Abstract
In recent years, access to complete genomic sequences, coupled with rapidly accumulating data related to RNA and protein expression patterns, has made it possible to determine comprehensively how genes contribute to complex phenotypes. However, for major crop plants, publicly available, standard platforms for parallel expression analysis have been limited. We report the conception and design of the new publicly available, 22K Barley1 GeneChip probe array, a model for plants without a fully sequenced genome. Array content was derived from worldwide contribution of 350,000 high-quality ESTs from 84 cDNA libraries, in addition to 1,145 barley (Hordeum vulgare) gene sequences from the National Center for Biotechnology Information nonredundant database. Conserved sequences expressed in seedlings of wheat (Triticum aestivum), oat (Avena strigosa), rice (Oryza sativa), sorghum (Sorghum bicolor), and maize (Zea mays) were identified that will be valuable in the design of arrays across grasses. To enhance the usability of the data, BarleyBase, a MIAME-compliant, MySQL relational database, serves as a public repository for raw and normalized expression data from the Barley1 GeneChip probe array. Interconnecting links with PlantGDB and Gramene allow BarleyBase users to perform gene predictions using the 21,439 non-redundant Barley1 exemplar sequences or cross-species comparison at the genome level, respectively. We expect that this first generation array will accelerate hypothesis generation and gene discovery in disease defense pathways, responses to abiotic stresses, development, and evolutionary diversity in monocot plants.
Microarray technology makes possible the parallel assessment of thousands of genes in a single experiment. As opposed to dissecting individual components of a biological system, system-wide analytical approaches can be pursued. However, for major crop plants, publicly available, standard platforms for parallel expression analysis have been limited. Largely because of the Interagency Plant Genome Initiative, cDNA and spotted oligomer microarrays are under development for maize (Zea mays), rice (Oryza sativa), soybean (Glycine max), tomato (Lycopersicon esculentum), Medicago truncatula, and others. Arguably, the most advanced technology is accessible for Arabidopsis, where cDNA arrays have been available from the Arabidopsis Functional Genomics Consortium (Maleck et al., 2000; Schenk et al., 2000), and new second generation oligo-based arrays based on the sequenced Arabidopsis genome are available from Affymetrix Inc. (22,700 sequences; Santa Clara, CA) and Agilent Technologies (21,500 sequences; Palo Alto, CA). Despite these advances, consistent data on parallel expression profiling from large-genome crop plants are infrequent; thus, examples of meta-analysis among several data sets across laboratories are rare or nonexistent.
Triticeae grain crops (barley [Hordeum vulgare], wheat [Triticum aestivum], and rye [Secale cereale]) are sown on 81 million acres in the United States with an average value of 9 billion dollars (USDA-National Agricultural Statistics Service, http://www.usda.gov/nass/pubs/agr02/acro02.htm). Barley is a true diploid and, despite its large genome (1C = 5.3 × 109; Arumuganathan and Earle, 1991), is well suited to genetic analysis with an extensive collection of mutants and over 150,000 accessions (von Wettstein-Knowles, 1992; http://www.ars-grin.gov/npgs/index.html; http://barley.ipk-gatersleben.de/ebdb/). Thousands of molecular, morphological, and quantitative trait markers have been mapped (Franckowiak, 1997; Ramsay et al., 2000; Kleinhofs and Graner, 2001; Kleinhofs and Han, 2001), and a bacterial artificial chromosome library covering 6.3 genome equivalents (Yu et al., 2000) is in use throughout the world. This report provides an overview of the new community-designed, 22K Barley1 GeneChip probe array, as a template for communities to engage in similar endeavors, particularly for plants without a fully sequenced genome.
Concept and Design
Over the course of several informal meetings at the International Plant and Animal Genome Conferences from 1998 through 2002, the barley research community came to a general consensus that a worldwide standard for parallel expression profiling was needed for Triticeae grain crops. During this period, funding was obtained for independent and sizeable barley expressed sequence tag (EST) sequencing projects in the United States, Germany, Japan, Scotland, and Finland. By the end of 2002, these projects had produced a combined total of approximately 350,000 high-quality barley ESTs originating from 84 cDNA libraries representing various developmental stages, in addition to abiotic and biotic stress treatments (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). From January to October 2002, each project transmitted their raw EST data to produce a collective resource for the purpose of designing a state-of-the-art genome array for parallel transcript profiling.
CAP3 Optimization
After the processing of raw EST sequence data from all groups (described below), high-quality EST sequences were assembled using the CAP3 program (Huang and Madan, 1999; http://deepc2.zool.iastate.edu/aat/cap/cap.html). Several CAP3 parameters were empirically tested with the intention of achieving full separation of paralogous genes. Clustering parameter “p” (overlap percentage identity) was tested over a range of 66 to 95 and clustering parameter “d” (maximum disagreements between high-quality base calls) was tested from 20 to 300. The settings of p = 95 and d = 60 with all other CAP3 settings at their default levels appeared satisfactory for a small set of test sequences. For example, 12 paralogs in the Dhn (dehydrin) multigene family (Choi et al. 1999, 2000) were entirely separated while maintaining some association between alleles. Two additional settings, “f” (maximum gap length) and “h” (maximum overhang percentage), were then adjusted to maintain separation of Dhn paralogs while adding tolerance for allelic variation, such as small InDels. As expected, differences in the number of contigs and singletons and, therefore, the total number of unique gene-oriented clusters or “UniGenes” resulted from varying these parameters. At values of p = 95, d = 60, and f = 100, a change in “h” from a value of 15 to 30 resulted in a 1.35% decrease in the number of contigs and a 16.2% decrease in the number of singletons. Similarly, a change to h = 65 resulted in a 1.5% reduction in the number of contigs and a 27.3% reduction in the number of singletons relative to h = 15. Settings of p = 95, d=60, f=100, and h=50 were found to give the most favorable results, yielding 26,634 contigs and 26,396 singletons for a sum of 53,030 tentative UniGenes. Note that the 53,030 UniGenes included substantial inflation because of over-separation of allelic sequences. This was the penalty paid for complete separation of paralogs among the genes used as a test case. However, for the purpose of array content, this result was appropriately offset by the Affymetrix probe design algorithm, which compensated for excessive redundancy by permitting probe sets from only one closely related sequence. Hence, in the final selection of Dhn genes, all 12 paralogs are represented, and five of these 12 are represented by probe sets targeting different alleles. Complete results of this iterative process can be viewed in HarvEST:Barley, which is publicly available Windows software downloadable at http://harvest.ucr.edu/Barley1.htm. Included in this assembly were 1,145 barley cDNA and gene sequences, including multiple alleles in some cases, from the National Center for Biotechnology Information (NCBI) nonredundant (nr) database. These nr sequences were included to aid in scaffolding the ESTs and to ensure representation of many genes and alleles of special interest (e.g. Mla, Rar1, Sgt1, and Rpg1) that might otherwise not have been represented on the array. Specific processing steps used for GeneChip sequences are summarized below.
The Phred program (Ewing and Green, 1998; Ewing et al., 1998; http://www.phrap.org/) was applied to source lab chromatograms to derive sequence and quality files.
Cross_match was applied to all sequence files to mask cloning vector and cloning system oligonucleotides.
An in-house script, “qvtrim,” was use to trim out low-quality regions (outside of a sliding window of Phred 17), reduce poly(T) and poly(A+) end lengths to a maximum of 17, and remove residual cloning system sequences that survived cross_match.
Sequences with less than 100 remaining bases after qvtrim, not counting terminal poly(A+) or poly(T) nucleotides, were discarded.
Barley gene sequences from the NCBI nr file were given uniform dummy quality values of 17.
Orientations were determined using information about the cloning system, sequencingprimer, high BLAST hit orientation, and presence of a poly(A+) or poly(T).
BLAST searches (Altschul et al., 1997; http://www.ncbi.nlm.nih.gov/BLAST/) were performed to identify and discard sequences from Escherichia coli, lambda, fungal, and human genomes, rRNA, and other repetitive sequences present in an enhanced Triticeae repeat element database (TREP; http://wheat.pw.usda.gov/ITMI/Repeats/index.shtml).
A low-complexity filter, based on consecutive four-nucleotide repeats, was applied to remove sequences that have high Phred scores but are the result of poor-quality sequencing reactions.
The Univec software (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html) was used to search for “wrong vector” sequences and identify cross-contamination from other libraries.
EST sequences that begin with poly(T) and ended with poly(A+) were discarded to eliminate chimeric cDNA clones.
An assembly of all remaining sequences was managed by the “tgicl” program from TIGR (http://www.tigr.org/tdb/tgi/software/). This entailed Mega BLAST followed by CAP3 (Huang and Madan, 1999), resulting in the production of contigs and their consensus sequence and singletons.
The orientation of every EST sequence was determined based on three criteria: sequencing primer used (all libraries were directional), orientation of high BLAST hit, and presence of poly(A+) or poly(T) on an end. The orientation of each contig was also determined, based on the ratio of forward and reverse EST sequences and the orientation of each EST used by CAP3.
Contigs containing consensus sequences starting with poly(T) and ending with poly(A+), or with unknown orientation, or with BLAST clusters spaced more than 800 bases apart, were manually examined to identify and remove chimeras.
Assembly and chimera removal from contigs was repeated several times.
Sequences with reliable 3′ ends were determined.
Reverse orientation consensus sequences and singletons were converted to their reverse complement forward-oriented sequences.
Multiple poly(A+) sites were located within the final 500 bases of consensus sequences, and such sequences were trimmed to their first poly(A+) site. The resulting consensus contigs (or singleton) were defined as “exemplars.”
“Housekeeping” genes (e.g. GAPDH, tubulin, EIF5A, and actin) were chosen as standard 3′- to 5′-labeling controls.
Common reporter gene sequences (for transgenics) were added.
Several barley resistance gene sequences were added.
Several “reliably not expressed” intergenomic regions were retrieved from two overlapping bacterial artificial chromosome sequences as nonspecific hybridization controls (GenBank accession no. AF427791; Wei et al., 2002).
A final collection of exemplar sequences was submitted to the Affymetrix chip design team to define probe sets.
NCBI nr
All GenBank records with the features “gene,” “exon,” “intron,” or “complete CDS” and species H. vulgare were downloaded from the NCBI nr data set. This set of records was then augmented by expert analysis for genes that managed to slip through this filter. The resulting collection was then filtered for duplication by excluding records that repeated Gen-Bank accession numbers. Records where exons could not be identified were removed. This resulted in a collection of 1,050 GenBank records for H. vulgare. From these, exons were extracted to serve as virtual ESTs for clustering. A total of 1,007 virtual ESTs were recovered in records with a single exon, and 138 were recovered for records with multiple exons for a total of 1,145 virtual ESTs. Hence, all previously cloned barley genes were utilized in the clustering process in an effort to represent characterized genes on the array.
Enhanced TREP
To create a file for pruning against known repeated sequences in grasses, all GenBank records were downloaded for Triticeae spp. and maize that contained the words transposon, retrotransposon, and repeat element in their features. Duplicate GenBank accessions were removed. The GenBank annotations then were used to extract repeat sequences when those sequences may have comprised only a part of the entire record. Resulting records with fewer than 20 bases of DNA were removed from the data set. These records were then compared with the TREP database (http://wheat.pw.usda.gov/ITMI/Repeats/index.shtml). Those records not appearing in the TREP database were added to those in the TREP database, and all records were put into FASTA format. As described above, this set of sequences was used at two stages. The first was to use BLAST to identify ESTs that were not cDNAs but instead appeared to be fragments of the barley genome. Second, the final list of 25-mer oligo probes was checked against this repeat dataset to avoid probes that hit known repeat elements—none were found. The data set contained 222 sequences derived from maize records, 255 Triticeae records not in the version of TREP available in June 2002, and 809 records from TREP.
Exemplar and Probe Set Selection
For organizational purposes, sequences were grouped into 11 sections as shown in Table I. A total of 25,459 exemplar sequences were submitted to Affymetrix for initial computation of probe sets. Several iterations later, the final 22,792 probe sets were approved for mask design in January 2003. After a 3-month final testing and validation period, the Barley1 GeneChip probe array was released for public distribution in June 2003.
Table I.
Classification of nonredundant exemplar sequences used for probe set design on the Barley1 GeneChip probe array
| Section | Content | No. with Unknown Function | No. with Known or Predicted Functiona | Total No. of Sequencesb |
|---|---|---|---|---|
| 1. | Specially sequences (Mla alleles; Rpg1) | 0 | 16 | 16 |
| 2. | Reporter genes (for investigation of transgenics) | 0 | 26 | 26 |
| Main assembly contigs | ||||
| 3. | Contains 3′ end read (including NCBI nr cDNA or gene) | 3,360 | 10,783 | 14,143 |
| 4. | No 3′-end read but has poly(A+) at terminal end | 706 | 577 | 1,283 |
| 5. | No 3′-end read or poly(A+) terminal but contains GenBank nr sequence | 3 | 76 | 79 |
| 6. | Chloroplast | 0 | 44 | 44 |
| 7. | Mitochondrion | 0 | 44 | 44 |
| Main assembly singletons | ||||
| 8. | NCBI nr cDNA or gene | 2c | 93 | 95 |
| 9. | Forward orientation and poly(A+) at terminal end | 1,237 | 279 | 1,516 |
| 10. | Reverse orientation and poly(T) at beginning | 2,514 | 869 | 3,383 |
| 11. | Reverse orientation but no poly(T) at beginning | 579 | 231 | 810 |
| Additions | Controls (eg., Affymetrix standard spiking controls: BioB, BioC, etc.; barley reliably nonexpressed sequences, housekeeping controls, etc.) | 0 | 48 | 48 |
| Total of sections 1 to 11 | 8,401 | 13,038 | 21,439 |
Defined by BLASTX cutoff of e-20
Totals in each section correspond to non-redundant probe sets. There are 1,524 probe sets that provide multiple representations of the same exemplar
The two unknowns in section 8 are above the e-20 cutoff, but both are putative hordeins
In general, probe sets on the Barley1 GeneChip are composed of 11 pairs of 25-mer oligonucleotides derived principally from the last 600 bases of each exemplar sequence or nr cloned gene (https://www.affymetrix.com/analysis/downloadonlogin.affx?onloadforward=/support/technical/other/custom_design_manual.pdf). The selected 600 bases most likely have probe pairs positioned in the 3′ end of the coding region and the 3′-untranslated region. This is in contrast to the Arabidopsis ATH1 genome array, which has a majority of its probes tiled from the annotated open reading frames. Each probe pair contains a perfect match oligonucleotide and a mismatch control containing a single substitution at the 13th base. The purpose of this feature is to help distinguish background from true low-level expression of particular genes in response to a particular treatment. Oligonucleotide placement within each represented exemplar sequence can be viewed on Windows computers using HarvEST:Barley (http://harvest.ucr.edu/Barley1.htm) or online at http://barleypop.vrac.iastate.edu/BarleyBase/probealign.php (within http://barleybase.org/). Exemplars represented on the GeneChip probe array can also be searched via BLAST at http://www.plantgdb.org/hordeum_vulgare.html and http://barleypop.vrac.iastate.edu/BarleyBase/content.php or compared with other cereal genes on the Gramene Rice Genome Browser (e.g. http://barleypop.vrac.iastate.edu/BarleyBase/barley1contig.php?exemplar-Barley1_05405).
Validation of the Barley1 GeneChip Probe Array
To perform initial validation of the GeneChip probe array, total RNA was isolated via a hot (60°C) phenol (pH 4.3)/guanidinium thiocyanate method. We have optimized this method for GeneChip sample labeling and routinely obtain high yields (approximately 500 μg RNA per gram fresh weight) with 260 to 280 ratios between 1.9 and 2.1. RNA purified further using the RNeasy Midi kit (Qiagen, Valencia, CA) yielded the most consistent cDNA synthesis and cRNA labeling among large numbers of samples. First strand cDNA synthesis (Invitrogen, Carlsbad, CA) was used to convert the mRNA into cDNA. The resulting cDNA was transcribed in vitro in the presence of biotinylated UTP and CTP to produce biotinylated target complementary RNA (cRNA). The labeled cRNA was purified, fragmented, and hybridized onto GeneChips. Probe labeling quality was verified at each step on an Agilent 2100 Bioanalyzer equipped with an RNA Nano LabChip (Agilent Technologies). All detailed protocols can be accessed online at http://barleypop.vrac.iastate.edu/BarleyBase/experiment_dataquery.php?class=protocol&name=any, within http://barleybase.org/.
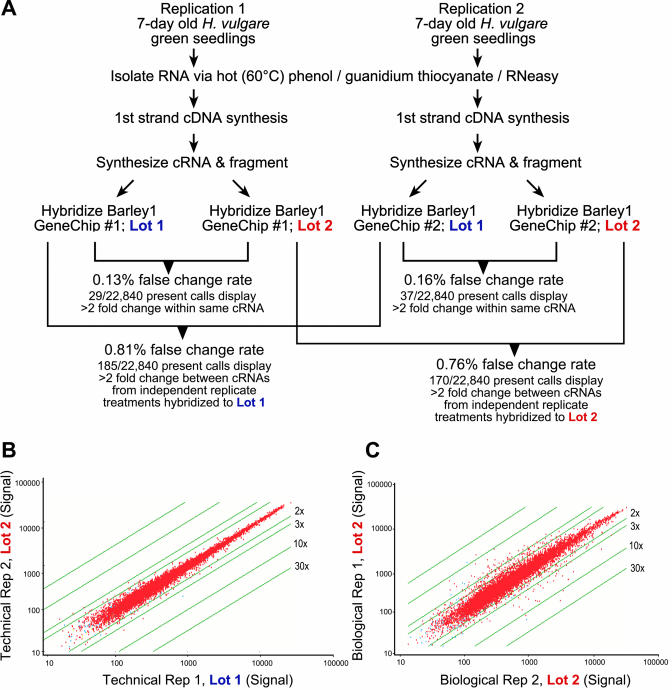
As illustrated in Figure 1, we assessed in parallel, chip-to-chip, lot-to-lot, and replication-to-replication comparisons to evaluate the false change rate for the Barley1 GeneChip probe array. RNA from 7-d-old barley leaves was used to generate the labeled cRNA. A single cRNA sample from each of two replications was hybridized to two individual GeneChip probe arrays, one from each of two separate lots. No significant signal was detected on the “reliably not expressed” intergenomic regions, indicating a low level of nonspecific hybridization. Expression data was scaled globally to a target intensity of 500 to allow comparisons between arrays. Detection of present calls with Affymetrix MAS 5.0 software was determined by at least eight of the 11 probe pairs within a probe set that exceeded the default probe pair threshold with a user-definable parameter Tau of 0.015. Comparisons of probe pair scores with the threshold Tau were summarized to calculate P values, and probe sets with P values less than 0.04 were considered reliably detected or “present.” Chip-to-chip and lot-to-lot comparisons resulted in 29 to 37 present calls of 22,840 probe sets that revealed greater than 2-fold change, translating to an overall false change rate of 0.13% to 0.16%, respectively. In addition to the 22,792 barley or reporter gene probe sets, these comparisons also included the 48 Affymetrix internal control sequences (Table I). Independent biological replicates from the same experiment resulted in an overall false change rate of 0.76% to 0.81%. These figures are consistent with previous results obtained from Arabidopsis and human GeneChips produced by Affymetrix (Zhu and Wang, 2000). This example and others (Jelinsky et al., 2000; http://www.lrgc.ca/?page=forms) support the interpretation that there is far greater variability between independent biological replicates than between GeneChips.
Figure 1.
A, Flow chart of experiments to assess false-change rate among Barley1 GeneChip probe arrays. B, Scatter plot illustrating the technical reproducibility and dynamic range of Barley1 arrays. C, Scatter plot illustrating the variation between independent biological replications. Expression level of each gene, as measured by quantitative estimate in the form of a signal log ratio, was plotted using MAS 5.0 software (Affymetrix). Two-, 3-, 10-, and 30-fold changes in expression are indicated by the parallel lines that flank the probe set data.
Even so, for more complex experiments with multiple treatments, additional biological replicates are necessary to increase statistical power and allow differentiation between minor, but significant, changes in expression (Kuehl, 2000; Nadon and Shoemaker, 2002). To minimize within-treatment variation, it is also beneficial to pool samples from a group of identically treated plants, but the pooled sample should represent a single biological replication. In general, exploratory screening of over 21,400 genes will require adjustments for a series of treatments (temperature, light, chemical, or pathogen) or changes in developmental stage. GeneChip experiments are similar to quantitative trait studies in the field; multiple testing coupled with proper statistical analysis is necessary to avoid excessive proportions of false positives, or false negatives, among identified genes (Li and Wong, 2001; Irizarry et al., 2003).
3′-/5′-Labeling Controls
Known housekeeping genes were tiled onto the Barley1 GeneChip probe array as cRNA labeling controls. Signal intensities of 3′ and 5′ probe sets for these genes can be used as a general indicator of the quality of cDNA synthesis and subsequent cRNA labeling. Ideally, the 3′ to 5′ ratio should be close to 1. Nevertheless, algorithm-derived oligomers within a particular probe set will produce inherently different signal intensities. Thus, for each newly developed GeneChip probe array, baseline 3′ to 5′ ratios must be determined empirically. In addition, deviations from the normal 3′ to 5′ ratios are sometimes because of transcript-related or image artifact problems and are not indications of the overall quality of the labeled sample.
As illustrated in Table II, results from 100 successful Barley1 hybridizations, representing various treatments, revealed that housekeeping gene controls produced average 3′ to 5′ signal ratios between 0.73 and 1.89. The average signal intensity for Contig333 (α-tubulin) was slightly higher in the 5′ region as compared with the 3′ region. In contrast, Contig865 (glyceraldehyde-3-phosphate dehydrogenase) produced consistently higher 3′ signal intensities as compared with the 5′ signal. Contig2580 has two probe sets (Contig2580_5_at and Contig 2580_5_s_at) targeted to the 5′ region. Of these two, Contig2580_5_s_at produced nearly the same signal as the 3′ probe set. Very low 5′ signal intensities were found for Contig1390 (actin). Thus, it is recommended that the signal intensity of the middle probe set for this contig be used for calculation with the 3′ probe set.
Table II.
Probe set signal intensities and 3′ to 5′ ratios of housekeeping genes for the Barley1 GeneChip probe array.
| Probe Set Name
|
Description (NCBI BLASTn Hit)
|
Signal Intensitya
|
Remarks
|
|||
|---|---|---|---|---|---|---|
| 5′ | Middle | 3′ | 3′ to 5′ Ratio ± sdb | |||
| Contig333 | Barley α-tubulin (tuba; 0.0) | 3,381 (333_5_at)c | 4,306 (333_M_at) | 2,334 (333_3_x_at) | 0.73 ± 0.15 | Similar signal intensity across all regions of the transcript |
| Contig865 | Barley glyceraldehyde-3-phosphate dehydrogenase (0.0) | 2,792 (865_5_s_at) | - | 5,336 (865_3_s_at) | 1.89 ± 0.30 | 5′ Region produced slightly lower signal intensity as compared with 3′ |
| Contig2580 | Rice translation initiation factor 5A (e-122) | 2,096 (2580_5_s_at) | - | 1,891 (2580_3_at) | 0.84 ± 0.16 | Of the two probe sets tiled for the 5′ region, contig2580_5_s_at showed signal similar to 3′ contig2580_3_at |
| Contig1390 | Barley actin (0.0) | 583 (1390_5_at) | 5,654 (1390_M_at) | 6,975 (1390_5_s_at) | 1.07 ± 0.22 (3′/middle) | Very little signal detected from 5′ probe set; recommend use of 3′ to middle ratio |
Average of 100 GeneChip probe arrays
Average 3′ to 5′ ratio ± sd based on 100 GeneChip probe arrays
Denotes specific probe set within designated contig
Genotype and Species Evaluation
To test the efficiency of Barley1 probe sets within H. vulgare, we subsampled the results from a larger experiment where 8- to 10-cm, first seedling leaves from three barley genotypes had been challenged with the fungus Blumeria graminis f. sp. hordei, the causal agent of powdery mildew disease. In each of six replications, 10 to 20 seedlings for each genotype were harvested and pooled for RNA preparation, labeling, and hybridization. As described above, expression data was scaled globally to a target intensity of 500, and detection of present calls with Affymetrix MAS 5.0 software was determined by at least eight of the 11 probe pairs within a probe set that exceeded the default probe pair threshold (Tau = 0.015). As presented in Table III, an average of 58% of the probe sets, as designated by the MAS 5.0 report file, produced present calls when hybridized with cRNA derived from barley seeding leaves. This percentage of present calls is equivalent to those observed with cRNA derived from Arabidopsis leaves on the Affymetrix ATH1 Genome Array (R. Caldo, S. Whitham, and R. Wise, unpublished data).
Table III.
Present calls on the Barley1 GeneChip probe array when hybridized with labeled cRNA from barley first seedling leaf after inoculation with powdery mildew
| Barley Accession (Feature) | Average No. of Present callsa | % of Total | Total Present Calls per Genotypeb | Present Calls Unique to a Specific Genotypeb |
|---|---|---|---|---|
| C.I. 16137 (Mla1c) | 13,343d ± 566 | 58.4e ± 2.5 | 11,322 | 243 |
| C.I. 16151 (Mla6) | 13,025 ± 496 | 57.1 ± 2.2 | 11,269 | 244 |
| C.I. 16155 (Mla13) | 13,518 ± 479 | 59.2 ± 2.1 | 11,703 | 438 |
Derived from the MAS 5.0 report file
Present in all six replications per genotype. There were 10,491 present calls in common among all three genotypes (18 hybridizations)
C.I. is the standard abbreviation for Cereal Introduction. Mla1, Mla6, and Mla13 are alleles conferring specific resistance to the powdery mildew fungus. Ten to 20 seedlings for each species were harvested and pooled for RNA preparation, labeling, and hybridization. Data were taken from six independent biological replications
Average ± sd of all present calls in each replication based on six independent RNA isolations. Seedlings harvested directly after inoculation with powdery mildew
Calculated as a percentage of baseline barley leaf probe sets that exceed the probe pair threshold (present calls)
To survey the Barley1 probe array for conservation of probe sets across grasses, we hybridized cRNA derived from first leaves of barley green seedlings (as a control) and the equivalent developmental stage of wheat, oat (Avena strigosa), rice, sorghum (Sorghum bicolor), and maize grown at the same time under identical conditions. The raw and normalized data from this cross-species test is publicly available for download as accession BB1 within the BarleyBase database (see below). Two independent biological replications were performed with pools of 10 to 20 seedlings per replication. In comparison to a baseline result of 9,972 present calls from barley leaves, 5,392 probe sets produced present calls when hybridized with cRNA derived from wheat leaves (Table IV). At first, this suggested that approximately 54% of the Barley1 probe sets called as present for barley leaf should produce diagnostic results when hybridized with labeled cRNA derived from wheat leaves. However, as shown in Table IV, closer inspection of the data revealed that additional probe sets, outside of those that produced present calls with cRNA from barley, identify patterns of conserved gene expression in the five other cereals. Most of these conserved probe sets represent genes of unknown function, whereas others have putative functions in metabolism, signaling mechanisms, regulation of gene expression, and oxidative stress. However, because probe set selection was generally based on the 3′-most 600 nucleotides of an EST contig or singleton, genes among various cereal species are likely to have divergent regions in the 3′-untranslated region, resulting in a bias toward barley-specific probe sets (Gu et al., 2002). In this regard, it is possible that many more probe pairs provide useful signal values than are called present by the MAS 5.0 software where, for this analysis, at least eight of the 11 probe pairs were needed to exceed the probe set threshold. Therefore, normalized signal intensity may be more useful in diversity studies where continuous values are preferable. MAS 5.0 present calls are discontinuous and indicate a consensus of probe pairs for detection and, therefore, whether the usage of the probe set is appropriate (G. Tanimoto, personal communication). Nevertheless, if seedling leaf can be viewed as a representative sample, one may predict that greater than 5,000 Barley1 probe sets may produce diagnostic results when used for any given wheat experiment or tissue type. Notably, these types of data sets provide the foundation for a common set of conserved gene sequences that can be used when designing other cereal probe arrays.
Table IV.
Diversity of present calls on the Barley1 GeneChip probe array when hybridized with labeled cRNA from seedling leaf of six cereal species
| Species
|
Accession (Feature)c
|
Haploid Chromosome No.
|
Overlapping Probe Setsa
|
Unique Probe Setsb
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Barley | Wheat | Oat | Rice | Sorghum | Maize | ||||
| Barley | C.I. 16151 (diploid) | 7 | 9,972 | - | - | - | - | - | 4,348 |
| Wheat | Wheaton (hexaploid) | 21 | 4,972 | 5,392 | (1,969) | (1,311) | (1,168) | (901) | 239 |
| Oat | C.I. 3815 (diploid A genome) | 7 | 2,309 | 2,075 | 2,645 | (1,064) | (984) | (803) | 139 |
| Rice | Lemont (diploid japonica) | 12 | 1,573 | 1,392 | 1,169 | 1,911 | (856) | (723) | 146 |
| Sorghum | BTX623 (diploid) | 10 | 1,393 | 1,253 | 1,089 | 962 | 1,709 | (781) | 123 |
| Maize | B73 (diploid) | 10 | 1,099 | 980 | 877 | 796 | 868 | 1,281 | 43 |
All totals represent detection in both of two replications. Ten to 20 seedlings for each replication were harvested and pooled for RNA preparation, labeling, and hybridization. Bold nos. are probe sets of an individual species. Nos. below the bold diagonal are probe sets detected in both species. Nos. in parentheses indicate probe sets detected in both species and barley
Probe sets unique to a given species but not detected in any other
C.I. is the standard abbreviation for Cereal Introduction. Mla6 is a gene conferring specific resistance to the powdery mildew fungus
Discussion and Future Directions
In recent years, access to complete genomic sequences, coupled with rapidly accumulating data related to RNA and protein expression patterns, has made it possible to envision new ways to understand how genes contribute to complex phenotypes. Today, greater than 890,000 ESTs are available for the Triticeae, and more than 2 million ESTs are publicly available for all cereals combined (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html; http://harvest.ucr.edu/). Coupled with the monocot reference genome sequence from rice (http://rgp.dna.affrc.go.jp/IRGSP/index.html), this infrastructure enables the functional analyses of complex pathways and gene families to be performed quickly with a high degree of precision.
Because the Barley1 genome array includes 22,792 probe sets derived from more than 84 libraries, representing at least 21,439 genes, it should be valuable to monitor transcripts for almost any biological comparison. In our experience, the CAP3 parameters of p = 95, d=60, f=100, and h=50 differentiated EST contigs extensively, often to the point of unraveling alleles; however, the degree of separation was appropriately counter balanced during the probe design process to restrict this discrimination to paralogs. As a result, the Barley1 array includes a high degree of specificity, facilitating independent details of highly similar genes, such as multiple members of gene families or alternatively spliced variants to be distinguished. Multiple probe pairs per gene sequence reduces false positives, enables statistical analysis to provide confidence and probability information, and makes possible the direct quantification of the level of expression of many transcripts in one sample (Lockhart et al., 1996; Wodicka et al., 1997). In addition, probe sets that represent conserved genes in metabolism, signaling and regulation of gene expression can be used for comparative studies among grasses. Nonetheless, among these commonly expressed genes, over one-half are of unknown function, underscoring the need for continued efforts in computational and cellular biology.
To enhance the usability of the data, BarleyBase (http://barleybase.org/), a new public functional genomics resource, features “click through” integration of expression profiling experiments from researchers worldwide. Contig alignments and oligo probe information from the Barley1 GeneChip are displayed using tools developed at PlantGDB (http://www.plantgdb.org/). In addition, Barley1 GeneChip exemplars are aligned on the Gramene rice genome browser (Ware et al., 2002; http://www.gramene.org/), enabling direct links to protein pages in Gramene. The protein pages will provide Barley-Base users with the rice annotations including, but not limited to, gene function, pathway, and cellular location based upon associations to gene ontology terms.
To ensure the interpretability of the results, as well as potential verification by third parties, BarleyBase conforms to the MIAME (minimum information about a microarray experiment) standards (http://www.mged.org/Annotations-wg/). These standards help solve the accessibility and uniformity by providing a core set of data and terms that will be recorded for any microarray experiment. BarleyBase stores four basic types of information: GeneChip and/or microarray structure data, experimental and labeling protocols, actual measured test data and annotations (e.g. DAT, CEL, and CHP files from the Affymetrix Scanner), and analysis data. User-contributed GeneChip data can be downloaded in batch files for further analysis. The database can also be queried with the user's gene of interest to discover under what conditions or experiments their target showed significant change. Probe set queries are integrated with analysis tools from Bioconductor software (Ihaka and Gentleman, 1996) such as hierarchical clustering, k-means partitioning, and multi-dimensional scaling analyses. The BarleyBase repository will be enhanced by adding the Plant Ontology and Gene Ontology controlled vocabularies from Gramene. The use of these terms will allow cross-species comparisons based upon the common identifiers and will facilitate interoperability between existing plant databases, enabling queries across species to determine genes that may have exhibited similar expression profiles. Meta analyses among several data sets will facilitate future comparative and functional analyses of cereal genes.
Acknowledgments
Content of the Barley1 probe array benefited greatly from EST sequence chromatograms and/or matching sequence and quality value files generously provided by Dorrie Main (Clemson University Genomics Institute, SC); Andreas Graner, Nils Stein, Winnie Weschke, and Hangning Zhang (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany); Kazuhiro Sato, Daisuke Saisho, and Kazuyoshi Takeda (Okayama University, Japan); Yuji Kohara and Tadasu Shin-I (National Institute of Genetics, Mishima, Japan); Robbie Waugh, David Marshall, and Linda Cardle (Scottish Crop Research Institute, Dundee, UK); Hans Bohnert (Uni-versity of Illinois, Urbana-Champaign); and Alan Schulman, Ari-Matti Sarén, and Jaakko Tanskanen (Institute of Biotechnology, University of Helsinki). Thanks also to David Matthews (USDA-Agricultural Research Service, Cornell University, Ithaca, NY) for sequences that were available only from the GrainGenes database and to Shibo Zhang and Peggy Lemaux (University of California, Berkeley) for help with common reporter gene sequences, Warren Krueger (University of Minnesota, St. Paul) for assistance with detection of fungal sequences, and Jan Svensson (University of California, Riverside) for assistance with chimera detection. Special thanks goes to Xiaoqiu Huang (Iowa State University, Ames) for providing customized versions of the CAP3 assembly program and answering many questions about the algorithms, Dan Nettleton (Iowa State University) for advice on experimental design, Qunfeng Dong (PlantGDB, Iowa State University) and Doreen Ware (Gramene Database, Cold Spring Harbor, NY) for facilitating linkage with BarleyBase, and the Affymetrix (Santa Clara, CA) team of Eric Schell, Gene Tanimoto, Alan Williams, Xue Mei Zhou, Lianne McLean, Paul Doran, Mike Mittman, Venu Valmeekam, Sandra Wells, Dawn Kerber, Curtis Fideler, Joe Prosser, and Kyle O'Connor.
This work was supported by the USDA (Initiative for Future Agriculture and Food Systems grant no. 01–52100–11346 to A.K., R.P.W., R.A.W., T.J.C., and G.J.M.; National Research Initiative grant nos. 99–35300–7694 to R.A.W., T.J.C., A.K., and R.P.W., 02–35300–12619 to J.A.D. and R.P.W., 02–35300–12548 to T.J.C., and 98–35300–6169 to R.P.W.; and Cooperative State Research, Education, and Extension Service North American Barley Genome Project funds to R.P.W. and D.A.A.).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 208-218 [Google Scholar]
- Choi DW, Zhu B, Close TJ (1999) The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allelic variation, chromosome assignments, and expression characteristics of 11 Dhn genes of cv. Dicktoo. Theor Appl Genet 98: 1234-1247 [Google Scholar]
- Choi DW, Koag MC, Close TJ (2000) Map locations of barley Dhn genes determined by gene-specific PCR. Theor Appl Genet 101: 350-354 [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred: II: Error probabilities. Genome Res 8: 186-194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred: I. Accuracy assessment. Genome Res 8: 175-185 [DOI] [PubMed] [Google Scholar]
- Franckowiak J (1997) Revised linkage maps for morphological markers in barley, Hordeum vulgare. Barley Genet Newslett 26: 9-21 [Google Scholar]
- Gu Z, Nicolae D, Lu HH-S, Li W-H (2002) Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet 18: 609-613 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA Sequence Assembly Program. Genome Res 9: 868-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5: 299-314 [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2: 249-264 [DOI] [PubMed] [Google Scholar]
- Jelinsky S, Estep P, Church G, Samson LD (2000) Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol 20: 8157-8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhofs A, Graner A (2001) An integrated map of the barley genome. In RL Phillips, IK Vasil, eds, DNA-Based Markers in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 187-199
- Kleinhofs A, Han F (2001) Molecular mapping of the barley genome. In JLM-CGA Slafer, JL Araus, R Savin, I Romagosa, eds, Barley Science: Recent Advances from Molecular Biology to Agronomy of Yield and Quality. Food Product Press, New York, pp 31-45
- Kuehl RO (2000) Design of Experiments: Statistical Principles of Research Design and Analysis, Ed 2. Duxbury Press, Pacific Grove, CA
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14: 1675-1680 [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis during systemic acquired resistance. Nat Genet 26: 215-218 [DOI] [PubMed] [Google Scholar]
- Nadon R, Shoemaker J (2002) Statistical issues with microarrays: processing and analysis. Trends Genet 18: 265-271 [DOI] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, Ivanissevich SD, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuvesson S, Morgante M, Massari A et al. (2000) A simple sequence repeat-based linkage map of barley. Genetics 156: 1997-2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655-11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein-Knowles P (1992) Cloned and mapped genes: current status. In PR Shewry, ed, Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology. C.A.B. International, Wallingford, UK, pp 73-98
- Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, Teytelman L, Schmidt SC, Zhao W, Chang K et al. (2002) Gramene, a tool for grass genomics. Plant Physiol 130: 1606-1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing R, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14: 1903-1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol 15: 1359-1367 [DOI] [PubMed] [Google Scholar]
- Yu Y, Tompkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA (2000) A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet 101: 1093-1099 [Google Scholar]
- Zhu T, Wang X (2000) Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol 124: 1472-1476 [DOI] [PMC free article] [PubMed] [Google Scholar]