Abstract
Cytokinesis completion in the budding yeast S. cerevisiae is driven by tightly regulated pathways, leading to actomyosin ring contraction coupled to plasma membrane constriction and to centripetal growth of the primary septum, respectively. These pathways can partially substitute for each other, but their concomitant inactivation leads to cytokinesis block and cell death. Here we show that both the lack of the functionally redundant FHA-RING ubiquitin ligases Dma1 and Dma2 and moderate Dma2 overproduction affect actomyosin ring contraction as well as primary septum deposition, although they do not apparently alter cell cycle progression of otherwise wild-type cells. In addition, overproduction of Dma2 impairs the interaction between Tem1 and Iqg1, which is thought to be required for AMR contraction, and causes asymmetric primary septum deposition as well as mislocalization of the Cyk3-positive regulator of this process. In agreement with these multiple inhibitory effects, a Dma2 excess that does not cause any apparent defect in wild-type cells leads to lethal cytokinesis block in cells lacking the Hof1 protein, which is essential for primary septum formation in the absence of Cyk3. Altogether, these findings suggest that the Dma proteins act as negative regulators of cytokinesis.
Keywords: Hof1, Cyk3, Tem1, budding yeast, cytokinesis
Introduction
Cytokinesis is the spatially and temporally regulated process by which, after chromosome segregation, eukaryotic cells divide their cytoplasm and membranes to produce 2 daughter cells. Although cytokinesis modes appear to be different among species, several cytokinesis factors and mechanisms are evolutionarily conserved. In particular, cytokinesis in the budding yeast Saccharomyces cerevisiae involves the same broadly conserved cleavage furrow mechanism that is found in metazoans, where contraction of an actomyosin ring leads to membrane furrowing and subsequent cytokinesis.1 The subcellular localization of the components of the budding yeast cytokinetic machinery is tightly controlled and follows a hierarchical order of assembly to the mother-bud neck during the cell cycle. The first step is the assembly of a rigid, hourglass shaped septin ring, which forms at the bud neck concomitantly with bud emergence as soon as cells enter S phase, thus marking the position where constriction between mother and daughter cell will take place at the end of mitosis. The septin ring, which appears to be essential for cytokinesis in both fungal and animal cells2 serves as a scaffold for recruiting other proteins at the bud neck. Among them, the chitin synthase Chs3 builds a chitin ring, which is important for the integrity of the division site around the neck of the emergent bud.3 Septins also recruit the only S. cerevisiae type II myosin, Myo1, which forms a ring at the presumptive bud site during early S phase, and whose localization depends on septins before cytokinesis, while it is controlled by the IQ-GTPase activating protein (GAP) Iqg1 during cytokinesis.4 The Myo1 ring persists at the mother-bud neck until the end of anaphase, when a coincident F-actin ring assembles. The resulting actomyosin ring (AMR) contracts symmetrically, after the rigid septin hourglass has split into two rings, thus accomplishing centripetal septum formation.5,6 After mitotic exit, chitin synthase Chs2 is recruited to the neck to build the primary septum (PS),7 which is mostly made of chitin. Once the cytokinetic apparatus is fully assembled, AMR contraction, membrane invagination, and PS synthesis all begin almost immediately. At either side of the completed PS structure, secondary septa are then synthetized, which are made of the same components as the cell wall.8 Once cytokinesis is complete, the separation between mother and daughter cell is accomplished by the action of endochitinase and glucanases that degrade the PS from the daughter side.9
The complex pathways leading to cytokinesis completion in budding yeast appear to be tightly interconnected and partially redundant. Indeed, the AMR is involved in constricting the plasma membrane at the division site to complete closure,5,6 and AMR contraction is coupled to the centripetal growth of the PS. However, AMR contraction is not essential for cytokinesis, and Myo1 is not essential for cell viability in some yeast strains.5 In its absence, cytokinesis can be completed by AMR-independent pathways, which likely lead to an asymmetric remedial septum deposition that allows cell separation and survival.5 While Iqg1 and the formins Bni1 and Bnr1 are all required for AMR construction, the Hof1 and Cyk3 factors appear to function in a parallel pathway that coordinates septum formation.10-12 Hof1, which is a member of the evolutionarily conserved PCH protein family, can associate with septin ring from G2 to anaphase; it is phosphorylated at telophase and colocalizes with the AMR during cytokinesis.12,13 Efficient AMR contraction and cell separation are allowed by subsequent degradation of Hof1 by the SCF (Grr1) complex, which thus functions as an adaptor linking the septum synthesis machinery to the actomyosin system.14 On the other hand, the SH3-domain protein Cyk3 was identified as a high-dosage suppressor of lethality caused by deletion of either IQG115 or MYO1,16 and Cyk3 has been recently shown to be a strong activator of Chs2 in vivo.17 Moreover, Hof1 and Cyk3 function redundantly in Myo1- and Iqg1-independent recruitment to the bud neck in telophase of the essential protein Inn1, which is required for chitin synthase activation, leading to PS formation.18-20 Notewhorthy, localization at the bud neck of Inn1, which is essential for cytokinesis completion, takes place independently of AMR in cells overexpressing CYK3,19 indicating a central role for Cyk3 in a cytokinesis rescue mechanism in the absence of functional AMR.
Cytokinesis is temporally coordinated with chromosome segregation and mitotic exit by a kinase cascade, which is called mitotic exit network (MEN) and ultimately activates the protein phosphatase Cdc14, thus allowing inactivation of the mitotic cyclin-dependent kinase complexes (Clb-Cdk1 in budding yeast), a key event for both mitotic exit and cytokinesis.21 Several MEN factors, which localize at the spindle poles during mitosis,22 are found at the bud neck after mitotic exit and likely have roles in cytokinesis regulation.17,23-29 In particular, the GTPase Tem1 appears to play a direct role in AMR contraction. In fact, cells carrying a tem1 mutant allele that allows anaphase progression cannot complete cytokinesis, because the actomyosin ring, which assembles normally, never contracts and persists even after the new bud appears.23 Moreover, Tem1 was shown to interact in vitro with the GAP-related domain of Iqg1,30 which is, in turn, essential for AMR contraction.
In all eukaryotic cells, cytokinesis failure can cause aneuploidy, a hallmark of cancer cells, due to errors in chromosome inheritance. In order to prevent these problems, mitotic events and cytokinesis must be tightly controlled and coordinated by still partially unknown mechanisms. A role in these controls can be envisaged for proteins of the FHA-RING ubiquitin ligase family, such as S. pombe Dma1, human Chfr and RNF8, and budding yeast Dma1 and Dma2, which all appear to control different aspects of the mitotic cell cycle, although several molecular details of their functions are still obscure.31 In particular, we have previously implicated the 58% identical and functionally redundant Dma proteins of S. cerevisiae in mitotic checkpoints32 and in regulation of septin ring dynamics and cytokinesis.32,33 Moreover, in vitro ubiquitin ligase activity has been described for these proteins,34 which were shown to be invoved in ubiquitylation of the Cdk1 inhibitor Swe135 as well as of septins,36 but their functions and targets in cytokinesis control are still undefined.
In this study, we provide evidence that budding yeast Dma proteins contribute to control both AMR contraction and PS deposition. In fact, by using moderate Dma2 overproduction conditions, we show that they cause inhibition of AMR contraction as well as asymmetric PS deposition. Moreover, the interaction between Tem1 and Iqg1, which is thought to be required for AMR contraction, is inhibited both in vitro and in a 2-hybrid assay by Dma2 excess that also causes mislocalization of the positive regulator of PS formation Cyk3. These defects, which are tolerated in otherwise wild-type cells, cause cytokinesis failure in Hof1-lacking cells, leading to their arrest as chains of cells with connected cytoplasms and no PS deposition. The role of the Dma proteins in cytokinesis is confirmed by the observation that their lack causes a partial defect in AMR contraction and Cyk3 localization, and that, accordingly, dma1Δ dma2Δ cells show an abnormal PS shape.
Results
Dma2 impairs a parallel cytokinesis pathway with respect to Hof1
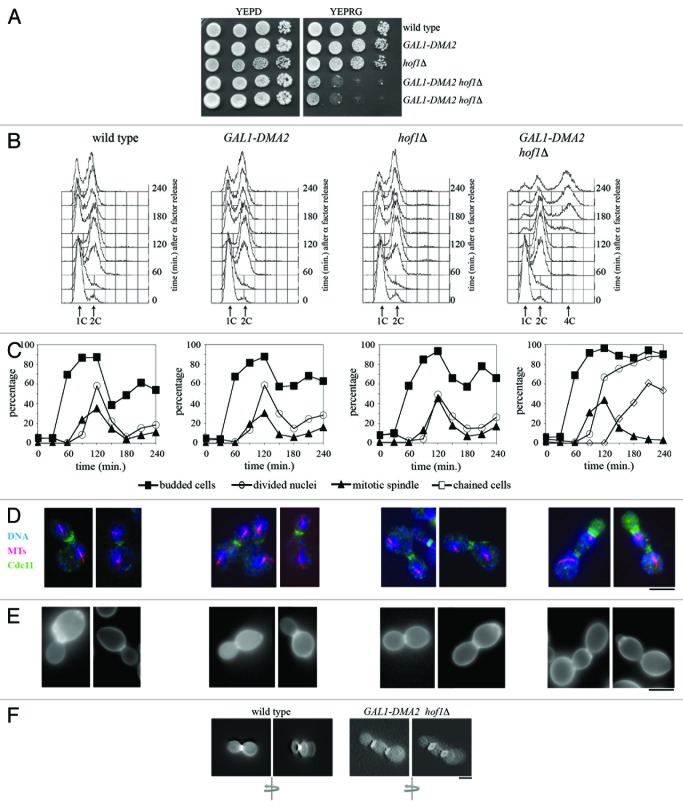
We previously implicated the Dma proteins in the control of mitotic progression. In order to gain insights into the molecular mechanism of this event, we studied genetic interactions between the Dma proteins and other factors involved in cytokinesis. The concomitant lack of DMA1 and DMA2, which does not cause growth defects in otherwise wild-type cells,32 did not affect viability or growth of meiotic segregants lacking either HOF1 or CHS2 or expressing the temperature sensitive (ts) allele iqg1-1 at the permissive temperature (Table 1). On the other hand, cyk3Δ dma1Δ dma2Δ meiotic segregants formed smaller colonies than cyk3Δ or dma1Δ dma2Δ meiotic segregants (Table 1), and this phenotype correlated with a partial cytokinesis defect (see below). In addition, galactose-induced DMA2 overexpression from a single copy of a GAL1-DMA2 fusion integrated in the genome, which we previously showed not to cause any growth defect in otherwise wild-type cells,32 was used to evaluate the possible effects in the same mutants described above of this moderate Dma2 overproduction. These same GAL1-DMA2 moderate overexpression conditions were used also for all the subsequent experiments. As shown in Table 1, GAL1-DMA2 overexpression did not affect the growth of chs2Δ and cyk3Δ meiotic segregants, but it caused poor growth of igq1-1 meiotic segregants at the permissive temperature, likely due to a partial cytokinesis defect (our unpublished observation). Moreover, overexpression of either GAL1-DMA2 or GAL1-DMA1 was lethal in the absence of the cytokinesis protein Hof1 (Table 1), further indicating redundancy between the 2 Dma proteins. We therefore performed more detailed analyses using only moderate GAL1-DMA2 overexpression. In agreement with the synthetic lethal effect observed in meiotic segregants, the ability of GAL1-DMA2 hof1∆ cells to form colonies on galactose-containing plates was strongly reduced compared with wild-type, GAL1-DMA2, or hof1∆ cells (Fig. 1A), which all formed colonies with similar efficiency under the same conditions. In order to reveal the terminal phenotype of GAL1-DMA2 hof1∆ cells, we synchronized wild-type, hof1∆, GAL1-DMA2, and GAL1-DMA2 hof1∆ cell cultures in G1, followed by release into cell cycle in the presence of galactose to induce Dma2 overproduction. After the release, the kinetics of DNA replication (Fig. 1B), budding, nuclear division, mitotic spindle assembly (Fig. 1C and D), and septin ring formation (Fig. 1D) showed very similar profiles in wild-type, hof1∆ and GAL1-DMA2 cells. By contrast, GAL1-DMA2 hof1∆ cells exited from mitosis, but then accumulated as chains of cells with more than 2C DNA content (Fig. 1B), divided nuclei, and disassembled spindles (Fig. 1C) and split the septin ring (Fig. 1D). Cells in the chains were not separable by zymolyase treatment (data not shown), indicating cytokinesis failure rather than cell separation defects. Indeed, Calcofluor White staining of chitin clearly showed that GAL1-DMA2 hof1∆ cell chains suffered primary septum deposition defects (Figs. 1E and F and 4A). In fact, they did not show the chitin disk structure that is typical of PS-containing cells, which was instead detectable in all control cells under the same conditions, but showed a chitin ring that is built by the chitin synthase Chs3 in early S phase8 (Fig. 1F). Thus, although Dma2 moderate overproduction does not alter cell cycle progression in otherwise wild-type cells, it leads to cytokinesis inhibition in the absence of Hof1, suggesting that Dma2 might impair a parallel cytokinesis pathway with respect to Hof1.
Table 1. Genetic interactions between the Dma proteins and other factors involved in cytokinesis.
| Genotype | Phenotype at 25 °C on YEPD | Phenotype at 25 °C on YEPRG |
|---|---|---|
| dma1Δ dma2Δ chs2Δ | healthy | ND |
| dma1Δ dma2Δ iqg1-1 | healthy | ND |
| dma1Δ dma2Δ cyk3Δ | sick/slow growth | ND |
| dma1Δ dma2Δ hof1Δ | healthy | ND |
| GAL1-DMA2 chs2Δ | healthy | healthy |
| GAL1-DMA2 iqg1–1 | healthy | sick/slow growth |
| GAL1-DMA2 cyk3Δ | healthy | healthy |
| GAL1-DMA2 hof1Δ | healthy | lethal |
| GAL1-DMA1 hof1Δ | healthy | lethal |
Strains carrying DMA1 and DMA2 deletions or expressing GAL1-DMA2 or GAL1-DMA1 were crossed with the chs2Δ, iqg1-1, cyk3Δ, and hof1Δ mutants. The resulting diploids were induced to sporulate and meiotic segregants were assayed for their ability to grow on rich medium containing glucose (YEPD) or galactose (YEPRG) at 25 °C. Only the phenotypes of meiotic segregants with the indicated genotypes are shown, the meiotic segregants with all the other possible genotypes all showed healthy phenotypes. ND, not determined.
Figure 1. Dma2 excess is toxic for cells lacking Hof1.(A) Serial dilutions of stationary phase cultures of strains with the indicated genotypes were spotted on YEPD or YEPRG plates, which were then incubated for 2 d at 25 °C. (B–F) Exponentially YEPR growing cultures of wild-type, GAL1-DMA2, hof1Δ, and GAL1-DMA2 hof1Δ cells were arrested in G1 by α-factor and released from G1 arrest in YEPRG at 25 °C (time 0). At the indicated times after release, cell samples were taken for FACS analysis of DNA contents (B), and for scoring budding, chained cells, nuclear division, and mitotic spindle formation (C). (D and E) Pictures were taken 120 min after release for wild-type, GAL1-DMA2, and hof1Δ cells, or 240 min after release for GAL1-DMA2 hof1Δ cells, to show in situ immunofluorescence analysis of nuclei (DNA), mitotic spindles (MTs) and septin ring deposition (Cdc11) (D), as well as chitin deposition analysis (E) by using Calcofluor White staining. (F) Three-dimensional reconstruction of pictures taken at different Z-stacks of wild-type and GAL1-DMA2 hof1Δ cells treated as in panel (E). The arrows indicate the direction of rotation. Wild-type cells show a complete chitin disk at the bud neck (left), while GAL1-DMA2 hof1Δ cells can only assemble a chitin ring (right). Bar, 5 μm.
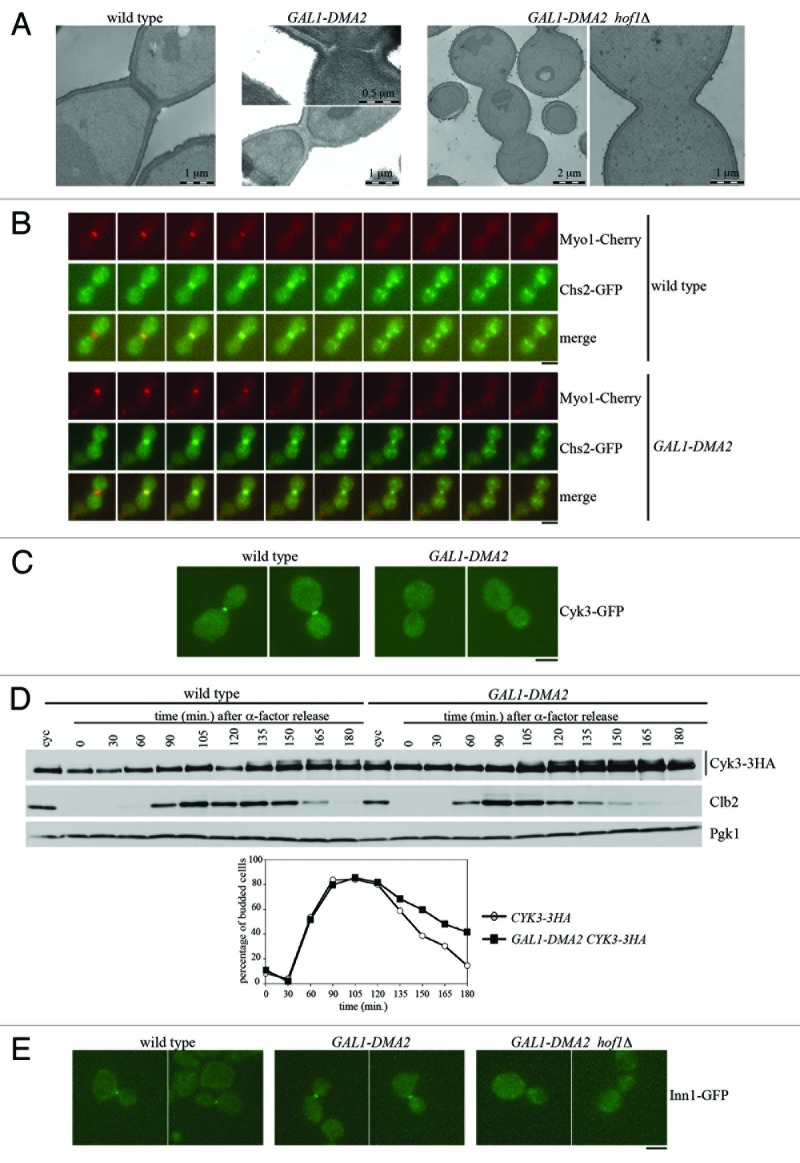
Figure 4. Dma2 overproduction affects PS formation and Cyk3 localization at the division site.(A) Samples for TEM analysis were taken from exponentially growing cultures of wild-type, GAL1-DMA2 and GAL1-DMA2 hof1Δ cells, which were pre-grown in YEPR medium and then incubated in YEPRG medium (1% galactose) for 4 h (wild-type and GAL1-DMA2) or 6 h (GAL1-DMA2 hof1Δ). The GAL1-DMA2 top and bottom panels show cells with either defective or completed PS formation, respectively. The GAL1-DMA2 hof1Δ panel on the right is a magnification of the bud neck in the cell of the adiacent panel.(B) Exponentially growing cultures of wild-type and GAL1-DMA2 cells, all expressing MYO1-Cherry and CHS2-GFP, were arrested in G1 by α-factor and released from G1 arrest at 25 °C in raffinose–galactose synthetic medium plates for time-lapse analysis. Pictures as shown in the panel were taken every 2 min for 5 h, and time-lapsed images were assembled to show Myo1 and Chs2 dynamics (Videos S3 and S4). Bar, 5 μm.(C) Exponentially growing cultures of CYK3-GFP and GAL1-DMA2 CYK3-GFP cells were arrested in G1 by α-factor and released from G1 arrest at 25 °C in YEPRG medium. Pictures were taken 120 min after release to show Cyk3-GFP localization. Bar, 5 μm.(D) Exponentially growing cultures of CYK3–3HA and GAL1-DMA2 CYK3-3HA cells were treated as in (B). At the indicated times, cell samples were taken for scoring budding (graph) and for determining Cyk3 and Clb2 levels by western blot analysis with anti-HA, anti-Clb2 and anti-Pgk1 (loading control) antibodies.(E) Exponentially growing cultures of INN1-GFP, GAL1-DMA2 INN1-GFP, and GAL1-DMA2 hof1Δ INN1-GFP cells were treated as in (B). Pictures were taken 165 min after release to show Inn1-GFP localization. Percentages of large budded binucleate cells with Cyk3 and Inn1 localization at the bud neck are reported in Table 2. Bar, 5 μm.
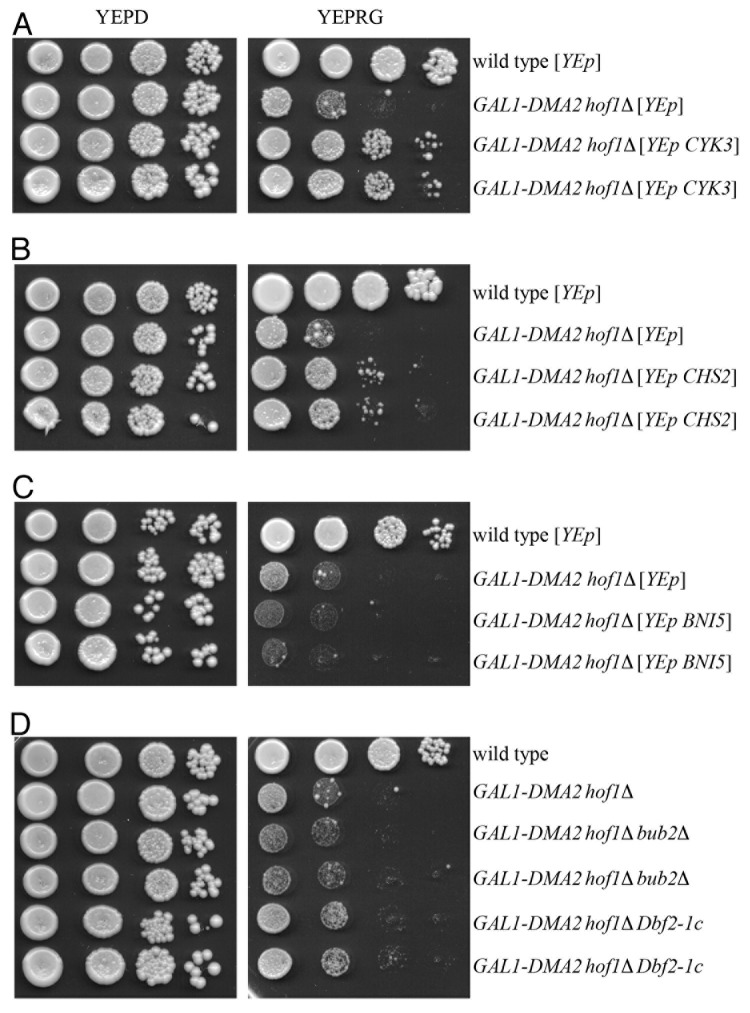
Budding yeast can tolerate either loss of AMR in myo1∆ cells or partial defects in PS formation in hof1∆ or cyk3∆ cells, but not both. Indeed, HOF1 deletion causes cell lethality in cells lacking either PS deposition proteins, like the chitin synthase Chs2 and its activator Cyk3, or proteins required for AMR formation, such as the septin regulator Bni5, the formin Bni1, and the myosin heavy chain Myo1.20 If the lethal effect of moderate Dma2 excess in the absence of Hof1 were due to Dma2-dependent inhibition of some cytokinesis factor(s) acting in parallel with Hof1, high levels of the same factor(s) might be expected to rescue this effect. Strikingly, both CYK3 and CHS2 overexpression from high copy number plasmids partially suppressed galactose-induced GAL1-DMA2 hof1∆ synthetic lethality (Fig. 2A and B), indicating that the cytokinesis defect of these cells can be rescued by forcing PS formation. Conversely, high levels of the septin-binding protein Bni5 did not increase viability of galactose-induced GAL1-DMA2 hof1∆ cells (Fig. 2C), which, on the other hand, did not exhibit defects in septin ring structure and dynamics (Fig. 1D). Based on these data, we did not assay the effects of septin overproduction. As MEN activity is required not only for mitotic exit but also for cytokinesis,23,24,37,38 we instead investigated the effects of MEN hyperactivation through either the lack of its inhibitor Bub2 or the Dbf2-1c hyperactive allele.39 As shown in Figure 2D, GAL1-DMA2 hof1∆ cell lethality on galactose-containing plates was not rescued by either condition of MEN hyperactivation, indicating that it is not likely due to MEN impairment. On the other hand, MEN targets for promoting cytokinesis are probably not available or not accessible in GAL1-DMA2 hof1∆ cells (see below).
Figure 2. Toxic effects of moderate Dma2 overproduction in cells lacking Hof1 are counteracted by high Chs2 and Cyk3 levels. (A–D) Serial dilutions of stationary phase cell cultures of strains with the indicated genotypes were spotted on YEPD or YEPRG plates, which were then incubated for 2 d at 25 °C. With the exception of the strains carrying the BUB2 deletion or the Dbf2-1c allele (D), which were grown in YEPD medium, cell cultures were grown to stationary phase in synthetic medium lacking leucine (A and C) or tryptophan (B) in order to maintain selective pressure for maintainance of the YEp high copy number plasmids.
Dma2 overproduction impairs AMR contraction and Tem1-Iqg1 interaction
The data above suggest that Dma2 excess can interfere with some cytokinesis-promoting pathway(s). Nonetheless, GAL1-DMA2 overexpression does not affect cell viability of otherwise wild-type cells32 (Fig. 1A), likely because budding yeast cytokinesis is driven by parallel and partially redundant pathways. As the localization of the cytokinetic machinery to the division site is crucial for its function, we analyzed the subcellular localization of important cytokinesis proteins in galactose-induced GAL1-DMA2 cells.
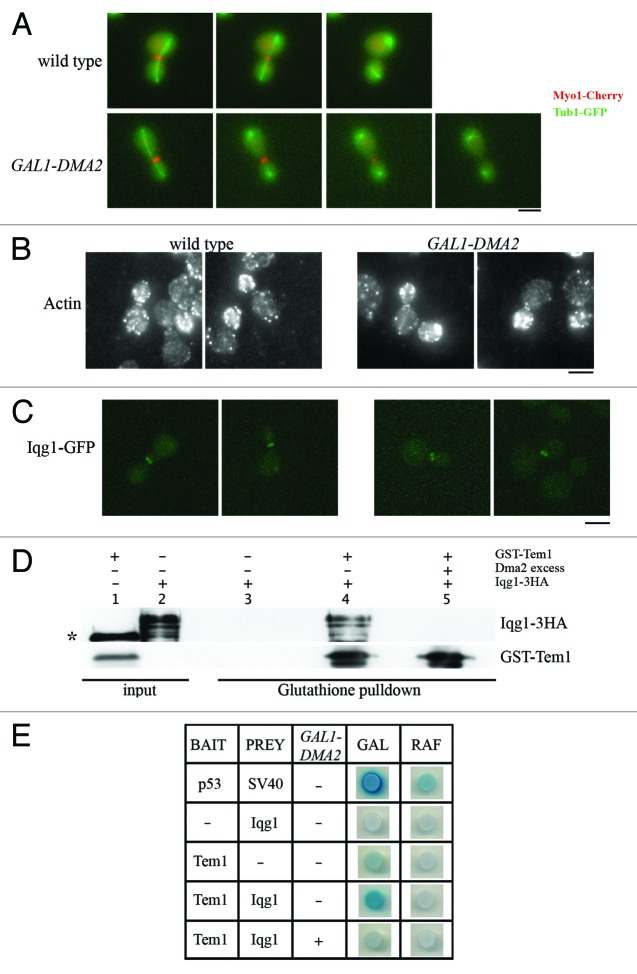
In order to analyze AMR formation and contraction, we generated wild-type and GAL1-DMA2 strains expressing a functional fluorescent variant of myosin I (Myo1-Cherry) to analyze its localization in synchronized cells. Preliminary analysis showed that Myo1-Cherry bars, which formed at the bud neck of both wild-type and GAL1-DMA2 cells, persisted longer in Dma2-overproducing cells than in wild-type cells, where they properly contracted into a single dot and disappeared (data not shown). In order to analyze this defect in detail, we performed live-cell imaging of wild-type and GAL1-DMA2 cells that expressed Myo1-Cherry and the mitotic spindle marker Tub1-GFP. After release from a G1 arrest, AMR contracted and disappeared from the bud neck of wild-type cells 5.6 min (± 1.88, in 76 cells) after mitotic spindle breakdown, while this time was significantly longer in Dma2-overproducing cells (11.2 min ± 4.4, in 98 cells) (Fig. 3A and Videos S1 and S2). The difference between the 2 strains is significant (P value obtained by Wilcoxon rank-sum test P = 7.6892 10−19). This phenomenon was not due to the lack of actin localization at the bud neck, as the actin distribution patterns were very similar in wild-type and GAL1-DMA2 cells (Fig. 3B). In addition, as shown in Figure 3C and summarized in Table 2, comparison of wild-type and GAL1-DMA2 synchronized cells revealed that localization of an Iqg1-GFP fusion at the bud neck of late anaphase cells (large budded, binucleate) was not significantly affected by GAL1-DMA2 overexpression. Defective AMR contraction in Dma2-overproducing cells did not appear to be due to hyperstabilization of the septin ring, because the same kinetics of AMR contraction was observed in galactose-induced GAL1-DMA2 and GAL1-DMA2 cdc12-1 cells, although the septin ring was destabilized in the latter cells due to the temperature-sensitive cdc12-1 allele (Fig. S1). We then hypothesized that defective AMR contraction in Dma2 overproducing cells might be due to specific inhibition of properly localized AMR components.
Figure 3. Dma2 excess affects AMR contraction and Tem1-Iqg1 interaction.(A) Exponentially growing cultures of wild-type and GAL1-DMA2 cells, all expressing fully functional MYO1-Cherry and TUB1-GFP fusions, were arrested in G1 by α-factor and released from G1 arrest at 25 °C in raffinose–galactose synthetic medium plates for time-lapse analysis. Pictures as shown in the panel were taken every 5 min for 5 h and time-lapsed images were assembled to show Myo1-Cherry ring contraction (Videos S1 and S2). Bar, 5 μm. (B and C) Exponentially growing cultures of IQG1-GFP and GAL1-DMA2 IQG1-GFP cells were arrested in G1 by α-factor and released from G1 arrest at 25 °C in YEPRG medium. Pictures were taken 150 min after release to show actin and Iqg1-GFP localization. Actin was visualized by in situ immunofluorescence analysis by using anti-actin antibodies. The percentage of late anaphase cells (large budded, binucleate) with Iqg1 localization at the bud neck is reported in Table 2.Bar, 5 μm.(D) Glutathione beads bound to GST-Tem1 were incubated with native protein extracts prepared from either wild-type or Dma2 overproducing cells, washed, and incubated with native protein extracts containing Iqg1-3HA (glutathione pull-down). Washed and boiled beads were subjected to SDS-PAGE followed by immunoblotting with anti-HA and anti-GST antibodies. Equivalent aliquots of the lysates were also loaded on the gel (input). The asterisk marks an aspecific band.(E) Plasmid pEG202-Tem1 carrying the lexADB-Tem1 fusion under the ADH promoter (bait) and plasmid pJG4-5 carrying the B42AD-Iqg1 fusion under the GAL1 promoter (prey) were cotrasformed with the β-galactosidase reporter plasmid pSH18-34 in the wild-type yeast strain EGY48 (see “Materials and Methods”). To assess 2-hybrid interaction, the indicated strains were spotted on 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-GAL) selective synthetic plates containing either raffinose (RAF, prey not induced) or 2% galactose (GAL, prey expressed). To assess the 2-hybrid interaction of Iqg1 and Tem1 in presence of Dma2 excess, we transformed the strain described above with a high copy number plasmid carrying the GAL1-DMA2 construct. As a positive control we used the strain carrying the p53 protein as the bait and the SV40 protein as the prey.
Table 2. Iqg1, Cyk3 and Inn1 localization at the division site of large budded binucleate cells.
| Strain genotypes | Minutes after α-factor release | Budded cells % | Binucleate cells % | Iqg1 at bud neck % ± SEM |
|---|---|---|---|---|
| IQG1-GFP | 150 | 66.7 | 29.2 | 31.8 ± 1.5 |
| GAL1-DMA2 IQG1-GFP | 150 | 75.7 | 40.0 | 43.4 ± 1.8 |
| Cyk3 at bud neck % ± SEM | ||||
| CYK3-GFP | 120 | 86.3 | 66.4 | 22.2 ± 1.2 |
| GAL1-DMA2 CYK3-GFP | 120 | 88.3 | 69.3 | 2.8 ± 0.9 |
| Inn1 at bud neck % ± SEM | ||||
| INN1-GFP | 165 | 63.2 | 53.2 | 19.9 ± 1.1 |
| GAL1-DMA2 INN1-GFP | 165 | 67.4 | 47.3 | 19.8 ± 1.2 |
| GAL1-DMA2 INN1-GFP hof1∆ | 165 | 82.9 | 61.3 | 1.0 ± 0.8 |
The percentages are the mean of the values obtained at the indicated time points in triplicates of the experiments described in Figure 3 (Iqg1) and Figure 4 (Cyk3 and Inn1). About 100 large budded binucleated cells were scored for protein localization in each cell culture for each experiment ± standard mean errors.
AMR contraction is brought about by Iqg1 interaction with the actomyosin ring, and Iqg1 has been shown to interact with Tem1 in vitro,30 leading to the proposal that its binding to Tem1 might be required for its activation. We therefore asked whether Tem1-Iqg1 complex formation was inhibited by Dma2 excess in the previously described in vitro assay.30 Glutathione pulldown analysis clearly showed Tem1-Iqg1 interaction in the absence (Fig. 3D, lane 4), but not in the presence of Dma2 excess (Fig. 3D, lane 5). In addition, Tem1 turned out to interact with Iqg1 by a 2-hybrid assay (see Material and Methods; Fig. 3E), and this interaction was impaired by GAL1-DMA2 overexpression. Thus, Dma2 overproduction appears to impair both AMR contraction and Tem1-Iqg1 interaction.
PS formation and Cyk3 localization at the bud neck are inhibited by Dma2 excess
As shown by trasmission electron microscopy (TEM), galactose-induced GAL1-DMA2 cells exhibited asymmetric PS deposition (Fig. 4A, middle top), which is typical of mutants impaired in AMR contraction.12 However, these cells ultimately formed a trilaminar septum (Fig. 4A, middle bottom), and indeed they were viable (Fig. 1A). PS is synthetized by Chs2, so we compared Chs2 dynamics at the bud neck of wild-type and GAL1-DMA2 cells that express Myo1-Cherry and Chs2-GFP by live-cell imaging on cells released from a G1 arrest. As expected, Chs2 accumulates at the bud neck after mitotic exit7 and remains there for 9–10 min in both strains (Videos S3 and S4). Interestingly, we observed that Chs2 contracts and disappears by endocytic removal within 4.17 min after AMR contraction in wild-type cells, and this phenomenon occurs earlier in GAL1-DMA2 cells (2.78 min.; P value of Wilcoxon rank-sum test P = 2.89 10−4). In addition, Chs2-GFP and AMR always constrict together and symmetrically in wild-type cells (28 of 28 cells), while they contract asymmetrically in GAL1-DMA2 cells (33 of 36 cells) (Fig. 4B; Videos S3 and S4), consistent with the asymmetric PS deposition observed in these cells (Fig. 4A, middle top).
We then analyzed galactose-induced GAL1-DMA2 cells for the subcellular localization of the Cyk3 and Inn1 proteins, which are both important for PS formation. To this end, we generated wild-type and GAL1-DMA2 strains expressing functional Cyk3-GFP and Inn1-GFP fusions that normally localized at the bud neck of both cell types in the absence of galactose (data not shown). Cell cultures of these strains were released from G1 arrest into galactose-containing medium to monitor DNA replication by FACS analysis (not shown), budding, and nuclear division (Table 2) as well as protein levels (not shown) and GFP localization (Table 2 and Fig. 4C and E). Strikingly, Cyk3 was found at the bud neck of only 2.8% of late anaphase GAL1-DMA2 cells, while it localized at the bud neck of 22.2% of late anaphase wild-type cells, as expected (Fig. 4C and Table 2). Western blot analysis showed similar Cyk3 total amounts in wild-type and GAL1-DMA2 cells throughout the experiment (data not shown), ruling out the possibility that mislocalization of Cyk3 could be due to its degradation. Moreover, wild-type and GAL1-DMA2 galactose-induced cell cultures released into cell cycle from G1 arrest contained very similar amounts not only of total Cyk3, but also of low electroforetic mobility Cyk3 species (Fig. 4D), indicative of Cyk3 phosphorylation,40 which is in turn required for Cyk3 recruitment to the bud neck.41 Thus Dma2 excess does not seem to inhibit phosphorylation of Cyk3, although it inhibits its bud neck localization.
The Inn1 protein is essential for cytokinesis, as both its lack and its defective localization lead to cytokinesis failure.18,20 This protein is targeted to the bud neck by the concerted action of Hof1 and Cyk3 and is properly localized in both hof1∆ and cyk3∆ cells, while concomitant Hof1 and Cyk3 lack causes Cyk3 mislocalization and cell death.19 Inn1-GFP localization was analyzed (Fig. 4E), and Inn1 was found at the bud neck of 19.9% late anaphase wild-type cells and of 19.8% late anaphase GAL1-DMA2 overexpressing cells (Table 2). On the contrary, Inn1-GFP bud neck localization dropped in GAL1-DMA2 hof1Δ cells (1% of late anaphase cells, Table 2). These data are consistent with the observation that cytokinesis is blocked in the absence of Hof1 by moderate Dma2 overproduction (Fig. 1), which also inhibits both AMR contraction and Cyk3 localization. The same Dma2 excess by itself might allow cytokinesis completion, likely because Hof1 can promote Inn1 localization and activation also under these conditions. On the other hand, Hof1 lack in the presence of Dma2 excess results in cytokinesis block, because all pathways that could promote the final step of cell division are inactive. Indeed, GAL1-DMA2 hof1∆ cells completely failed to build the primary septum (100% of chained cells), as exemplified by TEM pictures in Figure 4A.
Tem1 ubiquitylation and the Dma proteins
Dma1 and Dma2 act as E3 ubiquitin ligases in vitro34 are involved in Swe1 ubiquitylation in vivo35 and have been recently implicated in septins’ ubiquitylation,36 raising the possibility that they might regulate Tem1-Iqg1 interaction and Cyk3 localization by ubiquitylation. Iqg1 is known to be ubiquitylated by Cdh1/APC,16 while no ubiquitylation data are available for Tem1 and Cyk3. We first tested Iqg1 ubiquitylation in cdh1Δ cells, but we could not observe any residual ubiquitylation (data not shown). Moreover, we could not obtain reproducible results for Cyk3 ubiquitylation. In order to investigate Tem1 ubiquitylation, we produced wild-type, TEM1-3HA, and TEM1-3HA GAL1-DMA2 cells containing high copy number plasmids carrying a construct expressing 6xHis-tagged ubiquitin (YEp-CUP1-6xHIS-UBI4) or untagged ubiquitin (YEp-CUP1-UBI4) from the copper-inducible CUP1 promoter.42 After purification from crude extracts of the ubiquitylated proteins by Nickel (Ni) pulldown (see “Materials and Methods”), Tem1 ubiquitylated forms were detected in the pulldown eluates by western blot analysis with anti-HA andibody.
When Ni pulldowns were performed on crude extracts from samples taken at different time points after release from G1 arrest of TEM1-3HA cells expressing 6xHis-tagged ubiquitin, Tem1-ubiquitylated forms began to accumulate at 105 min after release (Fig. 5A), when 55% of cells were binucleate (Fig. 5B), and then decreased concomitantly with AMR contraction (Fig. 5B), suggesting that this event might require Tem1 deubiquitylation.
Figure 5. Tem1 undergoes cell cycle-dependent ubiquitylation. (A and B) TEM1–3HA cells, carrying high copy number plasmids with the CUP1-6xHIS-UBI4 construct and expressing the Myo1-Cherry fusion, were grown to log phase in glucose synthetic medium lacking tryptophan, arrested in G1 by α-factor and released in the cell cycle at 25 °C in YEPD medium containing 250 μM CuSO4 (time 0). At the indicated times after release, cell samples were taken for Ni pull-downs (A) and for scoring budding, nuclear division, mitotic spindle formation, and AMR assembly (B).(C) Wild-type, TEM1-3HA and GAL1-DMA2 TEM1-3HA cells, carrying high copy number plasmids with the CUP1-6xHIS-UBI4 construct or the CUP1-UBI4 construct, were grown to log phase in raffinose containing synthetic medium lacking tryptophan and incubated in the presence of 1% galactose for 30 min, followed by additional 3 h incubation in the presence of 250 μM CuSO4. Cells were then lysed and aliquots of the lysates were incubated with Ni-NTA beads to purify the proteins bound by 6xHis-ubiquitin (Ni pull-down), followed by SDS-PAGE and western blot analysis with anti-HA antibody of the bead eluates. Equivalent aliquots of the lysates were also loaded on the gel (input). The asterisk in (A and C) marks an aspecific band.
When a similar approach was used to compare Tem1 ubiquitylation in exponentially growing wild-type and GAL1-DMA2-overexpressing cells, the amount of ubiquitylated Tem1 was higher in the eluates from GAL1-DMA2-overexpressing cells (Fig. 5C, lane 12) than in wild-type eluates (Fig. 5C, lane 10). As total Tem1 levels in GAL1-DMA2 overproducing cells were not different from those found in wild-type cells (Fig. S2), Tem1 ubiquitylation in the presence of Dma2 excess does not appear to trigger its degradation. In a similar experiment, we found no difference in the amounts of Tem1 ubiquitylated forms in Ni pulldowns performed on wild-type or dma1Δ dma2Δ protein extacts (data not shown). Thus, although Dma2 excess increases the amount of ubiquitylated Tem1, we found no evidence for a role of the Dma proteins in Tem1 ubiquitylation under unperturbed conditions.
The lack of the Dma proteins impairs primary septum formation
As previously reported, dma1Δ dma2Δ cells progress normally through the cell cycle,32,33 and after exit from mitosis, they divide with kinetics similar to wild-type cells. As the effects of DMA2 overexpression suggested that the Dma proteins might act as inhibitors of AMR contraction and PS deposition, we investigated these processes in details in the absence of the same proteins.
We performed time-lapse analysis of AMR contraction in living cells by monitoring the Myo1-Cherry fusion in wild-type and dma1Δ dma2Δ cells that also expressed Tub1-GFP. AMR contracted and disappeared from the bud neck of wild-type cells 3.36 min (± 1.56, 132 cells) after mitotic spindle breakdown, while this time was reduced in cells lacking Dma1 and Dma2 (1.8 min ± 1.58, 100 cells) (Fig. 6A; Videos S5 and S6). The difference between the 2 strains is significant (P value obtained by Wilcoxon rank-sum test P = 2.6 10−15) and indicates that the Dma proteins are involved in maintaining proper AMR contraction timing with respect to mitotic exit.
Figure 6. The lack of Dma proteins partially impairs AMR contraction and PS deposition.(A) Exponentially growing cultures of wild-type and dma1Δ dma2Δ cells, all expressing MYO1-Cherry, and TUB1-GFP were plated on glucose synthetic medium plates at 25 °C for time-lapse analysis. Pictures as shown in the panel were taken every 3 min for 5 h, and time-lapsed images were assembled for showing Myo1-Cherry ring contraction (Video S5 and 6). Bar, 5 μm.(B) Exponentially growing cultures of CYK3-3HA and dma1Δ dma2Δ CYK3-3HA cells were arrested in G1 by α-factor and released from G1 arrest at 25 °C in YEPD medium (time = 0). At the indicated times, cell samples were taken for scoring budding and for determining Cyk3 levels by western blot analysis with anti-HA and anti-Cdc11 (loading control) antibodies.(C) Exponentially growing cultures of CYK3-GFP and GAL1-DMA2 CYK3-GFP cells were treated as in (B). Pictures were taken 120 min after release to show Cyk3-GFP localization. Bar: 5 μm.(D) Samples for TEM analysis were taken from exponentially growing cultures of wild-type and dma1Δ dma2Δ cells grown in YEPD medium at 25 °C.(E) FACS analysis of DNA content of the indicated strains exponentially growing in YEPD medium at 25 °C.
To evaluate Dma1 and Dma2 role in PS deposition, we analyzed the levels, phosphorylation, and subcellular localization of a functional Cyk3-3HA and Cyk3-GFP fusions in synchronized cell cultures. We found no differences in Cyk3-3HA total levels and phosphorylation between CYK3-3HA and CYK3–3HA dma1Δ dma2Δ strains during a synchronous cell cycle in standard YEPD medium (Fig. 6B). In addition, CYK3-GFP and CYK3-GFP dma1Δ dma2Δ synchronous cells showed very similar kinetics of recruitment of the Cyk3-GFP fusion to the bud neck (not shown). Nonetheless, the lack of Dma proteins caused Cyk3 mislocalization during cytokinesis in a small fraction of cells (6% ± 0.5 of 102 cells vs. 0% of 105 wild-type cells; Fig. 6C).
Thus, the lack of Dma proteins causes partial defects in the timing of AMR contraction and in Cyk3 localization. Accordingly, dma1Δ dma2Δ cells showed abnormally shaped PS (65% of 26 cells, vs. 10% of 20 wild-type cells), as judged by TEM analysis (Fig. 6D). However, these defects do not either block cell division or impair viability of dma1Δ dma2Δ cells, likely due to redundancy of cytokinesis controls. Indeed, the lack of the Dma proteins caused a cytokinesis defect in cyk3Δ cells, as cyk3Δ dma1Δ dma2Δ cells grew slowly (Table 1) and accumulated as chained cells (not shown) with more than 2C DNA content (Fig. 6E).
Discussion
The events leading to cytokinesis must be tightly controlled and coordinated with nuclear division in order for a single eukaryotic cell to generate 2 identical daughter cells at the end of the mitotic cell cycle. Genetic studies revealed the existence of 2 partially redundant pathways that allow cytokinesis completion in budding yeast. The first pathway leads to the assembly of a functional AMR and requires Tem1, Iqg1, Bni1, Myo1, and actin. The second pathway controls septum formation and involves a number of proteins, among which Cyk3, Hof1, and Inn1. This feature ensures that cytokinesis is completed even in the absence of some proteins important for one of the two pathways, thus allowing cell survival.
In previous studies, Dma1 and Dma2 were shown to have redundant roles in mitotic spindle orientation, mitotic checkpoints, regulation of septin ring dynamics,32,33 and septin organization and function.36 The results of this work indicate that the Dma proteins participate in both pathways that control cytokinesis (Fig. 7). In particular, their lack alters the timing of AMR contraction with respect to mitotic spindle disassembly, causes a partial cytokinesis defect in cyk3Δ cells, and impairs proper PS deposition. In addition, Dma2 appears to function as a cytokinesis inhibitor, as we show that its moderate overproduction impairs both symmetric AMR contraction and PS formation, likely by acting on specific targets in these 2 cytokinesis pathways. These findings are consistent with published data on orthologs of the Dma proteins in other organisms. In particular, human RNF8 overexpression caused a delay in cytokinesis,43 and ectopic expression of S. pombe Dma1 was shown to inhibit septum formation, leading to generation of elongated and multinucleate cells.44,45
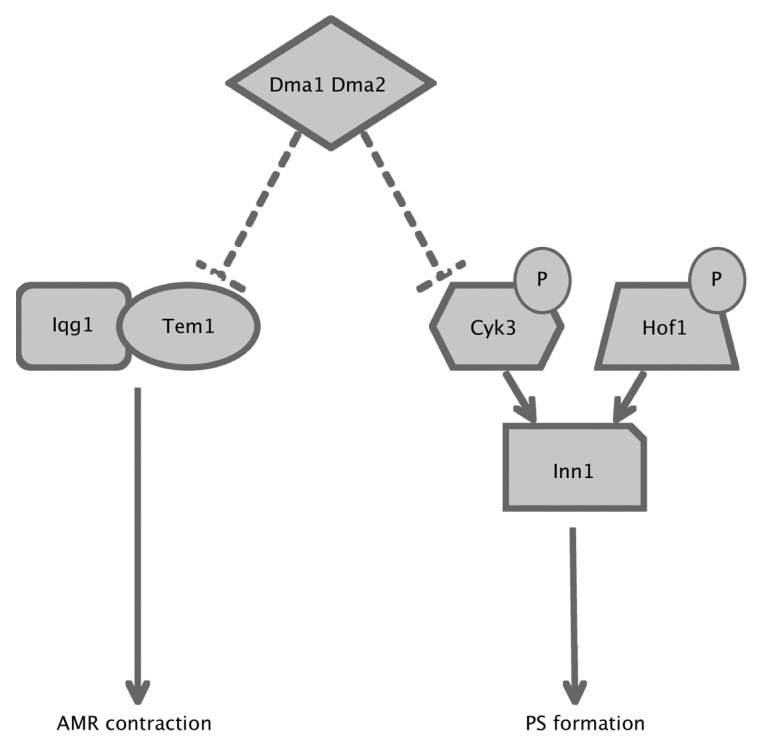
Figure 7. A model for the role of Dma proteins in cytokinesis control. Dma1 and Dma2 participate, directly or indirectly, in the control of Tem1-Iqg1 interaction and Cyk3 localization, which are critical events for AMR contraction and PS formation, respectively (see text for details).
Timely AMR contraction is brought about by a signaling network that includes Tem1-Iqg1 complex formation, which has been proposed to be an essential step.30 Both Tem1 and Iqg1 are subjected to several levels of regulation, but how Tem1-Iqg1 interaction is controlled is unknown. We show here that Tem1 undergoes cell cycle-regulated ubiquitylation, and that the amount of ubiquitylated Tem1 decreases concomitantly with cells undergoing AMR contraction. These data are very important per se, as Tem1 ubiquitylation has never been shown before. Unfortunately, we could not observe any effect of the absence of the Dma proteins on Tem1 ubiquitylation. On the other hand, the amounts of Tem1 ubiquitylated forms increase in the presence of Dma2 excess, which also inhibits Tem1-Iqg1 interaction, both in vitro and by 2-hybrid assay, and causes defective AMR contraction. Based on the mobility shift of its ubiquitylated forms, Tem1 is likely polyubiquitylated. However, this post-translational modification is unlikely to signal Tem1 degradation, as total Tem1 levels do not decrease in response to Dma2 overproduction, although the latter increases the amounts of Tem1 ubiquitylated forms. We cannot exclude that Tem1 polyubiquitylation might lead to a local degradation that cannot be detected by western blot analysis, but we favor the hypothesis that Tem1 ubiquitylation may regulate its activity, its subcellular localization, and/or its interaction with other cytokinesis factors. We could not evaluate whether Dma2 excess interferes with other possible Tem1 post-translational modifications, because none has been described in the literature so far. However, we observed that moderate Dma2 overproduction did not change the mobility of the Tem1-3HA doublet in SDS-PAGE (Fig. S2). In addition, we tried to analyze the Dma proteins’ role in Tem1 localization at the division site, but we could never visualize Tem1 at the bud neck even in wild-type cells, and indeed this localization has never been described in the literature. Anyhow, it must be pointed out that moderate Dma2 overproduction did not affect Tem1 activity either in promoting mitotic exit, septin dynamics, or SPB localization (our unpublished observations). Altogether, these findings tempt us to propose that ubiquitylation of Tem1 might inhibit Tem1 binding to Iqg1, which, in turn, will not be competent to promote AMR contraction. Subsequent Tem1 deubiquitylation would allow Tem1-Iqg1 interaction, thus promoting AMR contraction. Whether the Dma proteins play a physiological role in this control still remains unaddressed.
Proper PS formation requires bud neck recruitment and concerted action of Cyk3, Hof1, Inn1, and Chs2. We found that Dma2 excess causes asymmetric PS deposition, likely by interfering with Chs2 and Cyk3 dynamics at the bud neck. Indeed, we observed that the Chs2 ring contracts asymmetrically in GAL1-DMA2 cells. In addition, Dma2 can block Cyk3 localization at the bud neck without perturbing either Cyk3 total levels or phosphorylation, which is important for its recruitment at the division site.41 Unfortunately, we could not obtain reproducible results for Cyk3 ubiquitylation, and we can only speculate that the Dma proteins might regulate a still unknown factor(s) that controls Cyk3 function before its recruitment at the bud neck (Fig. 7). However Dma2 overproduction does not alter Inn1 bud neck localization, whose lack is known to cause a complete cytokinesis block,19 and indeed Dma2-overproducing cells are viable and form a PS, likely due to the presence of Hof1. In fact, the presence of Dma2 excess, which causes defective AMR contraction, lack of Cyk3 localization at the bud neck, and asymmetric PS deposition is lethal for cells that lack Hof1, and Dma2-overproducing hof1Δ cells cannot complete cytokinesis and do not localize Inn1 at the bud neck. These data are in agreement with the existence of partially overlapping cytokinesis pathways and with the observation that HOF1 or CYK3 deletions have no effect on AMR assembly, but their combination causes synthetic lethality,20 and each one leads to severe growth defect in cells lacking Myo1.12,15 As Hof1 and Cyk3 cooperate to recruit Inn1,19,20 the negative effect of Dma2 excess on Cyk3 might completely impede Inn1 recruitment to the bud neck in the absence of Hof1, thus causing cell lethality.
In agreement with the findings above, the lack of the Dma proteins results in partial defects in the timing of AMR contraction with respect to mitotic spindle breakdown as well as in Cyk3 localization at the bud neck, and causes abnormally shaped PS formation. However, these defects do not either block cell division or impair viability of dma1Δ dma2Δ cells, likely due to the redundancy of cytokinesis controls. Importantly, the lack of the Dma proteins causes a sick phenotype and cytokinesis defects in cyk3Δ cells. Indeed, improper AMR contraction might be toxic for Cyk3-lacking cells that are already impaired in PS formation, as expected based on the known central role for Cyk3 in a cytokinesis rescue mechanism acting in the absence of functional AMR.
In summary, our findings shed new light on the biological role of the Dma proteins and reveal a new level of complexity in cytokinesis regulation. In particular, the Dma proteins seem to control both AMR contraction and symmetric PS deposition, thus acting as new cytokinesis inhibitors and contributing to couple mitotic exit with the cytokinetic events. Since their lack is not essential for cytokinesis completion, their function is likely redundant with other factor(s) that control cytokinesis. A challenging goal for the future will be the identification of cytokinesis proteins directly controlled by Dma1 and Dma2.
Materials and Methods
Strains, media, and reagents, genetic manipulations
All yeast strains (Table S1) were derivatives of W303 (ade2-1, trp1-1, leu2-3,112, his3-11,15, ura3, ssd1), except yRF1457, yRF1650, yRF1651, yRF1652, yRF1667 that were derivatives of EGY48 (Mata his3 ura3 trp1 6lexAOP-LEU2; lexAOP-lacZ reporter on plasmid pSH18–34). Unless otherwise indicated, all the GAL1-DMA2 strains carry a single copy of the GAL1-DMA2 constuct32 integrated at the URA3 locus. Unless otherwise stated, α-factor was used at 2 μg/ml and cells were grown at 25 °C in either synthetic medium supplemented with the appropriate nutrients or YEP (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) medium supplemented with 2% glucose (YEPD) or 2% raffinose and 1% galactose (YEPRG).
Standard techniques were used for genetic manipulations.46,47 Gene deletions were generated by one-step gene replacement.48 One-step tagging techniques were used to create 3HA-tagged Tem1 variant.49 Iqg1-3HA-expressing strain was a kind gift from C Price (Lancaster University). Cyk3-3HA expressing strain was a kind gift from A Jendretzki (University of Osnabrück). GFP-Iqg1 and GFP-Cyk3 expressing strains were a kind gift of M Iwase (University of Pennsylvania School of Medicine). GFP-Inn1 and Myo1-Cherry expressing strains were a kind gift of G Pereira (German Cancer Research Center). The pRS315 CHS2-GFP plasmid was a kind gift of R Li (Stowers Institute for Medical Research). All gene replacements and tagging were controlled by PCR-based methods or Southern blot analysis.
The yeast 2-hybrid assay was performed using the B42/lexA system with strain EGY48 (Mata his3 ura3 trp1 6lexAOP-LEU2; lexAOP-lacZ reporter on plasmid pSH18-34) as the host strain.50 Bait plasmid pEG202-Tem1 (RF35) for the 2-hybrid assay, expressing a lexADB-Tem1 fusion, was obtained by amplifying the corresponding coding sequence of TEM1 gene from genomic DNA and ligating the resulting fragment into BamHI-digested pEG202. Prey plasmid pJG4-5-Iqg1 (RF44), expressing a B42AD-Iqg1 fusion, was obtained by amplifying the corresponding coding sequence of IQG1 from genomic DNA and ligating the resulting fragments into EcoRI-digested pJG4-5.
Fluorescence microscopy and live imaging
In situ immunofluorescence was performed on formaldehyde-fixed cells and performed as previously described.51 Nuclei were visualized by staining with DAPI 0.05 μg/ml. Visualization of septin rings was performed using anti-Cdc11 polyclonal antibodies (1:200, sc-7170 Santa Cruz) followed by indirect immunofluorescence with Alexa Fluor 488-conjugated anti-rabbit antibody (1:100, Invitrogen). To detect spindle formation and elongation, α-tubulin immunostaining was performed with the YOL34 monoclonal antibody (1:100, Serotec) followed by indirect immunofluorescence using rhodamine-conjugated anti-rat Ab (1:500, Pierce Chemical Co). To detect actin ring, actin immunostaining was performed using anti-actin polyclonal antibodies (1:1000, A2066, Sigma-Aldrich) followed by indirect immunofluorescence using rhodamine-conjugated anti-rabbit Ab (1:50, Invitrogen). To visualize Iqg1-GFP and Cyk3-GFP cells were fixed in cold ethanol and washed with Tris 50 mM pH8. To visualize Inn1-GFP and Myo1-Cherry, cells were fixed in 4% formaldeyde and washed with PBS. For chitin staining, cells were fixed in cold ethanol, washed with PBS, and treated with PBS containing 0.01 mg/ml Calcofluor White (purchased by Sigma-Aldrich). Digital images were taken with a Leica DC350F charge-coupled device camera mounted on a Nikon Eclipse 600 and controlled by the Leica FW4000 software or with the MetaMorph imaging system software on a fluorescent microscope (Eclipse 90i; Nikon), equipped with a charge-coupled device camera (Coolsnap, Photometrics) with an oil 100× 0.5–1.3 PlanFluor oil objective (Nikon). For three-dimensional reconstruction of chitin staining, 30 Z-stacks at 0.2 μm intervals were taken and projected onto a single image using MetaMorph imaging system software (Molecular Devices).
For time-lapse movies cells were grown in synthetic complete medium + 2% glucose (or 2% raffinose for GAL1-DMA2 experiment), then collected and imaged on agar in synthetic complete medium + 2% glucose (or 2% galactose for GAL1-DMA2 experiment) using a Delta Vision Elite imaging system (Applied Precision) based on an IX71 inverted microscope (Olympus) with a camera CoolSNAP HQ2 from Photometrics, and a UPlanApo 60× (1.4 NA) oil immersion objective (Olympus). Every 3 min (unless otherwise stated) 3 Z-stacks at 1.2 micron intervals were taken for each fluorescent channel and projected onto a single image per channel using Fiji software.52
Electron microscopy (TEM)
Samples were fixed by addition of glutaraldehyde (Sigma) directly to the culture medium to a final concentration of 2.5% for 10 min, centrifuged and incubated overnight at 4 °C. After washes (3 × 10 min) in distilled water, pellet of cells were incubated in 4% KMnO4 for 30 min, then washed in distilled water and transferred in a saturated solution of uranyl acetate for 30 min. After some washes in distilled water, samples were dehydrated in ethanol series, and, finally, in propylene oxide 100% for 20 min. Infiltration was subsequently performed with propylene–resin (Araldite-Epon, Sigma) mixture in volume proportions of 2:1 for 1 h, 1:1 overnight. The pellets were left for 1 h in volume proportions of 2:1 for 1 h. Then the pellets were left for 4 h in 100% pure resin and then shifted to 60 °C for polymerization. Ultrathin sections of 80 nm were made on an Ultracut E microtome (Reichert), post-stained in uranyl acetate and lead citrate and examined in a Leo912ab transmission electron microscope (Zeiss) at 80 KV. Digital images were acquired by Esivision CCD-BM/ 1K system.
Protein extracts and analysis
For monitoring Tem1 and Cyk3 levels during the cell cycle, total protein extracts were prepared by TCA precipitation as previously described.53 For western blot analysis, proteins were transferred to Protran membranes (Schleicher and Schuell) and probed with monoclonal anti-HA (1:3000), anti Pgk1 (1:40000) and anti-Cdc11 antibodies (1:2000) or with polyclonal anti-GST antibodies (1:2000) or anti-Clb2 antibodies (1:2000). Secondary antibodies were purchased from Amersham and proteins were detected by an enhanced chemioluminescence system according to the manufacturer.
Expression and purification of recombinant proteins and in vitro binding experiment
Tem1-Iqg1 binding assay was performed as described.30 Briefly, Glutathione S-transferase (GST)-Tem1 fusion protein was produced in Escherichia coli carrying pKT5 plasmid (kind gift of Rong Li, Stowers Institute for Medical Research), grown in LD broth containing ampicillin at 37 °C for 3 h, transferred to 14 °C for 1 h and induced with 0.1 mM IPTG for 15 h. Bacteria extract was prepared in lysis buffer (50 mM TRIS-HCl pH8, 200 mM NaCl, 2 mM MgCl2, 2 mM DTT, 1 mM GTP) supplemented with a cocktail of protease inhibitors. Cells were incubated with 1mg/ml lisozyme in ice for 30 min, placed at 37 °C for 5 min and sonicated at 4 °C. To prepare GST-Tem1 beads, the bacterial extracts containing GST-Tem1 were incubated with 40 µl glutathione agarose beads (GE Healthcare) for 1 h at 4 °C. As negative control we used 40 µl glutathione agarose beads incubated with an equal amount of lysis buffer. The beads were then washed 3 times in washing buffer (PBS containing 1 mM DTT, 1 mM GTP, and MgCl2 2 mM). Yeast extracts were prepared by using acid glass beads in UB buffer (0.05 M HEPES pH7.5, 0.1 M KCl, 3 mM MgCl2, 1 mM EGTA, 1 mM DTT) supplemented with a cocktail of protease inhibitors. For Tem1-Iqg1 binding assay, Tem1 beads were incubated for 1 h at 4 °C with either total extract of wild-type cells or total extract of GAL1-DMA2 cells induced for 2 h with 1% galactose. The beads were washed 3 times in washing buffer and were then incubated with total extract of IQG1-3HA cells for 1 h at 4 °C. The beads were washed 3 times in washing buffer, resuspended after drying in Laemmli buffer with β-mercaptoethanol and boiled for 3 min at 100 °C.
Detection of ubiquitin conjugates in vivo
For the experiments involving the expression of 6xHis-ubiquitin, cells were grown to log phase at 25 °C in selective medium (synthetic medium lacking tryptophan) and untagged ubiquitin or 6xHis-ubiquitin production was induced by addition of 250 μM CuSO4 to each culture. After 3 further hours, yeast cells were washed in water, and protein extracts were prepared under denaturing conditions as described,54 followed by resuspension of TCA precipitates in buffer A (6 M guanidium, 100 mM NaPO4 pH 8, 10 mM TRIS-HCl pH 8) and debris removal by centrifugation. Nickel pull-down was performed as described,35 using Ni-NTA agarose beads (Qiagen). For the experiments involving GAL1 induction, 1% galactose was added to the cell cultures 30 min before CuSO4 addition.
Statistical analyses and other techniques
Standard deviations (SD) and standard mean errors (SEM) were calculated using Microsoft Excel software. The significance of the differences between AMR contraction dynamics and Chs2 dynamics was statistically tested by the Wilcoxon signed-rank-sum test using MATLAB software, samples are significantly different if P value is P < 0.05.
Flow cytometric DNA quantification was performed according to Fraschini et al.55 on a Becton- Dickinson FACScan.
Supplementary Material
Acknowledgments
We are grateful Simonetta Piatti and Laura Merlini for carrying out preliminary experiments on Tem1 ubiquitylation with RF and to Erfei Bi, Charles Brenner, Judy Callis, Marco Geymonat, Jürgen J Heinisch, Masayuki Iwase, Arne Jendretzki, Rong Li, Achille Pellicioli, Gislene Pereira, Simonetta Piatti, and Clive Price for strains and plasmids. This work has been supported by grants from PRIN (Progetti di Ricerca di Interesse Nazionale) 2008 to RF. CC is supported by a fellowship from Fondazione Confalonieri. ER is supported by a fellowship from Fondazione Adriano Buzzati Traverso. EC is supported by a fellowship “Young Investigator Programme 2013” from Fondazione Veronesi.
Glossary
Abbreviations:
- AMR
actomyosin ring contraction
- MEN
mitotic exit network
- PS
primary septum
- TEM
transmission electron microscopy
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here:
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25869
References
- 1.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–6. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–19. doi: 10.1016/S0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 3.Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Durán A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–23. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191:1333–50. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–12. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–66. doi: 10.1083/jcb.140.2.355. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin CF, Bennett AM, Ma WK, Hall MC, Yeong FM. Dependence of Chs2 ER export on dephosphorylation by cytoplasmic Cdc14 ensures that septum formation follows mitosis. Mol Biol Cell. 2012;23:45–58. doi: 10.1091/mbc.E11-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesage G, Shapiro J, Specht CA, Sdicu AM, Ménard P, Hussein S, Tong AH, Boone C, Bussey H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeong FM. Severing all ties between mother and daughter: cell separation in budding yeast. Mol Microbiol. 2005;55:1325–31. doi: 10.1111/j.1365-2958.2005.04507.x. [DOI] [PubMed] [Google Scholar]
- 10.Bi E. Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct Funct. 2001;26:529–37. doi: 10.1247/csf.26.529. [DOI] [PubMed] [Google Scholar]
- 11.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–60. doi: 10.1083/jcb.143.7.1947. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–88. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 2005;24:1440–52. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korinek WS, Bi E, Epp JA, Wang L, Ho J, Chant J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol. 2000;10:947–50. doi: 10.1016/S0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- 16.Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, Pringle JR. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol Biol Cell. 2007;18:5139–53. doi: 10.1091/mbc.E07-05-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh Y, Chang KJ, Orlean P, Wloka C, Deshaies R, Bi E. Mitotic exit kinase Dbf2 directly phosphorylates chitin synthase Chs2 to regulate cytokinesis in budding yeast. Mol Biol Cell. 2012;23:2445–56. doi: 10.1091/mbc.E12-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Diaz A, Marchesi V, Murray S, Jones R, Pereira G, Edmondson R, Allen T, Labib K. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat Cell Biol. 2008;10:395–406. doi: 10.1038/ncb1701. [DOI] [PubMed] [Google Scholar]
- 19.Jendretzki A, Ciklic I, Rodicio R, Schmitz HP, Heinisch JJ. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol Genet Genomics. 2009;282:437–51. doi: 10.1007/s00438-009-0476-0. [DOI] [PubMed] [Google Scholar]
- 20.Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, et al. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J Cell Biol. 2009;185:995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–32. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 22.Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–88. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–86. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- 24.Hwa Lim H, Yeong FM, Surana U. Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast. Mol Biol Cell. 2003;14:4734–43. doi: 10.1091/mbc.E03-04-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bembenek J, Kang J, Kurischko C, Li B, Raab JR, Belanger KD, Luca FC, Yu H. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle. 2005;4:961–71. doi: 10.4161/cc.4.7.1798. [DOI] [PubMed] [Google Scholar]
- 26.Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci. 2000;113:3399–408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- 27.Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–83. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–98. doi: 10.1128/MCB.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu S, Huang HK, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10:329–32. doi: 10.1016/S0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- 30.Shannon KB, Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell. 1999;10:283–96. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks L, 3rd, Heimsath EG, Jr., Loring GL, Brenner C. FHA-RING ubiquitin ligases in cell division cycle control. Cell Mol Life Sci. 2008;65:3458–66. doi: 10.1007/s00018-008-8220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraschini R, Bilotta D, Lucchini G, Piatti S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol Biol Cell. 2004;15:3796–810. doi: 10.1091/mbc.E04-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merlini L, Fraschini R, Boettcher B, Barral Y, Lucchini G, Piatti S. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 2012;8:e1002670. doi: 10.1371/journal.pgen.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loring GL, Christensen KC, Gerber SA, Brenner C. Yeast Chfr homologs retard cell cycle at G1 and G2/M via Ubc4 and Ubc13/Mms2-dependent ubiquitination. Cell Cycle. 2008;7:96–105. doi: 10.4161/cc.7.1.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raspelli E, Cassani C, Lucchini G, Fraschini R. Budding yeast Dma1 and Dma2 participate in regulation of Swe1 levels and localization. Mol Biol Cell. 2011;22:2185–97. doi: 10.1091/mbc.E11-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chahwan R, Gravel S, Matsusaka T, Jackson SP. Dma/RNF8 proteins are evolutionarily conserved E3 ubiquitin ligases that target septins. Cell Cycle. 2013;12:1000–8. doi: 10.4161/cc.23947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menssen R, Neutzner A, Seufert W. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol. 2001;11:345–50. doi: 10.1016/S0960-9822(01)00095-1. [DOI] [PubMed] [Google Scholar]
- 38.Corbett M, Xiong Y, Boyne JR, Wright DJ, Munro E, Price C. IQGAP and mitotic exit network (MEN) proteins are required for cytokinesis and re-polarization of the actin cytoskeleton in the budding yeast, Saccharomyces cerevisiae. Eur J Cell Biol. 2006;85:1201–15. doi: 10.1016/j.ejcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Geymonat M, Spanos A, de Bettignies G, Sedgwick SG. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol. 2009;187:497–511. doi: 10.1083/jcb.200905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meitinger F, Petrova B, Lombardi IM, Bertazzi DT, Hub B, Zentgraf H, Pereira G. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123:1851–61. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- 42.Callis J, Ling R. Preparation, characterization, and use of tagged ubiquitins. Methods Enzymol. 2005;399:51–64. doi: 10.1016/S0076-6879(05)99004-6. [DOI] [PubMed] [Google Scholar]
- 43.Plans V, Guerra-Rebollo M, Thomson TM. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 2008;27:1355–65. doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- 44.Murone M, Simanis V. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 1996;15:6605–16. [PMC free article] [PubMed] [Google Scholar]
- 45.Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev Cell. 2002;3:779–90. doi: 10.1016/S1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- 46.Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY. 1992. [Google Scholar]
- 47.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-V. [DOI] [PubMed] [Google Scholar]
- 48.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 49.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–62. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 50.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-F. [DOI] [PubMed] [Google Scholar]
- 51.Fraschini R, D’Ambrosio C, Venturetti M, Lucchini G, Piatti S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol. 2006;172:335–46. doi: 10.1083/jcb.200507162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piatti S, Böhm T, Cocker JH, Diffley JF, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–31. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 54.Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984;81:4819–23. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraschini R, Formenti E, Lucchini G, Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–91. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.