Abstract
In order to resorb the mineralized bone extracellular matrix, the osteoclast relies on the generation of a resorption lacuna characterized by the presence of specific proteases and a low pH. Hence, bone resorption by osteoclasts is highly dependent on lysosomes, the organelles specialized in intra- and extracellular material degradation. This is best illustrated by the fact that multiple forms of human osteopetrosis are caused by mutations in genes encoding for lysosomal proteins. Yet, until recently, the molecular mechanisms regulating lysosomal biogenesis and function in osteoclasts were poorly understood. Here we review the latest developments in the study of lysosomal biogenesis and function in osteoclasts with an emphasis on the transcriptional control of these processes.
Keywords: PKCβ, RANKL, TFEB, bone resorption, lysosomal biogenesis, lysosome, osteoclast
Introduction
Bone remodeling is the physiological process by which bone extracellular matrix (ECM) is constantly renewed. It implies the coordinated activity of 2 highly specialized cell types: the osteoblast, responsible for de novo bone matrix formation,1 and the osteoclast, responsible for the resorption of the old matrix.2 An imbalance in the activity of these 2 cells can lead to severe bone diseases. In particular, an excess of bone resorption over bone formation is invariably associated with the most common bone disease, osteoporosis. Hence, it is essential to understand the molecular mechanisms regulating osteoclast activity in order to develop targeted therapies for this and other bone disorders. The molecular elucidation of various forms of osteopetrosis, a rare genetic disease caused by an absence of osteoclast or by a defect in their function,3 has highlighted the critical importance of lysosomal proteins to the osteoclast function.4,5 These molecular genetic studies of rare human diseases coupled with additional work performed in cell culture or in genetically modified mouse models have recently uncovered signaling pathways implicated in the regulation and in the biogenesis of lysosomes in osteoclasts. This review aims at giving an overview of the significance of the regulation of lysosomal biogenesis and function in this cell type.
Lysosomes and Osteoclast Physiological Functions
Osteoclasts are large multinucleated cells of hematopoietic origins specialized in the removal of the mineralized bone extracellular matrix (ECM). Osteoclast resorptive function is absolutely necessary for normal bone growth, i.e., for bone modeling,6 and for the renewing of bone mass in adult vertebrates, i.e., for bone remodeling.2 Bone resorption by osteoclasts is also required to maintain calcium and phosphate ions homeostasis.7 Additionally, it has been proposed that osteoclast resorptive function may play a role in the formation of hematopoietic stem cell niches in the bone marrow.8-13 Furthermore, it was recently shown that osteoclasts are implicated in the control of glucose metabolism and male fertility by regulating osteocalcin, a bone-derived hormone. Indeed, osteocalcin is activated by osteoclast through a non-enzymatic decarboxylation, which is dependent on the extracellular acid pH generated during bone resorption.14,15 Finally, a recent study has suggested that the bone resorptive activity of osteoclast was implicated in the regulation of bone formation by osteoblasts.16 Hence, several physiological functions depend on the resorptive capacity of this cell.
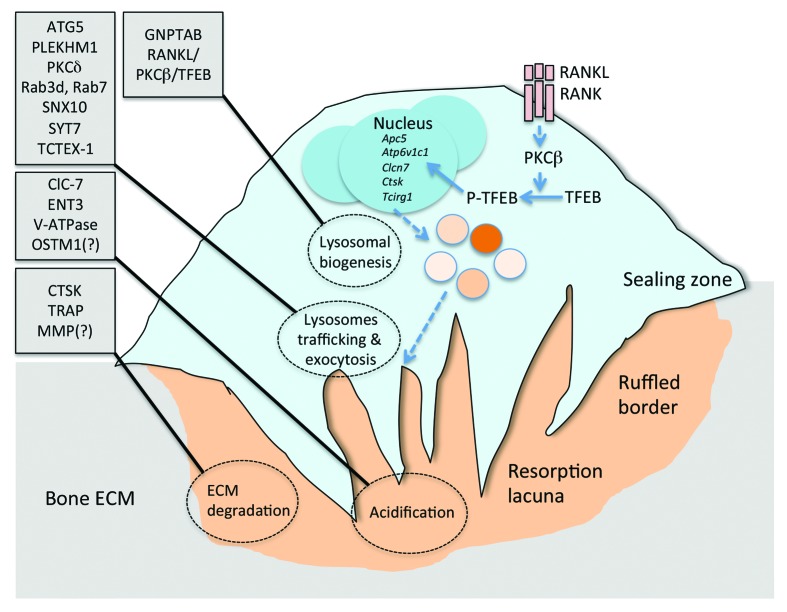
In order to resorb the bone ECM, the osteoclast attaches tightly to the bone surface, creating a sealing zone (see Fig. 1). This process, necessary for the activation and the polarization of the osteoclast, implicates integrin-dependent signaling and multiple cytoskeleton remodeling events.17 Following attachment and activation, a ruffled border is created within the sealing zone. This ruffled border is characterized by the presence of proton pumps and chloride channels that allow the acidification of the resorption lacuna, which then develops underneath the ruffled border.18 It is generally accepted that the formation of the ruffled border and thereby the resorption lacuna occurs through the simultaneous fusion and secretion of numerous intracellular vesicles.19 The presence of several lysosomal proteins at the ruffled border and in the lacuna and the acidic pH of this lacuna suggest that most of these vesicles must be of lysosomal origin.
Figure 1. Lysosomal function, regulation, and biogenesis in osteoclasts. A bone-resorbing osteoclast is schematized. Proteins implicated in lysosomal biogenesis, in lysosomes trafficking and exocytosis, in ECM acidification, and in ECM degradation are indicated on the left. The recently identified pathway linking RANKL signaling, PKCβ, and TFEB to transcriptional regulation of lysosomal biogenesis is also included. See text for more details.
A closer look at how bone resorption occurs explains better how a proper lysosomal function is playing a dual role in osteoclastic activity. First, through the generation of an acidic environment in the lacuna, lysosomes allow the dissolution of hydroxyapatite, the mineral component of the bone ECM, and the decarboxylation, i.e., the activation of osteocalcin.14 Second, lysosomes synthesize and secrete in the resorption lacuna proteases required for the degradation of the organic fraction of the bone ECM mostly composed of type I collagen. Before going into the details of lysosomal function regulation in osteoclasts, we will first present the main lysosomal proteins implicated in the resorptive activity of the osteoclast.
Lysosomal Proteins and Osteoclast Function
The lysosomal proteins required for osteoclast resorptive function can be subdivided into 2 classes (see Tables 1 and 2). The first one includes proteins implicated in the acidification of the resorption lacuna, while the second one comprises enzymes implicated in the degradation of the bone ECM. It should be noted that the proteolytic enzymes of the second group generally require an acidic environment to be activated and are thus dependent on the first group of proteins to function normally.
Table 1. List of lysosomal genes and proteins implicated in ECM acidification.
| Gene/protein name | Function in osteoclasts | Mouse mutant model(s) (phenotype[s]) | Human mutation(s) |
|---|---|---|---|
| ATP6AP1/AC45 | V-ATPase accessory subunit | Atp6ap1-/- (embryonic lethal) | None |
| ATP6V1C1/ATP6V1C1 | V-ATPase subunit | None | None |
| ATP6V0D2/ATP6V0D2 | V-ATPase subunit | Atp6v0d2−/− (high bone mass) | None |
| CLCN7/ClC-7 | Cl− channel Cl−/H+ antiporter(?) |
Clcn7−/− (osteopetrosis) Clcn7unc/unc (mild osteopetrosis) |
Autosomal recessive osteopetrosis Autosomal dominant osteopetrosis |
| OSTM1/OSTM1 | ClC-7 β subunit Other functions (?) |
gl/gl (osteopetrosis) |
Autosomal recessive osteopetrosis with neural involvement |
| SLC29A3/ENT3 | Lysosomal nucleoside transport (?) |
Slc29a3−/− (No bone phenotype described) |
Dysosteosclerosis |
| TCIRG1/ATP6a3 | V-ATPase regulatory subunit |
oc/oc Tcirg1−/− Tcirg1+/R740S (osteopetrosis) |
Infantile autosomal recessive osteopetrosis Autosomal dominant osteopetrosis |
Table 2. List of lysosomal genes and proteins implicated in ECM protein degradation.
| Gene/protein name | Function in osteoclasts | Mouse mutant model (phenotype) | Human mutation(s) |
|---|---|---|---|
| ACP5/TRAP | Man6P dephosphorylation (?) Osteopontin dephosphorylation (?) |
Acp5−/− (Mild osteopetrosis) |
Spondyloenchondrodysplasia |
| CTSK/CTSK | Collagen proteolysis |
Ctsk−/− (Mild osteopetrosis) |
Pycnodysostosis |
|
MMP9/MMP9 MMP13/MMP13 Other MMPs(?) |
Collagen proteolysis (?) |
Mmp13−/− Mmp9−/−;Mmp13−/− (High bone mass, decreased bone remodeling) |
Metaphyseal anadysplasia or sponduloepimetaphyseal dysplasia (no bone resorption defect reported) |
Lysosomal Proteins Implicated in the Lacuna Acidification
The largest group of proteins implicated in the acidification of the resorption lacuna is formed by different subunits of the vacuolar-type H+ATPase (V-ATPase). V-ATPases function to acidify particular organelles, such as lysosomes, by coupling ATP hydrolysis to proton transport across membranes. In osteoclasts, V-ATPase activity is enriched at the ruffled border and is essential for bone resorption. This is best illustrated by the fact that mutations in TCIRG1, a gene encoding for the subunit a3 of the V-ATPase complex (also called OC116), is mutated in about 50% of the cases of autosomal recessive osteopetrosis in human.5,20 A mutation in the same gene was found to cause osteopetrosis in the oc/oc mice.21 Although the number of multinucleated osteoclasts is increased in the patients with mutations in TCIRG1 and in oc/oc mice, their capacity to resorp bone ECM is deficient. It is generally believed that the a3 subunit is responsible for the specific and unique localization of the V-ATPase complex at the ruffled border in osteoclasts. This notion is supported by the highly restricted expression of TCIRG1 to osteoclasts.20 Interestingly, in mice, a Tcirg1 dominant-negative mutation (R740S) results not only in an increased lysosomal pH, but also in a cell-autonomous defect of osteoclastogenesis in vitro caused by an inhibition of the nuclear translocation of the transcription factor NFATc1.22 These results suggest that lysosomal pH may be important for osteoclast differentiation.
Other V-ATPase subunits have been implicated in osteoclast activity. One of them, the ATP6V0D2, an isoform of the d subunit of the V-ATPase, is highly expressed in osteoclasts, interacts with the a3 subunit, and is required for osteoclast fusion and for extracellular acidification in cell culture.23 Interestingly, mice deficient in Atp6v0d2 have increased bone mass, but this phenotype appears to be secondary to a decrease in osteoclast fusion rather than a defect in osteoclast function. It should be noted, however, that the Atp6v0d2−/− mice are characterized, in addition to this osteoclast defect, by an increased bone formation.24 ATP6V1C1, a c subunit of the V-ATPase, also appears to be important for lysosome acidification in osteoclasts and co-localizes with the a3 subunit at the ruffled border in vitro.25 Finally, cell culture experiments have demonstrated that ATP6AP1 (AC45), an accessory subunit of the V-ATPase,26 is required for extracellular acidification in osteoclasts, possibly by playing a role in the proper localization of the V-ATPase complex.27
Since there is a massive transport of protons toward the lacuna during bone resorption, it was assumed that a parallel transport of anions (Cl−) must take place simultaneously in order to maintain electroneutrality within the osteoclast. This function is achieved in osteoclasts by the chloride channel ClC-7 (CLCN7). Mice lacking this gene develop a severe osteopetrotic phenotype, and mutations in CLCN7 cause recessive and dominant forms of osteopetrosis in humans.28-30 In both mice and humans, the absence of CLCN7 results in a defective resorption by osteoclasts despite a normal differentiation process.29 Recent findings have challenged the classical view of ClC-7 as a Cl− channel, and rather suggest that this protein acts as a Cl−/H+ antiporter.31,32 This was supported by 2 observations. The first one is that the lysosomal pH is normal in Clcn7−/− neurons despite a lysosomal storage disorder. The second one is that a Clcn7 mutation preventing proton exchange without affecting Cl− transport (Clcn7unc/unc) is sufficient to cause osteopetrosis and neuronal disorders as observed in the null mice.31 However, acidification of the resorption lacuna is defective in Clcn7−/− osteoclasts,29 and the osteopetrotic phenotype is milder in Clcn7unc/unc mice than in Clcn7−/− mice.31 This suggests that ClC-7 Cl− transport function may be more important than its Cl−/H+ antiporter function in osteoclasts.
Another chloride channel family member ClC-3, which may also acts as a Cl−/H+ antiporter, have been reported to be required for proper intracellular acidification in osteoclasts in cell culture.33 However, it is not clear that the defects observed in Clcn3-deficient osteoclasts in vitro decrease bone resorption in vivo.
OSTM1 is a transmembrane protein localized in lysosomes in osteoclasts and neurons,34,35 which was found to be mutated in the osteopetrotic gl mice and in several patients suffering from the recessive form of osteopetrosis, some of them with neural involvement.36,37 It was proposed that OSTM1 might function as a β subunit of ClC-7, allowing its proper localization and its stabilization in neurons and osteoclasts.35 Interestingly, transgenic expression of Ostm1 in mature gl/gl osteoclasts using the TRAP promoter cannot rescue the osteopetrotic phenotype of gl/gl mice, while OSTM1 forced expression in myeloid progenitors using PU.1 regulatory sequences fully rescue their osteoclast defect in vivo.38 In contrast, expression of ClC-7 in Clcn7−/− mice using the same TRAP regulatory sequences rescued the osteopetrosis in these mice.39 These results suggest that either OSTM1 has additional functions besides being a ClC-7 subunit, within the osteoclast lineage, or that it acts in a non-cell-autonomous manner and independently of ClC-7 in other hematopoietic cell types affecting osteoclast function.38
More recently, dysosteosclerosis, a form of osteopetrosis associated with skin manifestation, such as histiocytosis, was found to be caused by mutations in SLC29A3 a gene encoding the equilibrative nucleoside transporter ENT3.40 SLC29A3 is expressed in primary osteoclasts, and osteoclasts lacking ENT3 have decreased capacity to demineralize calcium surface. Interestingly, in macrophages, ENT3 is required for the proper clearance of the nucleosides from the lysosomes, and in Ent3−/− macrophages intralysosomal pH is elevated and phagocytic function altered.41 Whether nucleoside transport may also be important for lysosomal acidification in osteoclasts remains to be tested.
Lysosomal Proteins Implicated in the Degradation of the Bone ECM
Once the acidic environment of the resorption lacuna has dissolved the mineralized bone ECM, the organic components of bone become exposed and can be degraded by proteolytic enzymes (see Fig. 1 and Table 2). These enzymes are active at low pH, given the acidic condition of the lacuna. Among them, cathepsin K (CTSK), a lysosomal cysteine protease, may be the most important. It is CTSK’s ability to catabolize collagen type I fibers that degrades the organic phase of the bone ECM. Mutations in CTSK are responsible for pycnodysostosis, a recessive form of osteosclerosis in human.42 Osteoclasts from pycnodysostosis patients and from mice deficient in Ctsk differentiate normally, form a ruffled border, and can even dissolve the mineral matrix, but they are unable to degrade the collagenous bone matrix.43,44
Other lysosomal proteases possibly implicated in bone resorption include matrix metalloproteinases (MMPs). In cell culture MMPs may contribute to the degradation of the bone ECM and mediate the release of the C-terminal telopeptide of type I collagen (CTX) fragment in the absence of cathepsin K activity.45 Interestingly, mice lacking MMP13 or both MMP9 and MMP13 display bone remodeling defects and increased trabecular bone density.46 Loss of function and dominant point mutations in MMP9 and MMP13, respectively, were found to cause autosomal recessive and dominant metaphyseal anadysplasia in human.47 However, it has not been shown that osteoclast activity is altered in metaphyseal anadysplasia or in MMPs-null mice. Thus, whether MMPs contribute to bone resorption in vivo remains to be established.
The tartrate acid-resistant phosphatase (TRAP or ACP5), which is commonly used as a specific marker of differentiated osteoclasts, is another lysosomal enzyme implicated in osteoclast resorptive function. TRAP is a glycosylated metallophosphatase that is activated by proteolytic cleavage, most likely involving cathepsin K activity.48 TRAP is abundant at the ruffled border and in intracellular secretory organelles, including lysosomes.49 Mice deficient in Acp5 develop only mild osteopetrosis,50 but further studies have suggested that another acid phosphatase, lysosomal acid phosphatase (LAP), can compensate for the absence of TRAP in osteoclasts in mice.51 More recently, mutations in ACP5 were identified as being responsible for spondyloenchondrodysplasia, a rare skeletal dysplasia in humans.52,53 Nonetheless, the exact function of TRAP in osteoclasts resorptive activity remains debated. It was proposed that TRAP dephosphorylates specific proteins present in the bone ECM, including osteopontin, which then could contribute to the migration of osteoclasts on the resorbed bone matrix.54 In addition, recent findings in other cell types suggest that TRAP might remove the mannose 6-phosphate (Man6P) moiety on proteins targeted for lysosomal degradation, thereby contributing to the hydrolysis and removal of degradation products by lysosomes.55,56
Regulation of Lysosomal Function in Osteoclasts
Given its critical role in osteoclast function, the activity of the lysosomes must be tightly regulated in these cells. This is achieved through the action of a growing number of proteins and pathways (see Fig. 1 and Table 3). Although most of these proteins are implicated in the trafficking and exocytosis of the lysosomal vesicles, some new players were recently identify that control alternative pathways regulating the proper localization of lysosomal enzymes or lysosomal biogenesis.
Table 3. List of genes and proteins implicated in lysosomal regulation in osteoclasts.
| Gene/protein name | Function in osteoclasts | Mouse mutant model (phenotype) | Human mutation(s) |
|---|---|---|---|
| Atg5/Autophagy related 5 | Regulates the secretion of lysosomes |
Atg5fl/fl;LysM-Cre (Decreased bone resorption) |
None |
| Dynlt1b/TCTEX-1 | Regulates vesicular trafficking via Rab3d | None | None |
| GNPTAB/GlcNAc-1-phosphotransferase α, β-subunits | Regulates the targeting of Cathepsin K and TRAP to secretory lysosomes | Gnptab−/− (Reduced bone growth) | Mucolipidosis type II α/β |
| PLEKHM1/PLEKHM1 | Lysosome transport and acidification (?) | None (Mutated in Incisor absent osteopetrotic rat) | Intermediate recessive osteopetrosis |
| Prkcb/PKCβ | Regulates lysosomal biogenesis through the phosphorylation of TFEB |
Prkcb−/− (Decreased osteoclast activity, increased bone mass) |
None |
| Prkcd/PKCδ | Regulation of cathepsin K exocytosis |
Prkcd−/− (Decreased osteoclast activity, increased bone mass) |
None |
| Rab3d/RAB3D | Lysosome trafficking | Rab3d−/− (Osteosclerosis) | None |
| Rab7/RAB7 | Vesicular trafficking | Rab7−/− (Embryonic lethal) | None |
| SNX10/SNX10 | Sorting nexin Endosomes trafficking (?) |
None | Infantile autosomal recessive osteopetrosis |
| Syt7/Synaptotagmin VII | Regulates the secretion of lysosomes |
Syt7−/− (Decreased osteoclast and osteoblast activity, low bone mass) |
None |
| Tfeb/TFEB | Transcriptional regulation of lysosomal biogenesis |
Tfebfl/fl;Ctsk-Cre (Decreased osteoclast activity, increased bone mass) |
None |
Among the proteins implicated in the trafficking of the lysosomes in osteoclasts are the 2 small GTPases, Rab3d and Rab7. Rab7, which was known to be associated with the late endosomes in other cell types, is highly expressed and localized at the ruffled border in osteoclasts. Downregulation of Rab7 in osteoclasts results in a defective polarization, an absence of ruffled border, and an impaired bone resorptive capacity in vitro.57,58 Rab3d is also abundant in osteoclasts and localized to a lysosomal subset of the post-trans-Golgi network (TGN) vesicles. Rab3d deficiency in mice results in osteoclasts with underdeveloped ruffled borders, leading ultimately to an osteosclerosis phenotype, suggesting that Rab3d functions at the post-TGN to regulate the traffic of lysosomal vesicles implicated in ruffled border formation.59 The GTPase activity of Rab proteins is dependent on their post-translational prenylation by Rab geranylgeranyl transferase (RGGT). The gunmetal (gm/gm) mouse bears a mutation in the catalytic subunit of RGGT causing hypoprenylation of several Rabs in various cell types, including osteoclasts.60,61 gm/gm osteoclasts differentiate normally, but have underdeveloped ruffled borders and reduced capacity to resorb bone ex vivo,61 confirming the importance of Rabs proteins for osteoclast activity.
Synaptotagmin VII, a member of a family of proteins that mediate calcium-dependent regulation of membrane trafficking in synaptic transmission, is associated with Rab7-positive lysosomes in osteoclasts. Absence of synaptotagmin VII in mice inhibits ruffled border formation, and cathepsin K secretion in osteoclasts in vitro and results in reduced bone resorption in vivo,62 suggesting that this protein is required for lysosome exocytosis in osteoclasts. Plekhm1, a RUN and pleckstrin containing protein, is another Rab7-interacting protein that was identified recently as an important regulator of vesicular trafficking in osteoclasts.63 PLEKHM1 is mutated in a type of recessive human osteopetrosis and in the osteopetrotic rat strain incisors absent.64 PLEKHM1-deficient osteoclasts differentiate normally but fail to form a ruffled border and to resorb the bone ECM.64 In addition, a dominant mutation in PLEKHM1 has been identified in a patient with generalized osteopenia with focal sclerosis. In this case, however, the defect appeared to be at the level of endosomal acidification.65 Finally, proteins implicated in autophagy such as Atg5 and LC3 have been shown to localize at the ruffled border in resorbing osteoclasts. Osteoclasts lacking Atg5 fail to localize Rab7-positive lysosomes to the ruffled border and have reduced resorptive capacity.66
Tctex-1, a dynein light chain protein, was identified as an interacting partner of Rab3d in a yeast 2-hybrid screen.67 The same group demonstrated that Tctex-1 co-localizes with Rab3d on lysosomal vesicles and that Tctex-1 downregulation in osteoclasts result in mislocalization of these vesicles and in impaired bone resorption capacity. This study and more recent work implicating the dynein–dynactin complex proteins and LIS1, a microtubule regulator in the resorptive capacity of osteoclasts, suggest that the active transport of vesicular cargo along microtubules is essential to osteoclasts function.68,69 Moreover, nocodazole, a microtubule-depolymerizing reagent, inhibits the bone resorptive activity of differentiated osteoclasts,70 illustrating the importance of microtubule networks in osteoclast function.
Recently, mutations in SNX10, the gene encoding the sorting nexin 10, a phox (PX) domain containing protein, were found in a subset of patients with autosomal infantile recessive osteopetrosis.71-73 SNX10 in osteoclasts is important for endosomal trafficking, TRAP exocytosis, and for the resorptive capacity of osteoclasts.73,74 Additional studies in other cell types or organisms have shown that SNX10 is interacting with the V-ATPase complex and may be important for its proper localization.75
The protein kinase C-delta (PKCδ) seems to be an important and specific regulator of cathepsin K exocytosis in osteoclasts. PKCδ−/− osteoclasts form a normal ruffled border and have proper trafficking of the V-ATPase containing lysosomes. However, they fail to secrete cathepsin K, because PKCδ normally phosphorylates and modulates the actin bundling protein myristoylated alanine-rich C-kinase substrate (MARCKS).76 Thus, different pathways may control V-ATPase and cathepsin K containing lysosomes in osteoclasts. The differential localization of proteins to specific sub-type of lysosomes may implicate the mannose 6-phosphate (Man-6-P)-targeting pathway. Indeed, osteoclasts deficient in GlcNAc-1-phosphotransferase (Gnptab−/−), which lack a functional Man-6-P targeting pathway, show increased cathepsin K and TRAP exocytosis, but impaired secretory lysosome formation.77
Transcriptional Control of Lysosomal Biogenesis in Osteoclast
Although many proteins have been implicated in lysosomal biogenesis and function, the transcriptional programs controlling these biological functions in osteoclasts are still unclear. In contrast, many transcription factors have been shown to be involved in the differentiation of osteoclasts from hematopoietic myeloid progenitors. For instance, PU.1, c-FOS, NFκB, and PPARγ act early in the differentiation process,78-81 while NFATc1, MITF, and TFE3 affect later aspects of osteoclast maturation.82,83 The transcriptional machinery deployed at different times to regulate osteoclast differentiation and maturation is under the control of RANKL and M-CSF-dependent signaling pathways.2 Therefore, an important question to ask is whether lysosome biogenesis is under the control of the same pathway(s) regulating osteoclastogenesis, or if this process requires a different mechanism. Interestingly, it was observed more than a decade ago that RANKL signaling can modulate the ability of mature osteoclasts to resorb bone ECM,84,85 but the downstream mediator(s) of this effect remained elusive. Our recent work has linked lysosomal biogenesis to this function of RANKL in differentiated osteoclasts.86
Stimulation of fully differentiated osteoclasts with RANKL increases the size and number of lysosomes in these cells because of the activation of a specific transcriptional program increasing the expression of lysosomal genes implicated in bone resorption. Importantly, this occurs without affecting osteoclasts number.86 The dissociation of the role of RANKL in osteoclasts differentiation from the one it has in lysosomal biogenesis suggested that RANKL might control osteoclasts differentiation and activity through distinct mechanisms.
TFEB, a recently identified transcriptional regulator of lysosomal biogenesis,87 is a basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor, member of the MITF/TFE subfamily including MITF, TFE3, and TFEC.88 Many of the genes encoding for proteins implicated in ECM acidification and degradation by osteoclasts (i.e., ACP5, ATP6V0D1, ATP6V0D2, ATP6V1C1, CTSK, CLCN7, and OSTM1), have been shown to be regulated by TFEB in HeLa cells, mouse embryonic fibroblasts (MEFs), and/or hepatocytes.87,89 Therefore, we hypothesized that TFEB might be mediating RANKL-dependent lysosomal biogenesis in osteoclasts.
TFEB is highly expressed in osteoclasts, and mice lacking Tfeb specifically in this cell type show increased bone mass with normal osteoclasts number, suggesting that Tfeb might regulate osteoclasts activity once they are fully differentiated. Indeed, loss of Tfeb in osteoclasts caused a reduction in their capacity to resorb bone or a mineralized substrate in an in vitro assay without affecting their differentiation capacity. Since Tfeb−/− osteoclasts contain fewer and smaller lysosomes, it appears that this functional defect is caused by reduced lysosomal biogenesis. In addition, the vacuolar pH and the pH of the resorption lacuna are increased in Tfeb−/− osteoclasts. The lysosomal biogenesis dysfunction observed in Tfeb−/− osteoclasts is probably caused by a direct effect on lysosomal gene transcription, since TFEB binds the promoter region of several lysosomal genes (Atp6v1c1, Clcn7, and Ctsk), and loss of Tfeb in osteoclasts is characterized by lower expression levels of Acp5, Atp6v1c1, Clcn7, Ctsk, and Tcirg1.86 Altogether, these data point to TFEB as a critical regulator of lysosomal biogenesis directly modulating the expression of key effectors in this process.
Furthermore, our recent work has shown that TFEB fulfills this function downstream of RANKL. Stimulation of osteoclasts by RANKL provokes an accumulation of TFEB protein independently of TFEB transcription and translation. This stabilization of TFEB following RANKL stimulation depends on the activity of the protein kinase C β (PKCβ). PKCβ directly phosphorylates TFEB on specific serine residues (S468, S461/S462, and/or S465/S466), an absolutely necessary step for TFEB capacity to induce lysosomal gene expression and to increase lysosomes size and number. Accordingly, inactivation or inhibition of PKCβ prevents TFEB stabilization in osteoclasts, decreases lysosomal gene expression, lysosomes size and numbers, and eventually increases bone mass in mice.86 Although PKC family members have been previously proposed to be involved in osteoclasts function,76,90-92 this study provided the first in vivo evidence that PKCβ regulates bone resorption by controlling lysosomal biogenesis.
Previous studies have shown that TFEB phosphorylation on S142 and/or S211 by ERK or mammalian target of rapamycin (mTOR) in the presence of nutrients prevents TFEB from entering the nucleus and decreases lysosomal biogenesis.89,93,94 In addition, TFEB induces its own expression through an autoregulatory feedback loop in response to starvation in hepatocytes.95 It was also reported that mTOR-dependent cytosolic-nuclear TFEB transport is controlled by a serine-rich region within TFEB C-terminal domain.96 Of note, this domain also contains the serine residues implicated in TFEB accumulation in osteoclasts. Our study shows that TFEB-dependent lysosomal biogenesis does not require TFEB cytosolic-nuclear transport and mTOR, but instead relies on a novel post-translational regulation of TFEB downstream of PKCβ86. Thus, although TFEB acts as a master regulator of lysosomal biogenesis in different tissues, the signaling pathways and mechanisms regulating TFEB function differ depending on the cell type involved. Interestingly, it was recently shown that the transcription factor ZKSCAN3 inhibits lysosomal biogenesis in HeLa cells.97 Whether a pathway counteracting TFEB also exists in osteoclasts is not known, but one can speculate that the tight control of the balance between this putative pathway and TFEB will be critical to bone mass maintenance.
MITF and TFE3, 2 other members of the MITF/TFE subfamily, are implicated in osteoclast maturation. Loss of function of either Mitf or Tfe3 does not affect osteoclasts, while Mitf−/−;Tfe3−/− mice show severe osteopetrosis due to a defect in pre-osteoclast fusion,83 demonstrating that these 2 genes act redundantly to regulate osteoclast maturation in vivo. In contrast, forced MITF expression in either osteoclasts or HeLa cells do not induce lysosomal biogenesis, and Mitf−/−;Tfeb+/− mice do not show any osteoclasts dysfunction,83,86,87 suggesting that Mitf and Tfeb do not fulfill the same function in osteoclasts.
Conclusion
The understanding of lysosomal biogenesis and functions in osteoclasts has progressed in recent years, yet important questions remain to be addressed. One of them is to determine if additional transcriptional and non-transcriptional pathways are implicated in lysosomal biogenesis in osteoclasts. Defining more precisely how the vesicular trafficking network is controlled in osteoclasts will be critical to identify osteoclast-specific lysosomal proteins and pathways. This important information will help to define pharmacological targets for the treatment of diseases characterized by excessive or defective bone resorption.
Acknowledgments
We thank Dr Jean Vacher for his critical reading of the manuscript. This work was supported by the Canada Research Chair program (MF) and by the National Institutes of Health (GK grant P01AG032959).
Glossary
Abbreviations:
- ATG5
autophagy related 5
- c-FOS
FBJ osteosarcoma oncogene
- ClC-7
chloride channel 7
- CTSK
cathepsin K
- ECM
Extracellular matrix
- GNPTAB
GlcNAc-1-phosphotransferase α, β-subunits
- LIS1
lissencephaly 1 protein
- MITF
microphthalmia-associated transcription factor
- MMP
matrix metalloproteinase
- NFATc1
nuclear factor of activated T cells
- cytoplasmic
calcineurin dependent 1
- OSTM1
osteopetrosis associated transmembrane protein 1
- PKC
protein kinase C
- PLEKHM1
pleckstrin homology domain-containing family M member 1
- PU.1
transcription factor PU.1
- RAB3D
member RAS oncogene family 3D
- RAB7
member RAS oncogene family 7
- RANK
receptor activator of NF-kappa-B
- RANKL
Receptor activator of NF-kappa-B ligand
- SLC29A3
solute carrier family 29 member 3
- SNX10
sorting nexin 10
- SYT7
synaptotagmin VII
- TCIRG1
T cell, immune regulator 1
- TCTEX-1
T-complex testis-specific protein 1 homolog
- TFE3
transcription factor binding to IGHM enhancer 3
- TFEB
transcription factor EB
- TGN
trans-golgi network
- TRAP
tartrate-resistant acid phosphatase
- V-ATPase
vacuolar-type H+ATPase
- ZKSCAN4
zinc finger with krab and scan domain 4
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25825
References
- 1.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–4. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 2.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–8. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 3.Tolar J, Teitelbaum SL, Orchard PJ. Osteopetrosis. N Engl J Med. 2004;351:2839–49. doi: 10.1056/NEJMra040952. [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–49. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 5.Segovia-Silvestre T, Neutzsky-Wulff AV, Sorensen MG, Christiansen C, Bollerslev J, Karsdal MA, Henriksen K. Advances in osteoclast biology resulting from the study of osteopetrotic mutations. Hum Genet. 2009;124:561–77. doi: 10.1007/s00439-008-0583-8. [DOI] [PubMed] [Google Scholar]
- 6.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–48. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 7.Schinke T, Schilling AF, Baranowsky A, Seitz S, Marshall RP, Linn T, Blaeker M, Huebner AK, Schulz A, Simon R, et al. Impaired gastric acidification negatively affects calcium homeostasis and bone mass. Nat Med. 2009;15:674–81. doi: 10.1038/nm.1963. [DOI] [PubMed] [Google Scholar]
- 8.Mansour A, Wakkach A, Blin-Wakkach C. Role of osteoclasts in the hematopoietic stem cell niche formation. Cell Cycle. 2012;11:2045–6. doi: 10.4161/cc.20534. [DOI] [PubMed] [Google Scholar]
- 9.Mansour A, Abou-Ezzi G, Sitnicka E, Jacobsen SE, Wakkach A, Blin-Wakkach C. Osteoclasts promote the formation of hematopoietic stem cell niches in the bone marrow. J Exp Med. 2012;209:537–49. doi: 10.1084/jem.20110994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lymperi S, Ersek A, Ferraro F, Dazzi F, Horwood NJ. Inhibition of osteoclast function reduces hematopoietic stem cell numbers in vivo. Blood. 2011;117:1540–9. doi: 10.1182/blood-2010-05-282855. [DOI] [PubMed] [Google Scholar]
- 11.Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein RM, Goichberg P, Spiegel A, et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12:657–64. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto K, Yoshida S, Kawasumi M, Hashimoto K, Kimura T, Sato Y, Kobayashi T, Miyauchi Y, Hoshi H, Iwasaki R, et al. Osteoclasts are dispensable for hematopoietic stem cell maintenance and mobilization. J Exp Med. 2011;208:2175–81. doi: 10.1084/jem.20101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takamatsu Y, Simmons PJ, Moore RJ, Morris HA, To LB, Lévesque JP. Osteoclast-mediated bone resorption is stimulated during short-term administration of granulocyte colony-stimulating factor but is not responsible for hematopoietic progenitor cell mobilization. Blood. 1998;92:3465–73. [PubMed] [Google Scholar]
- 14.Ferron M, Wei J, Yoshizawa T, Del Fattore A, DePinho RA, Teti A, Ducy P, Karsenty G. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142:296–308. doi: 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oury F, Ferron M, Huizhen W, Confavreux C, Xu L, Lacombe J, Srinivas P, Chamouni A, Lugani F, Lejeune H, et al. Osteocalcin regulates murine and human fertility through a pancreas-bone-testis axis. J Clin Invest. 2013;123:2421–33. doi: 10.1172/JCI65952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lotinun S, Kiviranta R, Matsubara T, Alzate JA, Neff L, Lüth A, Koskivirta I, Kleuser B, Vacher J, Vuorio E, et al. Osteoclast-specific cathepsin K deletion stimulates S1P-dependent bone formation. J Clin Invest. 2013;123:666–81. doi: 10.1172/JCI64840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teitelbaum SL. The osteoclast and its unique cytoskeleton. Ann N Y Acad Sci. 2011;1240:14–7. doi: 10.1111/j.1749-6632.2011.06283.x. [DOI] [PubMed] [Google Scholar]
- 18.Coxon FP, Taylor A. Vesicular trafficking in osteoclasts. Semin Cell Dev Biol. 2008;19:424–33. doi: 10.1016/j.semcdb.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Patrick Ross F. Mechanisms of osteoclastic secretion. Ann N Y Acad Sci. 2007;1116:238–44. doi: 10.1196/annals.1402.058. [DOI] [PubMed] [Google Scholar]
- 20.Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–6. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 21.Scimeca JC, Franchi A, Trojani C, Parrinello H, Grosgeorge J, Robert C, Jaillon O, Poirier C, Gaudray P, Carle GF. The gene encoding the mouse homologue of the human osteoclast-specific 116-kDa V-ATPase subunit bears a deletion in osteosclerotic (oc/oc) mutants. Bone. 2000;26:207–13. doi: 10.1016/S8756-3282(99)00278-1. [DOI] [PubMed] [Google Scholar]
- 22.Voronov I, Ochotny N, Jaumouillé V, Owen C, Manolson MF, Aubin JE. The R740S mutation in the V-ATPase a3 subunit increases lysosomal pH, impairs NFATc1 translocation, and decreases in vitro osteoclastogenesis. J Bone Miner Res. 2013;28:108–18. doi: 10.1002/jbmr.1727. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Xu G, Li YP. Atp6v0d2 is an essential component of the osteoclast-specific proton pump that mediates extracellular acidification in bone resorption. J Bone Miner Res. 2009;24:871–85. doi: 10.1359/jbmr.081239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–9. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- 25.Feng S, Deng L, Chen W, Shao J, Xu G, Li YP. Atp6v1c1 is an essential component of the osteoclast proton pump and in F-actin ring formation in osteoclasts. Biochem J. 2009;417:195–203. doi: 10.1042/BJ20081073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen EJ, Martens GJ. Novel insights into V-ATPase functioning: distinct roles for its accessory subunits ATP6AP1/Ac45 and ATP6AP2/(pro) renin receptor. Curr Protein Pept Sci. 2012;13:124–33. doi: 10.2174/138920312800493160. [DOI] [PubMed] [Google Scholar]
- 27.Yang DQ, Feng S, Chen W, Zhao H, Paulson C, Li YP. V-ATPase subunit ATP6AP1 (Ac45) regulates osteoclast differentiation, extracellular acidification, lysosomal trafficking, and protease exocytosis in osteoclast-mediated bone resorption. J Bone Miner Res. 2012;27:1695–707. doi: 10.1002/jbmr.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleiren E, Bénichou O, Van Hul E, Gram J, Bollerslev J, Singer FR, Beaverson K, Aledo A, Whyte MP, Yoneyama T, et al. Albers-Schönberg disease (autosomal dominant osteopetrosis, type II) results from mutations in the ClCN7 chloride channel gene. Hum Mol Genet. 2001;10:2861–7. doi: 10.1093/hmg/10.25.2861. [DOI] [PubMed] [Google Scholar]
- 29.Kornak U, Kasper D, Bösl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 2001;104:205–15. doi: 10.1016/S0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 30.Waguespack SG, Koller DL, White KE, Fishburn T, Carn G, Buckwalter KA, Johnson M, Kocisko M, Evans WE, Foroud T, et al. Chloride channel 7 (ClCN7) gene mutations and autosomal dominant osteopetrosis, type II. J Bone Miner Res. 2003;18:1513–8. doi: 10.1359/jbmr.2003.18.8.1513. [DOI] [PubMed] [Google Scholar]
- 31.Weinert S, Jabs S, Supanchart C, Schweizer M, Gimber N, Richter M, Rademann J, Stauber T, Kornak U, Jentsch TJ. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl- accumulation. Science. 2010;328:1401–3. doi: 10.1126/science.1188072. [DOI] [PubMed] [Google Scholar]
- 32.Graves AR, Curran PK, Smith CL, Mindell JA. The Cl-/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature. 2008;453:788–92. doi: 10.1038/nature06907. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto F, Kajiya H, Toh K, Uchida S, Yoshikawa M, Sasaki S, Kido MA, Tanaka T, Okabe K. Intracellular ClC-3 chloride channels promote bone resorption in vitro through organelle acidification in mouse osteoclasts. Am J Physiol Cell Physiol. 2008;294:C693–701. doi: 10.1152/ajpcell.00251.2007. [DOI] [PubMed] [Google Scholar]
- 34.Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat Med. 2003;9:399–406. doi: 10.1038/nm842. [DOI] [PubMed] [Google Scholar]
- 35.Lange PF, Wartosch L, Jentsch TJ, Fuhrmann JC. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature. 2006;440:220–3. doi: 10.1038/nature04535. [DOI] [PubMed] [Google Scholar]
- 36.Maranda B, Chabot G, Décarie JC, Pata M, Azeddine B, Moreau A, Vacher J. Clinical and cellular manifestations of OSTM1-related infantile osteopetrosis. J Bone Miner Res. 2008;23:296–300. doi: 10.1359/jbmr.071015. [DOI] [PubMed] [Google Scholar]
- 37.Pangrazio A, Poliani PL, Megarbane A, Lefranc G, Lanino E, Di Rocco M, Rucci F, Lucchini F, Ravanini M, Facchetti F, et al. Mutations in OSTM1 (grey lethal) define a particularly severe form of autosomal recessive osteopetrosis with neural involvement. J Bone Miner Res. 2006;21:1098–105. doi: 10.1359/jbmr.060403. [DOI] [PubMed] [Google Scholar]
- 38.Pata M, Héraud C, Vacher J. OSTM1 bone defect reveals an intercellular hematopoietic crosstalk. J Biol Chem. 2008;283:30522–30. doi: 10.1074/jbc.M805242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasper D, Planells-Cases R, Fuhrmann JC, Scheel O, Zeitz O, Ruether K, Schmitt A, Poët M, Steinfeld R, Schweizer M, et al. Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. EMBO J. 2005;24:1079–91. doi: 10.1038/sj.emboj.7600576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campeau PM, Lu JT, Sule G, Jiang MM, Bae Y, Madan S, Högler W, Shaw NJ, Mumm S, Gibbs RA, et al. Whole-exome sequencing identifies mutations in the nucleoside transporter gene SLC29A3 in dysosteosclerosis, a form of osteopetrosis. Hum Mol Genet. 2012;21:4904–9. doi: 10.1093/hmg/dds326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu CL, Lin W, Seshasayee D, Chen YH, Ding X, Lin Z, Suto E, Huang Z, Lee WP, Park H, et al. Equilibrative nucleoside transporter 3 deficiency perturbs lysosome function and macrophage homeostasis. Science. 2012;335:89–92. doi: 10.1126/science.1213682. [DOI] [PubMed] [Google Scholar]
- 42.Gelb BD, Shi GP, Chapman HA, Desnick RJ. Pycnodysostosis, a lysosomal disease caused by cathepsin K deficiency. Science. 1996;273:1236–8. doi: 10.1126/science.273.5279.1236. [DOI] [PubMed] [Google Scholar]
- 43.Li CY, Jepsen KJ, Majeska RJ, Zhang J, Ni R, Gelb BD, Schaffler MB. Mice lacking cathepsin K maintain bone remodeling but develop bone fragility despite high bone mass. J Bone Miner Res. 2006;21:865–75. doi: 10.1359/jbmr.060313. [DOI] [PubMed] [Google Scholar]
- 44.Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, Moritz JD, Schu P, von Figura K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci U S A. 1998;95:13453–8. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henriksen K, Sørensen MG, Nielsen RH, Gram J, Schaller S, Dziegiel MH, Everts V, Bollerslev J, Karsdal MA. Degradation of the organic phase of bone by osteoclasts: a secondary role for lysosomal acidification. J Bone Miner Res. 2006;21:58–66. doi: 10.1359/JBMR.050905. [DOI] [PubMed] [Google Scholar]
- 46.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–95. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lausch E, Keppler R, Hilbert K, Cormier-Daire V, Nikkel S, Nishimura G, Unger S, Spranger J, Superti-Furga A, Zabel B. Mutations in MMP9 and MMP13 determine the mode of inheritance and the clinical spectrum of metaphyseal anadysplasia. Am J Hum Genet. 2009;85:168–78. doi: 10.1016/j.ajhg.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ljusberg J, Wang Y, Lång P, Norgård M, Dodds R, Hultenby K, Ek-Rylander B, Andersson G. Proteolytic excision of a repressive loop domain in tartrate-resistant acid phosphatase by cathepsin K in osteoclasts. J Biol Chem. 2005;280:28370–81. doi: 10.1074/jbc.M502469200. [DOI] [PubMed] [Google Scholar]
- 49.Reinholt FP, Widholm SM, Ek-Rylander B, Andersson G. Ultrastructural localization of a tartrate-resistant acid ATPase in bone. J Bone Miner Res. 1990;5:1055–61. doi: 10.1002/jbmr.5650051009. [DOI] [PubMed] [Google Scholar]
- 50.Hayman AR, Jones SJ, Boyde A, Foster D, Colledge WH, Carlton MB, Evans MJ, Cox TM. Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disrupted endochondral ossification and mild osteopetrosis. Development. 1996;122:3151–62. doi: 10.1242/dev.122.10.3151. [DOI] [PubMed] [Google Scholar]
- 51.Suter A, Everts V, Boyde A, Jones SJ, Lüllmann-Rauch R, Hartmann D, Hayman AR, Cox TM, Evans MJ, Meister T, et al. Overlapping functions of lysosomal acid phosphatase (LAP) and tartrate-resistant acid phosphatase (Acp5) revealed by doubly deficient mice. Development. 2001;128:4899–910. doi: 10.1242/dev.128.23.4899. [DOI] [PubMed] [Google Scholar]
- 52.Briggs TA, Rice GI, Daly S, Urquhart J, Gornall H, Bader-Meunier B, Baskar K, Baskar S, Baudouin V, Beresford MW, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43:127–31. doi: 10.1038/ng.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lausch E, Janecke A, Bros M, Trojandt S, Alanay Y, De Laet C, Hübner CA, Meinecke P, Nishimura G, Matsuo M, et al. Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat Genet. 2011;43:132–7. doi: 10.1038/ng.749. [DOI] [PubMed] [Google Scholar]
- 54.Ek-Rylander B, Andersson G. Osteoclast migration on phosphorylated osteopontin is regulated by endogenous tartrate-resistant acid phosphatase. Exp Cell Res. 2010;316:443–51. doi: 10.1016/j.yexcr.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Sun P, Sleat DE, Lecocq M, Hayman AR, Jadot M, Lobel P. Acid phosphatase 5 is responsible for removing the mannose 6-phosphate recognition marker from lysosomal proteins. Proc Natl Acad Sci U S A. 2008;105:16590–5. doi: 10.1073/pnas.0807472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makrypidi G, Damme M, Müller-Loennies S, Trusch M, Schmidt B, Schlüter H, Heeren J, Lübke T, Saftig P, Braulke T. Mannose 6 dephosphorylation of lysosomal proteins mediated by acid phosphatases Acp2 and Acp5. Mol Cell Biol. 2012;32:774–82. doi: 10.1128/MCB.06195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H, Laitala-Leinonen T, Parikka V, Väänänen HK. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J Biol Chem. 2001;276:39295–302. doi: 10.1074/jbc.M010999200. [DOI] [PubMed] [Google Scholar]
- 58.Sun Y, Büki KG, Ettala O, Vääräniemi JP, Väänänen HK. Possible role of direct Rac1-Rab7 interaction in ruffled border formation of osteoclasts. J Biol Chem. 2005;280:32356–61. doi: 10.1074/jbc.M414213200. [DOI] [PubMed] [Google Scholar]
- 59.Pavlos NJ, Xu J, Riedel D, Yeoh JS, Teitelbaum SL, Papadimitriou JM, Jahn R, Ross FP, Zheng MH. Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Mol Cell Biol. 2005;25:5253–69. doi: 10.1128/MCB.25.12.5253-5269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Detter JC, Zhang Q, Mules EH, Novak EK, Mishra VS, Li W, McMurtrie EB, Tchernev VT, Wallace MR, Seabra MC, et al. Rab geranylgeranyl transferase alpha mutation in the gunmetal mouse reduces Rab prenylation and platelet synthesis. Proc Natl Acad Sci U S A. 2000;97:4144–9. doi: 10.1073/pnas.080517697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor A, Mules EH, Seabra MC, Helfrich MH, Rogers MJ, Coxon FP. Impaired prenylation of Rab GTPases in the gunmetal mouse causes defects in bone cell function. Small GTPases. 2011;2:131–42. doi: 10.4161/sgtp.2.3.16488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao H, Ito Y, Chappel J, Andrews NW, Teitelbaum SL, Ross FP. Synaptotagmin VII regulates bone remodeling by modulating osteoclast and osteoblast secretion. Dev Cell. 2008;14:914–25. doi: 10.1016/j.devcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, Yoshimori T. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell. 2010;21:4162–72. doi: 10.1091/mbc.E10-06-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Wesenbeeck L, Odgren PR, Coxon FP, Frattini A, Moens P, Perdu B, MacKay CA, Van Hul E, Timmermans JP, Vanhoenacker F, et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest. 2007;117:919–30. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Del Fattore A, Fornari R, Van Wesenbeeck L, de Freitas F, Timmermans JP, Peruzzi B, Cappariello A, Rucci N, Spera G, Helfrich MH, et al. A new heterozygous mutation (R714C) of the osteopetrosis gene, pleckstrin homolog domain containing family M (with run domain) member 1 (PLEKHM1), impairs vesicular acidification and increases TRACP secretion in osteoclasts. J Bone Miner Res. 2008;23:380–91. doi: 10.1359/jbmr.071107. [DOI] [PubMed] [Google Scholar]
- 66.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21:966–74. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavlos NJ, Cheng TS, Qin A, Ng PY, Feng HT, Ang ES, Carrello A, Sung CH, Jahn R, Zheng MH, et al. Tctex-1, a novel interaction partner of Rab3D, is required for osteoclastic bone resorption. Mol Cell Biol. 2011;31:1551–64. doi: 10.1128/MCB.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng PY, Cheng TS, Zhao H, Ye S, Sm Ang E, Khor EC, Feng HT, Xu J, Zheng MH, Pavlos NJ. Disruption of the dynein-dynactin complex unveils motor-specific functions in osteoclast formation and bone resorption. J Bone Miner Res. 2013;28:119–34. doi: 10.1002/jbmr.1725. [DOI] [PubMed] [Google Scholar]
- 69.Ye S, Fowler TW, Pavlos NJ, Ng PY, Liang K, Feng Y, Zheng M, Kurten R, Manolagas SC, Zhao H. LIS1 regulates osteoclast formation and function through its interactions with dynein/dynactin and Plekhm1. PLoS One. 2011;6:e27285. doi: 10.1371/journal.pone.0027285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okumura S, Mizoguchi T, Sato N, Yamaki M, Kobayashi Y, Yamauchi H, Ozawa H, Udagawa N, Takahashi N. Coordination of microtubules and the actin cytoskeleton is important in osteoclast function, but calcitonin disrupts sealing zones without affecting microtubule networks. Bone. 2006;39:684–93. doi: 10.1016/j.bone.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Pangrazio A, Fasth A, Sbardellati A, Orchard PJ, Kasow KA, Raza J, Albayrak C, Albayrak D, Vanakker OM, De Moerloose B, et al. SNX10 mutations define a subgroup of human autosomal recessive osteopetrosis with variable clinical severity. J Bone Miner Res. 2013;28:1041–9. doi: 10.1002/jbmr.1849. [DOI] [PubMed] [Google Scholar]
- 72.Mégarbané A, Pangrazio A, Villa A, Chouery E, Maarawi J, Sabbagh S, Lefranc G, Sobacchi C. Homozygous stop mutation in the SNX10 gene in a consanguineous Iraqi boy with osteopetrosis and corpus callosum hypoplasia. Eur J Med Genet. 2013;56:32–5. doi: 10.1016/j.ejmg.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 73.Aker M, Rouvinski A, Hashavia S, Ta-Shma A, Shaag A, Zenvirt S, Israel S, Weintraub M, Taraboulos A, Bar-Shavit Z, et al. An SNX10 mutation causes malignant osteopetrosis of infancy. J Med Genet. 2012;49:221–6. doi: 10.1136/jmedgenet-2011-100520. [DOI] [PubMed] [Google Scholar]
- 74.Zhu CH, Morse LR, Battaglino RA. SNX10 is required for osteoclast formation and resorption activity. J Cell Biochem. 2012;113:1608–15. doi: 10.1002/jcb.24029. [DOI] [PubMed] [Google Scholar]
- 75.Chen Y, Wu B, Xu L, Li H, Xia J, Yin W, Li Z, Shi D, Li S, Lin S, et al. A SNX10/V-ATPase pathway regulates ciliogenesis in vitro and in vivo. Cell Res. 2012;22:333–45. doi: 10.1038/cr.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cremasco V, Decker CE, Stumpo D, Blackshear PJ, Nakayama KI, Nakayama K, Lupu TS, Graham DB, Novack DV, Faccio R. Protein kinase C-delta deficiency perturbs bone homeostasis by selective uncoupling of cathepsin K secretion and ruffled border formation in osteoclasts. J Bone Miner Res. 2012;27:2452–63. doi: 10.1002/jbmr.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Meel E, Boonen M, Zhao H, Oorschot V, Ross FP, Kornfeld S, Klumperman J. Disruption of the Man-6-P targeting pathway in mice impairs osteoclast secretory lysosome biogenesis. Traffic. 2011;12:912–24. doi: 10.1111/j.1600-0854.2011.01203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, Teitelbaum SL. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature. 1997;386:81–4. doi: 10.1038/386081a0. [DOI] [PubMed] [Google Scholar]
- 79.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–8. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 80.Iotsova V, Caamaño J, Loy J, Yang Y, Lewin A, Bravo R. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat Med. 1997;3:1285–9. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 81.Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496–503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- 82.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/S1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 83.Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci U S A. 2002;99:4477–82. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–76. doi: 10.1016/S0092-8674(00)81569-X. [DOI] [PubMed] [Google Scholar]
- 85.Burgess TL, Qian Y, Kaufman S, Ring BD, Van G, Capparelli C, Kelley M, Hsu H, Boyle WJ, Dunstan CR, et al. The ligand for osteoprotegerin (OPGL) directly activates mature osteoclasts. J Cell Biol. 1999;145:527–38. doi: 10.1083/jcb.145.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, Ballabio A, Karsenty G. A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–69. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 88.Fisher DE, Carr CS, Parent LA, Sharp PA. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991;5(12A):2342–52. doi: 10.1101/gad.5.12a.2342. [DOI] [PubMed] [Google Scholar]
- 89.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sørensen MG, Karsdal MA, Dziegiel MH, Boutin JA, Nosjean O, Henriksen K. Screening of protein kinase inhibitors identifies PKC inhibitors as inhibitors of osteoclastic acid secretion and bone resorption. BMC Musculoskelet Disord. 2010;11:250. doi: 10.1186/1471-2474-11-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williams JP, Thames AM, 3rd, McKenna MA, McDonald JM. Differential effects of calmodulin and protein kinase C antagonists on bone resorption and acid transport activity. Calcif Tissue Int. 2003;73:290–6. doi: 10.1007/s00223-002-0012-2. [DOI] [PubMed] [Google Scholar]
- 92.Lee SW, Kwak HB, Chung WJ, Cheong H, Kim HH, Lee ZH. Participation of protein kinase C beta in osteoclast differentiation and function. Bone. 2003;32:217–27. doi: 10.1016/S8756-3282(02)00976-6. [DOI] [PubMed] [Google Scholar]
- 93.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–58. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J 2011; On-line. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]