycosyl transfer. Considering that α-l-Ara is structurally identical to β-d-Gal except for the absence of the C-6 hydroxymethyl group, a function of MUR3 homologs as inverting l-arabinosyltransferases is also a strong possibility. One likely function for MUR3 homolog(s) is the attachment of Gal to the central Xyl residue within the XXXG core structure of XyG, i.e. the conversion of XXXG to XLXG and/or the conversion of XXLG to XLLG. There are also numerous β-d-galactosyl and α-l-arabinosyl linkages within the arabinogalactan chains of RG-I and AGPs, which may be formed by the MUR3-like AtGT gene products or other members of GT47.
The cell type-specific expression patterns within certain organs (Figs. 5 and 6) may reflect differences in the wall structures of certain cell types that have been observed via methods of immunocytochemistry (for review, see Knox, 1997) but may not be detectable by carbohydrate analysis of entire seedlings. Alternatively, isoforms of the same enzyme may be expressed in a tissue-specific manner. One argument against the latter possibility is that none of the AtGT gene products are very closely related to each other with the possible exception of AtGT12/AtGT13 and AtGT16/AtGT20 (see below).
RT-PCR results indicated that AtGT16 and AtGT17 are expressed in all major plant organs (Fig. 4); however, promoter:GUS fusions did not reveal significant reporter gene expression in inflorescences (Fig. 6). This may reflect insufficient sensitivity of the GUS assay or the presence of regulatory sequences that were not included in the reporter gene constructs. Similar considerations apply to AtGT12 and AtGT20, where GUS expression was limited to pollen grains and hydathodes, respectively (Figs. 5 and 6). In this case, RT-PCR results revealed amplification products only in RNA from stems and flowers at the highest number of cycles, which suggests that both genes are only weakly expressed. It is interesting in this context that AtGT12 shows 67% amino acid sequence identity to AtGT13, and AtGT20 shows 73% amino acid sequence identity to AtGT16 (Table I). Furthermore, AtGT12/AtGT13 are the only AtGT genes that are arranged as a tandem repeat indicating a recent duplication event. These results suggest that AtGT20 may be a cell type-specific isoform of AtGT16, and AtGT12 may be a cell type-specific isoform of AtGT13.
As a first step to elucidate the biochemical function of the AtGT gene products, we analyzed the monosaccharide composition of cell wall material from insertion mutants of AtGT13 and AtGT18. Leaf material was chosen for this purpose because both RT-PCR results and data from promoter:GUS fusions indicated expression in this organ. Both mutant lines showed a significant decrease in the Gal content of total cell wall material, which suggests that both genes encode galactosyltransferases, although more complex explanations cannot be ruled out. For instance, the atgt13 and atgt18 mutations may affect the synthesis of the acceptor substrate for galactosyltransferase(s), which would lead to a reduced Gal content as a secondary effect. One example for this scenario is the 50% reduction in Fuc content in Arabidopsis mur3. Although the MUR3 gene encodes a galactosyltransferase in XyG biosynthesis, the most dramatic alteration in cell wall monosaccharide composition is the relative amount of Fuc rather than Gal because the acceptor substrate for the AtFUT1 fucosyltransferase is missing in mur3 plants (Madson et al., 2003). We hope that linkage composition analyses and fractionation experiments on atgt13- and atgt18-derived cell wall material will permit us to narrow down the alterations to a single cell wall component. The altered polysaccharide then can be used as an acceptor substrate for glycosyltransferase assays to obtain data on the biochemical function(s) of the AtGT gene products.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants were grown in an environmental chamber at 23°C and 60% to 70% humidity under continuous fluorescent light (60-70 μmol m-2 s-1). Arabidopsis plants of the Columbia ecotype were used for transformation and isolation of DNA and RNA. T-DNA-mutagenized seeds of ecotypes Columbia and Wassilewskija were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Seeds were planted on either ProMix BX potting mixture or on nutrient agar plates (Haughn and Somerville, 1986).
Nucleic Acid and Protein Sequence Analysis
To identify coding regions within the AtGT genes, genomic sequences within the MAtDB database at the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/thal/db/index.html) were evaluated by the GlimmerM algorithm (http://www.tigr.org/software/glimmerm/) that has been trained for Arabidopsis genes. Derived protein sequences were evaluated for transmembrane domains using the TMHMM algorithm (http://www.cbs.dtu.dk/services/TMHMM/). Prediction of signal peptides and signal anchor sequences employed the SignalP-HMM method (Nielsen and Krogh, 1998; http://www.cbs.dtu.dk/services/SignalP-2.0/).
RT-PCR
For expression analysis, leaves from 3-week old plants and stems, flowers, and roots from 1-month old plants were harvested and frozen immediately in liquid nitrogen. Approximately 100 mg of tissue samples was ground in liquid nitrogen, and total RNA was extracted with the RNeasy plant mini kit (Qiagen, Valencia, CA) according to the instructions of the manufacturer. The crude RNA preparations were treated with 10 units each of RNase-free DNase I (Promega, Madison, WI) and further purified according to the RNeasy plant mini kit protocol. RT-PCR was carried out by using the OneStep RT-PCR kit (Qiagen). One microgram of DNA-free RNA was used as template. Reverse transcription was performed at 50°C for 30 min, followed by activation of HotStar Taq DNA polymerase within the reaction mixture at 95°C for 15 min. PCR amplification was conducted for up to 40 cycles using the following thermal profile: denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and polymerization at 72°C for 2 min, with a 10-min terminal extension step at 72°C. To determine whether comparable amounts of RNA had been used for RT-PCR from the different tissues, the EF1αA4 gene was used as a control (Nesi et al., 2000). Control reactions without RT were used to rule out contamination by genomic DNA. Primers used for RT-PCR were as follows: MUR3, 5′-ATGGAGAAGGGAAATGG-3′ and 5′-CTGTGTCTTATCTCTCTG-3′; AtGT11, 5′-CTGGGCATTGTTGTTGTACTTCCA-3′ and 5′-GTGTCTTATCTCTCTGCTCACTCT-3′; AtGT12, 5′-CAACTTCCTCATCTCCTTCGTTGCTTCTT-3′ and 5′-TCATTAACAATGTCTTAAGGCTTAAGCAG-3′; AtGT13, 5′-CAACACCTTTGTTCTCCATGCTCTTTCTG-3′ and 5′-TGCATCTTCAATCTTTTCCGGTCCAACCT-3′; AtGT14, 5′-CTCTGCTCTCTTCACCGACACAGA-3′ and 5′-ACTACCATCTTTCCCTTGTCTCATC-3′; AtGT15, 5′-CTTCTATTCTCTCTAGCACTC-3′ and 5′-TGACATGGATCATCAAATCAAG-3′; AtGT16, 5′-AAACCAGGCAAAAGCTCATGAACAA-3′ and 5′-GTTGAGACGACCAAGAACGTCCAAA-3′; AtGT17, 5′-CTCTTGCTTGCTTAGCTAGTG-3′ and 5′-AAACCGTCAAGTTGACACTTC-3′; AtGT18, 5′-AACATCGAGACTGTAAAGTAAGTAGGAAT-3′ and 5′-TCATTTAACTCGCTTCACTCGGTCGGAAA-3′; AtGT19, 5′-CTATACATAACTCTAGGCATCATCTCTA-3′ and 5′-ATCTCAGCCGTTGATTCAGAATGCACCAA-3′; and AtGT20, 5′-GTCTCCATATACCAATCCAGGCAAA-3′ and 5′-TTCAAAGAGAAAGTCAAGAACT-3′.
In the case of AtGT11, AtGT12, AtGT15, and AtGT16, additional primers were used to determine whether the coding regions predicted by GlimmerM were transcribed in their entirety. Primer sequences used for this purpose were as follows: AtGT11, 5′-CATCCTTCAGATCCAGAAATCA-3′ and 5′-CGATCGAAGTAAGGATCCC-3′; AtGT12, 5′-CTTCTCCTTAAGATTACATTTATTAA-3′ and 5′-TTAAGCAGTTTTTTCATGTATAAT-3′; AtGT15, 5′-GAGAGCCATACTACTCTGAC-3′ and 5′-CTAATCAGTTTTGAATTCGTTTC-3′; and AtGT16, 5′-ATGTCCCTATCAAAACATCTA-3′ and 5′-TTATACAGTTTTTGCAATCTTC-3′.
Plasmid Constructs and Cloning
For promoter:GUS constructs, approximately 2.5 kb upstream of the predicted ATG start codons were PCR amplified with gene-specific oligonucleotides containing a BamHI site engineered into the upstream primer, and an NcoI site was engineered into the downstream primer. This primer design was used for all genes except AtGT16, where a combination of SacI and SpeI was used. The sequences of the individual primers were as follows (engineered restriction sites are underlined): AtGT11, 5′-CAACAGGATCCACTCCCAATTTTGGCTTTCAGTTCTCAAGCA-3′ and 5′-AACGTCCATGGTCATTGCCATTGTTCCTCCTCTCCTATGTG-3′; AtGT12, 5′-CAACAGGATCCATGTATGGACGAGATCATTTCTTTGTCACG-3′ and 5′-AACGTCCATGGTCATCATCTTCATTTACTTGTGAGAAAACATTGG-3′; AtGT13, 5′-CAACAGGATCCTATTGGTACCCACTGATCTATTTCATTAGTTTG-3′ and 5′-AACGTCCATGGTCATTTTGATTATGGAGAAACGAAAGTGGTACTA-3′; AtGT14, 5′-CAACAGGATCCAACCATAAACGGTGTTACATCATATTAAAACCATA-3′ and 5′-AACGTCCATGGTCATCTGAGAATAATTCTTGGGTCGCATCAAA-3′; AtGT15, 5′-CAACAGGATCCACTGCCTGCGTTTCAAGGAACAGTTTATAATC-3′ and 5′-AACGTCCATGGTCATGGTGTTCTTGATTTGATGATCCATGTCA-3′; AtGT16, 5′-CCACAGAGCTCTCAGAGACGACGAAGAGAACCCTGCCTGGC-3′ and 5′-AACGTACTAGTCATCTGGACATGCACTCACGTCGTCACATAATTG-3′; AtGT17, 5′-CAACAGGATCCGAAGCTAGGAATAGAAGTCTAGTAGCTAGGTTA-3′ and 5′-AACGTCCATGGTCATGGTAAATGGAGAGAGAGAGAGAAATATG-3′; AtGT18, 5′-CAACAGGATCCTGGTGTATAAAATACTGCAGTCTATTGAACTA-3′ and 5′-AACGTCCATGGTCATAGTTTATAATTAGCTGAAAATGAGATTA-3′; AtGT19, 5′-CAACAGGATCCTGGAGAAGACAATGAGACAATTTTGGTTAGTA-3′ and 5′-AACGTCCATGGTCATTGTTATGGATGTTGTCGGAGTGAGA-3′; and AtGT20, 5′-CAACAGGATCCGGTTAGTGCTATAACTCTTTCCATCTCTACTG-3′ and 5′-AACGTCCATGGTCATTTTTCTGACTTGGTCCTTTACTTCTTCAC-3′.
PCR reactions were carried out on total chromosomal DNA with TaKaRa EX Taq polymerase (PanVera, Madison, WI) under the following conditions: an initial denaturation step at 96°C for 1 min followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, polymerization at 72°C for 2.5 min, and a final extension at 72°C for 10 min.
After cleavage with the appropriate restriction enzymes, PCR products were cloned in frame with the GUS reporter gene into the pCAMBIA1301 plant transformation vector (CAMBIA, Canberra, Australia), except for the AtGT16-derived PCR product, which was cloned into pCAMBIA1303. To verify the integrity of the constructs, the vector-insert junctions were sequenced before transformation into Arabidopsis. Primers used for sequence analysis were gene-specific sense primers and the pCAMBIA1301 vector primer 5′-AAATAGATCAGTTTAAAGAAAGATCAAAGCT-3′. In case of the AtGT16:GUS construct, the gene-specific primer 5′-TCTGCTTCGTTTTCTTCTCGTATA-3′ was used to verify the junction sequence between the promoter region and the pCAMBIA1303 vector. All plasmids were introduced into Arabidopsis by Agrobacterium tumefaciens-mediated transformation according to the method of Bechtold et al. (1993). A. tumefaciens strain GV3101 (pMP90) was used in all cases.
Histochemical GUS Assays
Arabidopsis plants transformed with the promoter:GUS constructs were selected on one-half-strength Murashige and Skoog media (Sigma, St. Louis) containing 0.8% (w/v) Bacto agar (Difco Laboratories, Detroit), 2% (w/v) Suc, 50 μg mL-1 hygromycin B (Calbiochem, La Jolla, CA), and 500 μg mL-1 vancomycin (Wako Pure Chemical Industries, Ltd., Osaka). Resistant T1 seedlings were transferred to soil to produce T2 seeds. GUS activities of transgenic plants were analyzed by using a protocol adapted from Jefferson et al. (1987).
Mutant Screening
The collection of T-DNA insertion mutants at the University of Wisconsin (Sussman et al., 2000) was screened by PCR using the AtGT18 gene-specific primers 5′-CAATTTTACCTAGAGTAATCAACGTGTCA-3′, and 5′-GACGGATTCTAATCTCATTTTCAGCTAAT-3′, in combination with the T-DNA-specific primers JL-202 and JL-270 according to the instruction manual at http://www.biotech.wisc.edu/Arabidopsis. PCR products were analyzed by Southern-blot and DNA sequence analysis as specified by the knockout facility. A search of the database at the Salk Institute Genome Analysis Laboratory (SIGnAL; http://signal.salk.edu) revealed a T-DNA insertion within the coding region of AtGT13 (mutant line Salk_053593). The precise location of the insert was determined by DNA sequence analysis of a PCR amplification product that was obtained with the AtGT13 sense primer 5′-ACCACTTTCGTTTCTCCATAATCAA-3′ and the T-DNA left border primer LBb1 as specified by the SIGnAL instruction manual. PCR amplification was carried out with 0.2 mm dNTPs, 0.4 μm of each primer, 1× EX-Taq buffer, and 1 unit of TaKaRa EX Taq polymerase (PanVera) in a volume of 25 μL. The amplification program consisted of an initial denaturation at 96°C for 1 min followed by 35 to 40 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, polymerization at 72°C for 90 s, and a final extension at 72°C for 10 min. DNA sequencing was carried out by with a CEQ 2000 Dye Terminator Cycle Sequencing Kit and a CEQ 2000XL DNA sequencer (Beckman, Fullerton, CA) under the conditions specified by the vendor. Homozygous insertion mutants in AtGT13 were identified by PCR amplification with gene-specific primers (sense primer as described above and the reverse primer 5′-CCTAGCCACTGCAATTTCAAATGCATCTT-3′). In case of AtGT18, the two gene-specific primers described above were used. PCR was carried out with 0.2 mm dNTPs, 0.4 μm each of the primers, 1× Advantage 2 PCR buffer, and 1 unit of Advantage 2 DNA polymerase mix (BD Biosciences CLONTECH, Palo Alto, CA) in a volume of 25 μL. The amplification program consisted of an initial denaturation at 96°C for 1 min followed by 36 cycles of denaturation at 94°C for 30 s, annealing and extension at 68°C for 1 min kb-1, and a final extension at 68°C for 10 min.
Determination of Cell Wall Composition
To determine the cell wall composition of wild-type and mutant lines, plants were grown for 3 to 4 weeks, and two leaves from 15 plants per line were harvested and analyzed separately (n = 15). Hydrolysis of leaf material and quantification of monosaccharides via gas-liquid chromatography of alditol acetates were carried out as described by Reiter et al. (1993). The experiments were repeated several times with similar results.
Acknowledgments
We thank the Center for the Application of Molecular Biology to International Agriculture for plant transformation vectors and the Arabidopsis Biological Resource Center for seed stocks.
This work was supported by the National Science Foundation (grant no. IBN-0215535) and by a fellowship from the Danish Agricultural and Veterinary Research Council (grant no. SJVF 23000237 to M.M.).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717-720 [DOI] [PubMed] [Google Scholar]
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 14. Academic Press, New York, pp 297-371 [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194-1199 [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter W-D (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose 4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA 94: 2085-2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget EG, Verma R, Mølhøj M, Reiter W-D (2003) The biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgilocalized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15: 523-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351-362 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Cassab GI (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49: 281-309 [DOI] [PubMed] [Google Scholar]
- Delmer DP (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245-276 [DOI] [PubMed] [Google Scholar]
- Edwards ME, Dickerson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG (1999) Molecular characterization of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J 19: 691-697 [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB (2002) Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71: 435-471 [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refrégier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an α-xylosyltransferase involved in XyG biosynthesis. Proc Natl Acad Sci USA 99: 7797-7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Bar-Peled M, DeRocher AE, Zeng WQ, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K (2000) Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem 275: 15082-15089 [DOI] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ (2001) The complex structures of arabinogalactan-proteins and the journey toward understanding function. Plant Mol Biol 47: 161-176 [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430-434 [Google Scholar]
- Henrissat B, Coutinho PM, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55-72 [PubMed] [Google Scholar]
- Henrissat B, Davies GJ (2000) Glycoside hydrolases and glycosyltransferases: families, modules, and implications for genomics. Plant Physiol 124: 1515-1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H, Masaoka N, Ishii T, Satoh S (2002) A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci USA 99: 16319-16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP (1997) The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int Rev Cytol 171: 79-120 [DOI] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter W-D (2003) The MUR3 gene of Arabidopsis thaliana encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662-1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Roberts K (1991) Architecture of the primary cell wall. In CW Lloyd, ed, The Cytoskeletal Basis of Plant Growth and Form. Academic Press, London, pp 109-129
- McNeil M, Darvill A, Fry SC, Albersheim P (1984) Structure and function of the primary cell wall of plants. Annu Rev Biochem 53: 625-663 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863-1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Krogh A (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6). AAAI Press, Menlo Park, CA, pp 122-130 [PubMed]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637-12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, Jia Z, Wagner TA, O'Neill MA, Sarria R, York WS, Raikhel NV, Keegstra K (2003) Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol 132: 768-778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM (2001) Cellulose: how many cellulose synthases to make a plant? Curr Biol 11: R213-R216 [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976-1979 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple C, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335-345 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple C, Somerville CR (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032-1035 [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124: 495-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley BL, O'Neill M, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929-967 [DOI] [PubMed] [Google Scholar]
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra K, Raikhel NV (2001) Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase 1. Plant Physiol 127: 1595-1606 [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529-2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769-780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K (2002) Fucosylated arabinogalactan-proteins are required for full root elongation in Arabidopsis. Plant J 32: 105-113 [DOI] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter W-D (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340-3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Hocart CH (2001) Cellulose synthesis: mutational analysis and genomic perspectives using Arabidopsis thaliana. Cell Mol Life Sci 58: 1475-1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Müller B, Darvill AG, Albersheim P (1995) Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107: 1129-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]