During meiosis, chromosomes undergo the remarkable choreography of developmentally programmed breakage, pairing with their homologous partners and subsequent DNA exchange. The goal of this elaborate process is to halve chromosome content, thereby producing haploid germ cells (sperm and eggs). Self-inflicted DNA damage in meiosis comes in the form of DNA double-stranded breaks (DSBs), and its subsequent repair is the prerequisite for the stable pairing of homologous chromosomes in mammals and many other organisms studied to date.
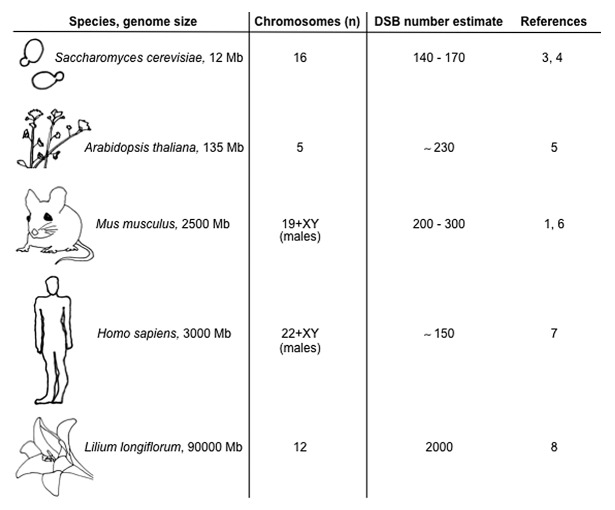
Meiotic DSBs are catalyzed by the protein SPO11, a topoisomerase-like enzyme that is essential for fertility. The number of SPO11-induced DSBs in meiotic cells is astonishingly high: on average, more than 200 DSBs per nucleus are formed in mice (Fig. 1) (for comparison, 1 Gy of ionizing radiation causes approximately 20–40 DSBs, and 2 Gy can be lethal to somatic cells). Ensuring that at least one crossover forms per chromosome pair is one reason meiotic DSBs form in such great numbers.1 However, only about 10% of DSBs mature into crossovers in mice, while the vast majority is resolved as non-crossovers,1 so crossover assurance is unlikely to be the only reason. Surprisingly little attention has focused on the other essential (pre-crossover) function of DSBs, namely their role in promoting homolog pairing.
Figure 1. DSB numbers in different species, listed by genome size. Note that there are organisms, not listed here and exemplified by Caenorhabditis elegans and Drosophila melanogaster, where homolog synapsis can be achieved in the absence of DSBs.
Recently, we explored the DSB numerical requirements for chromosome pairing in mammals by taking advantage of a transgenic mouse model with reduced SPO11 levels.2 Compared with control animals, spermatocytes of these mice formed approximately half the number of DSBs, as judged by quantitative analysis of cytological DSB markers and SPO11-oligonucleotide complexes, a by-product of meiotic DSB formation. This reduced DSB level led to severe defects in homolog pairing, which manifested as a mix of illegitimate synapsis between nonhomologous chromosomes combined with outright failure of pairing/synapsis. Mice were sterile, apparently as a consequence of these defects.2
We also uncovered intriguing differences between chromosomes with respect to sensitivity to DSB reduction: large autosomes were less prone to pairing defects than the X chromosome and small autosomes.2 The propensity of the X chromosome to illegitimately synapse with nonhomologous partners may be explained by its lack of pairing partner in male meiosis: pairing and recombination are restricted to the tiny pseudoautosomal region, the only area with homology with the Y chromosome. When fewer than normal DSBs are formed, autosomal pairing is compromised in a stochastic manner that varies from cell to cell. In contrast, the long unpaired segment of the X is available for aberrant behavior in every cell, and could in fact trigger a chain reaction of illegitimate pairing involving a subset of autosomes.
What might be the absolute lower limit for DSB numbers that still can support successful chromosome synapsis? Our data do not provide a direct answer, but one important finding was that the DSB dose that small autosomes receive is a limiting factor. In light of these results, it is interesting to compare DSB numbers across species in which homologous pairing is known or inferred to be dependent on recombination (Fig. 1). Clearly, neither physical genome size nor chromosome number nor karyotype alone can explain these phylogenetic differences. Instead, constraints are likely posed by the combination of all three. Budding yeast and mice, for instance, make similar numbers of DSBs despite vastly different genome sizes. Perhaps in yeast, as in mice, the low-end threshold for DSB numbers is dictated by the smallest chromosomes that need to receive just enough breaks. One must further take into account inter-species differences in the ability to cope with consequences of having “too few” DSBs, for example the propensity to non-homologous synapsis and potential back-up mechanisms for promoting disjunction of achiasmate chromosomes. Therefore, observations cannot be extrapolated readily across species, and direct experimental approaches in each organism continue to uncover previously unappreciated features of meiotic chromosome behavior.3-8
Glossary
Abbreviations:
- DSBs
double-stranded breaks
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26079
References
- 1.Cole F, et al. Nat Cell Biol. 2012;14:424–30. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kauppi L, et al. Genes Dev. 2013;27:873–86. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen SY, et al. Dev Cell. 2008;15:401–15. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancera E, et al. Nature. 2008;454:479–85. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vignard J, et al. PLoS Genet. 2007;3:1894–906. doi: 10.1371/journal.pgen.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plug AW, et al. Proc Natl Acad Sci U S A. 1996;93:5920–4. doi: 10.1073/pnas.93.12.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barlow AL, et al. EMBO J. 1997;16:5207–15. doi: 10.1093/emboj/16.17.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terasawa M, et al. Genes Dev. 1995;9:925–34. doi: 10.1101/gad.9.8.925. [DOI] [PubMed] [Google Scholar]