Abstract
Dihydroflavonol-4-reductase (DFR; EC1.1.1.219) catalyzes a key step late in the biosynthesis of anthocyanins, condensed tannins (proanthocyanidins), and other flavonoids important to plant survival and human nutrition. Two DFR cDNA clones (MtDFR1 and MtDFR2) were isolated from the model legume Medicago truncatula cv Jemalong. Both clones were functionally expressed in Escherichia coli, confirming that both encode active DFR proteins that readily reduce taxifolin (dihydroquercetin) to leucocyanidin. M. truncatula leaf anthocyanins were shown to be cyanidin-glucoside derivatives, and the seed coat proanthocyanidins are known catechin and epicatechin derivatives, all biosynthesized from leucocyanidin. Despite high amino acid similarity (79% identical), the recombinant DFR proteins exhibited differing pH and temperature profiles and differing relative substrate preferences. Although no pelargonidin derivatives were identified in M. truncatula, MtDFR1 readily reduced dihydrokaempferol, consistent with the presence of an asparagine residue at a location known to determine substrate specificity in other DFRs, whereas MtDFR2 contained an aspartate residue at the same site and was only marginally active on dihydrokaempferol. Both recombinant DFR proteins very efficiently reduced 5-deoxydihydroflavonol substrates fustin and dihydrorobinetin, substances not previously reported as constituents of M. truncatula. Transcript accumulation for both genes was highest in young seeds and flowers, consistent with accumulation of condensed tannins and leucoanthocyanidins in these tissues. MtDFR1 transcript levels in developing leaves closely paralleled leaf anthocyanin accumulation. Overexpression of MtDFR1 in transgenic tobacco (Nicotiana tabacum) resulted in visible increases in anthocyanin accumulation in flowers, whereas MtDFR2 did not. The data reveal unexpected properties and differences in two DFR proteins from a single species.
Flavonoids represent a large group of plant secondary metabolites with diverse biological activities, and the biochemical and genetic investigations of flavonoid biosynthesis have been well documented (Hahlbrock and Grisebach, 1975; Harborne, 1988; Stafford, 1990; Holton and Cornish, 1995; Dixon and Steele, 1999; Winkel-Shirley, 2001). Flavonoids are divided into several structural classes, including anthocyanins, which provide flower and leaf colors, catechins, and condensed tannins (CTs; proanthocyanidins), which contribute to resistance to microbes, and other derivatives with diverse roles in plant development and interactions with the environment (Harborne, 1988; Dixon and Paiva, 1995; Paiva, 2000). Several flavonoids are active ingredients in herbal medicines and appear to confer health benefits to humans when consumed regularly. Particular attention has been placed on the anthocyanins, catechins, and proanthocyanidins because of their antioxidant activities and their interactions with proteins (Tanner et al., 1994; Scalbert and Williamson, 2000; Ross and Kasum, 2002; Hou, 2003).
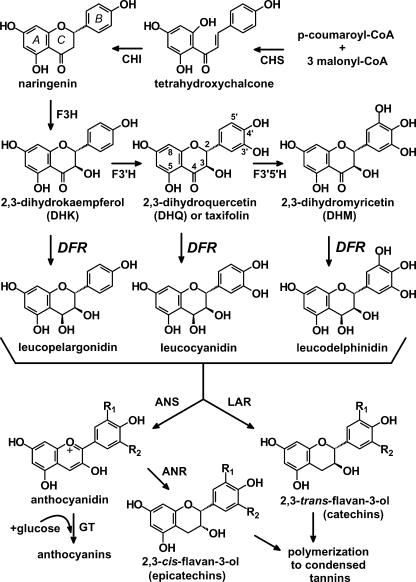
The common precursors in the biosynthesis of all classes of flavonoids are malonyl-CoA and p-coumaroyl-CoA, condensed into chalcone intermediates by the action of chalcone synthase (CHS; Fig. 1). Single genes and multigene families encoding CHS, chalcone isomerase, and flavanone 3-hydroxylase have been studied extensively from many plant species (Holton and Cornish, 1995). Specific isoforms of these enzymes may assemble as macromolecular complexes producing specific classes of flavonoids (Burbulis and Winkel-Shirley, 1999). When present, flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase (Fig. 1) hydroxylate the B ring, which in turn determines flower color by controlling the ratio of pelargonidin (red to orange), cyanidin (red to violet), and delphinidin (violet to blue) anthocyanins (Harborne, 1988; Holton and Cornish, 1995; Winkel-Shirley, 2001).
Figure 1.
Biosynthetic relationship of DFR to anthocyanidins, leucoanthocyanidins, catechins, and condensed tannins (CTs). CHI, Chalcone isomerase; F3H, (2S)-flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′,5′H, flavonoid 3′,5′-hydroxylase; ANS, anthocyanidin synthase (also known as leucoanthocyanidin dioxygenase); GT, anthocyanidin glucosyl transferase; LAR, leucoanthocyanidin reductase. A to C on the naringenin structure indicate the standard nomenclature assigned to the three flavonoid rings. After 3-O-glucosylation of anthocyanidins to form anthocyanins by GT, anthocyanins may be further modified, undergoing additional glycosylation, methylation, and acylation. The pathway differs from that in recent reviews of flavonoid biosynthesis to account for the recent discovery that anthocyanin reductase (ANR or BAN) produces 2,3-cis-flavan-3-ols such as epicatechin (3′,4′ B ring hydroxylated) via the NADPH-dependent reduction of anthocyanidins, not leucoanthocyanidins (Xie et al., 2003).
Dihydroflavonol 4-reductase (DFR; EC1.1.1.219) is a later key enzyme controlling flux into biosynthetic pathway branches leading to anthocyanins and CTs. DFR preparations from several plants catalyze the reduction of the three dihydroflavonols dihydrokaempferol (DHK), dihydroquercetin (DHQ), and dihydromyricetin (DHM) into leucoanthocyanidins, which are common precursors for anthocyanins and CT synthesis (Fig. 1). DFR proteins in certain species such as petunia (Petunia hybrida) and Cymbidium hybrida do not accept the monohydroxylated DHK and, therefore, cannot produce the corresponding monohydroxylated pelargonidin anthocyanins (Meyer et al., 1987; Johnson et al., 1999, 2001). Induction of DFR activity in plants has been linked to an increase in CT accumulation, which may be important for defense against herbivores (Peters and Constabel, 2002).
Anthocyanidin synthase catalyzes the conversion of leucoanthocyanidins to anthocyanidins as the first step of anthocyanin (anthocyanidin-3-O-glucoside) biosynthesis (Nakajima et al., 2001; Fig. 1). Leucoanthocyanidin reductase originally was proposed as the key branch point enzyme for CT synthesis in plants, converting leucoanthocyanidins to 2,3-trans-flavan-3-ols such as catechin (dihydroxylated B ring; Stafford, 1990; Tanner and Kristiansen, 1993; Fig. 1). A gene encoding leucoanthocyanidin reductase was isolated recently and successfully expressed in Escherichia coli (Tanner et al., 2003). However, many flavan-3-ols show an alternative 2,3-cis-stereochemistry, such as the epi-catechin repeating units in many CT polymers, including those isolated from alfalfa (Medicago sativa) seed coats (Harborne, 1988; Koupai-Abyazani et al., 1993). Recently a cDNA from Medicago truncatula was shown to encode anthocyanidin reductase (ANR; Fig. 1), a novel enzyme catalyzing the NADPH-dependent reduction of anthocyanidins to 2,3-cis-flavan-3-ols (Xie et al., 2003). The factors controlling the condensation of trans- and cis-flavan-3-ols monomers to form CT polymers are still unknown.
The genetics and regulation of DFR have been studied extensively in several plant species, where it has usually been found as a single gene or a small gene family. DFR was reported as a single gene in the genomes of barley (Hordeum vulgare), Arabidopsis, tomato (Lycopersicon esculentum), grape (Vitis vinifera), snapdragon (Antirrhinum majus), and rice (Oryza sativa; Kristiansen and Rohde, 1991; Winkel-Shirley et al., 1992; Bongue-Bartelsman et al., 1994; Sparvoli et al., 1994; Holton and Cornish, 1995; Chen et al., 1998). In Arabidopsis, the mutation of the single DFR structural gene at the tt3 locus resulted in the tt (transparent testa) phenotype because of the lack of anthocyanin accumulation in seed coats (Winkel-Shirley et al., 1992). Several Arabidopsis mutants with modified transcription factors resulting in altered DFR expression have also been identified, including tt8, ttg1, and tt2 (Winkel-Shirley et al., 1995; Walker et al., 1999; Nesi et al., 2000, 2001). Snapdragon DFR is encoded by the single pallida locus, but gene expression is regulated by at least three transcription factors (Delila, Eluta, and Rosea; Almeida et al., 1989; Holton and Cornish, 1995). Examples of plants verified to contain multiple genes encoding catalytically active DFR proteins expressed in a single tissue are rare. Petunia harbors three DFR genes, but only dfrA appears to be expressed in petunia flower limbs (Beld et al., 1989; Gerats et al., 1990; Huits et al., 1994; Holton and Cornish, 1995). Japanese morning glory (Ipomoea nil) and common morning glory (Ipomoea purpurea) each contain three tandem DFR genes, but mutations in only DFR-B will block anthocyanin production in flowers (Inagaki et al., 1999; Hoshino et al., 2001). Southern analysis suggests the presence of multiple DFR genes in Gerbera hybrida, but only GDFR1 was shown to be catalytically active and expressed in flowers, and analysis of the corresponding promoter region (gdfr1) indicates a complex expression pattern correlating well with anthocyanin accumulation (Helariutta et al., 1993; Elomaa et al., 1998).
Knowledge of the details of DFR biochemistry is very important to understanding aspects of flavonoid biosynthesis, especially how plants regulate CT and anthocyanin biosynthesis and composition and different stereochemical features of flavan-3-ols and related compounds. Early in vitro enzyme assays using cell-free extracts of Gingko biloba and Douglas fir (Pseudotsuga menziesii) showed that DFR converted DHQ to leucocyanidin and DHM to leucodelphinidin (Stafford and Lester, 1982, 1984, 1985). Subsequent data regarding DFR substrate specificity has resulted mainly from combinations of genetic analysis of mutant or transgenic plants followed by phytochemical analysis (Meyer et al., 1987; Helariutta et al., 1993; Johnson et al., 1999, 2001). Limited information was provided recently by heterologous expression of DFR in yeast (Saccharomyces cerevisiae) or E. coli (Martens et al., 2002; Peters and Constabel, 2002; Fischer et al., 2003). Little has been reported regarding other biochemical properties of DFR isoenzymes, and the role of isoforms in the regulation of specific branches of flavonoid pathways is unclear.
M. truncatula is a popular model legume species for which many cDNA and genomic sequence databases are being developed (Cook, 1999; Bell et al., 2001) but which only recently is being extensively characterized at the phytochemical and enzyme levels. M. truncatula is also closely related to alfalfa, the fifth largest crop in the United States, and analysis of gene function in M. truncatula may aid future alfalfa improvement. From M. truncatula, we isolated two cDNAs encoding DFR isoenzymes, MtDFR1 and MtDFR2, and determined their expression profiles in anthocyanin- and CT-accumulating tissues. Biochemical characterization of the two recombinant DFR enzymes revealed differences in their catalytic properties in vitro, including differing relative substrate preferences. Furthermore, we show here that both DFR isoenzymes catalyze the reduction of the 5-deoxydihydroflavonols fustin and dihydrorobinetin (DHR), substances that have been identified in other legumes but not M. truncatula. Transgenic tobacco (Nicotiana tabacum) plants overexpressing the two M. truncatula DFR cDNAs revealed that only MtDFR1 could interact with tobacco flavonoid pathway enzymes in vivo to alter flower color.
RESULTS
Characterization of M. truncatula DFR cDNA Clones
By searching the BLASTX results (Bell et al., 2001) from expressed sequence tags (ESTs) generated from a M. truncatula young seed (YS) cDNA library, we identified two different cDNA clones with high similarity to DFR, designated MtDFR1 (NF001D05Y-S1F1046) and MtDFR2 (NF004H10YS1F1092). The MtDFR1 cDNA contained 1,331 nucleotides, including a full-length open reading frame (ORF) encoding 334 amino acids (GenBank accession no. AY389346). The second DFR cDNA clone contained a truncated ORF, but alignment with overlapping EST sequences in public databases allowed the design of PCR primers to generate full-length clones. Through PCR using vector and N-terminal MtDFR2 primers and the YS cDNA library as the template, we obtained a clone containing the full-length MtDFR2 ORF (encoding 339 amino acids) and the 3′-untranslated region totaling 1,242 nucleotides (GenBank accession no. AY389347). Alignment of the predicted translations of the MtDFR1 and MtDFR2 clones indicated a 79.1% amino acid sequence identity. The similarity of amino acid sequences is much higher in the N-terminal halves of the proteins than in the C-terminal halves (Fig. 2).
Figure 2.
Alignment of the amino acid sequences encoded by MtDFR1 and MtDFR2. Residues that are identical in the two sequences are marked with an asterisk.
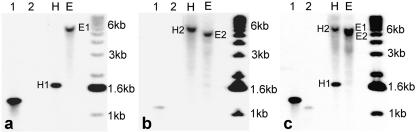
Despite the high DNA sequence similarities (79.6% identical at the nucleotide level in the coding regions when aligned using ClustalW), the two cDNA clones differ in their restriction maps, including a HindIII restriction site present only in MtDFR1 [near the poly(A+) tail] and an EcoRI restriction site present only in MtDFR2 (position 451 in the coding region). Although the nucleotide sequences are highly conserved in the 5′ end of the coding regions, the sequence similarity is sufficiently low in 3′ regions (data not shown) to allow the design of gene-specific hybridization probes, and these regions were selectively amplified using PCR. Southern hybridization showed that each gene specific probe only hybridized with the corresponding cDNA clone, and no cross hybridization occurred (Fig. 3, A and B, first and second lanes). Each probe hybridized to only a single band in EcoRI- or HindIII-digested M. truncatula genomic DNA, indicating that MtDFR1 and MtDFR2 are each present as a single copy in the M. truncatula cv Jemalong line A-17 genome (Fig. 3, A and B). Although the sizes of the bands detected in the EcoRI-digested DNA are similar with both MtDFR1 and MtDFR2 probes, simultaneous hybridization with both probes confirmed that these were two distinct bands (Fig. 3C). When either the MtDFR1 or MtDFR2 complete coding regions were used as probes, no additional hybridizing bands were observed, indicating that no additional DFR genes are present in this species. The “DFR-like” ANR (Xie et al., 2003; also know as BAN) from M. truncatula does not cross-hybridize to DFR probes under these conditions (data not shown).
Figure 3.
Southern-blot hybridization of M. truncatula genomic DNA with MtDFR1 and MtDFR2 gene-specific probes. A, MtDFR1-specific probe (225 bp). B, MtDFR2-specific probe (173 bp). C, MtDFR1 and MtDFR2 probes combined. Lane 1, MtDFR1 cDNA insert. Lane 2, MtDFR2 cDNA insert. Genomic DNA was digested with EcoRI (E) or Hind III (H). The hybridizing bands in the EcoRI lane in C are resolvable upon extended electrophoresis.
Functional Expression in E. coli and in Vitro Biochemical Characterization
Although several DFR genes have been identified by genetic studies, and DFR cDNAs have been characterized and expressed in plants, little has been published regarding the biochemical characterization of the proteins encoded by these clones, especially multiple DFR enzymes from the same species. To functionally characterize the two M. truncatula DFR enzymes, we subcloned the coding regions of MtDFR1 and MtDFR2 into pSE380 (Brosius, 1989) and expressed the recombinant proteins in E. coli strain BL21. Soluble protein extracts from isopropylthio-β-D-thio-galactoside-induced cells were prepared and assayed for enzyme activity using methods previously used for isoflavone reductase (Paiva et al., 1991) and vestitone reductase (VR; Guo and Paiva, 1995), two NADPH-dependent reductases from alfalfa. The only modification made was that reducing agents (such as β-mercaptoethanol and dithioerythritol) were omitted from the lysis buffer because of reports that thiols can form adducts with the DFR reaction products (Stafford and Lester, 1984), interfering with quantitative enzyme assays. The most common dihydroflavonol in plants is DHQ (or taxifolin) with two hydroxyls in the B ring (Fig. 1). Thus, we chose taxifolin to develop an in vitro enzyme assay. Because of reports of instability of DFR in vitro during purification (Stafford, 1990), we used crude bacterial protein extracts as the enzyme source to minimize enzyme manipulations.
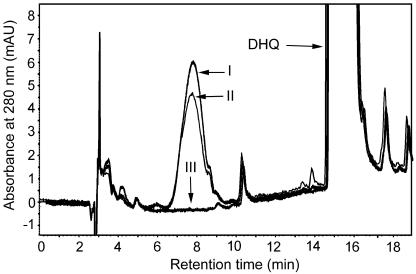
Enzyme extracted from cultures expressing either MtDFR1 or MtDFR2 protein converted (±)-taxifolin to a product eluting earlier in the HPLC system, whereas this product did not accumulate in reactions carried out with protein extracted from cultures harboring the empty pSE380 expression vector (Fig. 4). The relative retention time (Stafford and Lester, 1984, 1985) and maximum A280 were consistent with the product being leucocyanidin. LC-MS analysis confirmed that the product had the correct Mr (mass-to-charge ratio [m/z] = 306), exactly 2 mass units higher than the taxifolin substrate (data not shown). Product formation was almost 2-fold higher when (+)-taxifolin was substituted for (±)-taxifolin in assays with MtDFR1 or MtDFR2 proteins, indicating that the enzymes are stereospecific for (+)-taxifolin, a (2R,3R)-2, 3-trans-dihydroflavonol that is typical for DFR enzymes from other species (Harborne, 1988; Stafford, 1990). In multiple repeated trials, the maximum activity of fresh MtDFR2 extracts was always slightly higher (10%–20%) than MtDFR1 extracts (data not shown) under the standard assay conditions.
Figure 4.
HPLC chromatograms of DFR enzyme assay extracts. Assay mixtures contained (±)-taxifolin as substrate (marked DHQ), NADPH, and protein extracts from E. coli harboring pSE380-MtDFR2 (line I), pSE380-MtDFR1 (line II), or the empty expression vector pSE380 (line III). Chromatograms were recorded at the UV absorbance wavelength of 280 nm. The identity of the leucocyanidin product (early eluting peak in I and II) was confirmed based on relative retention time, UV spectra, and mass spectrum (via liquid chromatography [LC]-mass spectrometry [MS]).
The activities of MtDFR1 and MtDFR2 were assayed at (±)-taxifolin and (+)-taxifolin concentrations ranging from 0 to 200 μg reaction-1 (0–1.32 mm final assay concentration) in an attempt to estimate the binding constants for the substrates. Plots of reaction velocity versus substrate concentration and corresponding double-reciprocal plots from three independent determinations (data not shown) revealed patterns inconsistent with classical Michaelis-Menton kinetics, in some cases indicating inhibition of DFR activity by high substrate and/or product concentrations. Similar inhibition was observed for VR with structurally similar isoflavanone substrates (Guo et al., 1994). Although an accurate determination of the Km values could not be performed, the data indicate that the reaction catalyzed by MtDFR1 is saturated at lower substrate concentrations than MtDFR2, suggesting a higher substrate affinity (lower Km) for taxifolin. The reaction rate catalyzed by MtDFR1 was half-maximal at approximately 0.33 mm for both (±)-taxifolin and (+)-taxifolin, whereas MtDFR2 was half-maximal at approximately 0.65 mm. Using radioactive taxifolin as substrate, a Km value of about 37 μm for (±)-taxifolin was estimated for partially purified DFR extracted from Douglas fir tissue cultures, but instability of the leucocyanidin product was reported to add uncertainty to the DFR Km calculations (Stafford and Lester, 1984).
The activity of MtDFR1 and MtDFR2 was also assayed at NADPH concentrations from 0 to 4 mm. MtDFR1 and MtDFR2 exhibited Km values of approximately 0.8 and 1 mm NADPH, respectively. No reduction products were observed when NADH was substituted for NADPH, as was previously reported for some DFR preparations (Stafford, 1990).
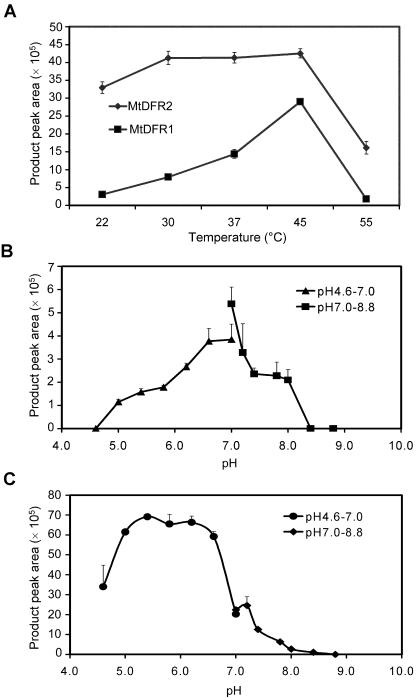
Dramatic differences were observed in the temperature and pH dependence of MtDFR1 and MtDFR2. MtDFR1 activity exhibited a sharp temperature optimum at 45°C, whereas the activity of MtDFR2 was maximal over the broad range of 30°C to 45°C (Fig. 5A). The optimum pH values for MtDFR1 were from 6.6 to 7.0 (Fig. 5B), whereas the optimum pH values for MtDFR2 were in the range from 5.4 to 6.2 (Fig. 5C).
Figure 5.
DFR reaction velocity versus temperature and pH for MtDFR1 and MtDFR2. A, To determine the effect of temperature on the rate of product formation, enzyme assays were conducted at temperatures from 22°C to 55°C for 30 min at pH 7.0, 1 mm NADPH, and 0.66 mm (±)-taxifolin. B and C, To determine the effect of pH on the rate of product formation, enzyme assays were conducted at 30°C and the above conditions, except that for pH 4.6 to pH 7.0, citrate/phosphate buffer was substituted for Tris-HCl buffer. B, MtDFR1. C, MtDFR2.
Relative Substrate Preferences of Recombinant MtDFR1 and MtDFR2
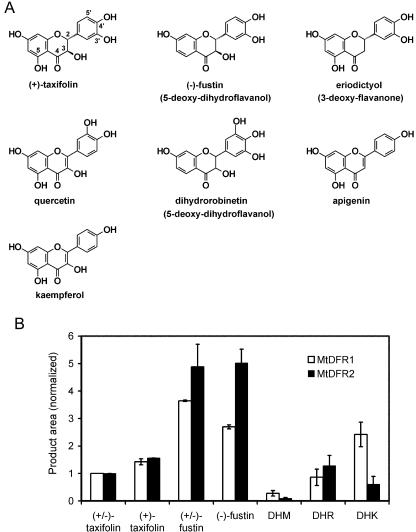
In addition to taxifolin, DFR proteins present in different plant species also catalyze the reduction of DHK or DHM, although in some cases multiple DFR proteins or isoforms may be present in individual cells or enzyme extracts (Stafford, 1990). Therefore, the rates of reduction of a number of potential substrates were compared for MtDFR1 and MtDFR2, relative to the rate at which the respective enzymes reduced (±)-taxifolin. The substrates included typical 5-hydroxy-dihydroflavonols known to be anthocyanin precursors [(±)-taxifolin, (+)-taxifolin, DHK, and DHM], rarer 5-deoxy-dihydroflavonols [(-)-fustin, (±)-fustin, and DHR], and other substrate analogs including flavonols (quercetin and kaempferol), a flavanone (eriodictyol), and a flavone (apigenin; Figs. 1 and 6A). The enzyme activities were assayed at their respective optimum pH values (MtDFR1, pH 7.0; and MtDFR2, pH 6.2), 2 mm NADPH, and 30°C, with all substrates at 400 μg mL-1. The extent of conversion of each substrate was quantitated by HPLC and then normalized against the value obtained for (±)-taxifolin for the same enzyme.
Figure 6.
Relative substrate preferences of MtDFR1 and MtDFR2. A, Structures of representative chemicals tested as substrates for MtDFR1 and MtDFR2, shown in comparison to taxifolin. Additional substrates are shown in Figure 1. B, Relative activity of MtDFR1 and MtDFR2 on selected dihydroflavonols. The enzyme activities were assayed at their respective optimum pH values (pH 7.0 for MtDFR1 and pH 6.2 for MtDFR2),2mm NADPH, and 30°C, with all substrates at a final concentration of 400 μg mL-1. [Differences in Mrs of the substrates only cause a variation of at most 10% (for DHM) in the millimolar concentrations of the substrates, and most concentrations are within ±5% of the (±)-taxifolin concentrations.] For each enzyme, product peak areas were normalized relative to the (±)-taxifolin product area to enable a comparison of the activities relative to a common substrate. Thus, the (±)-taxifolin value is set at 1.0 for both MtDFR1 and MtDFR2 proteins. All calculations assumed that molar extinction coefficients of the products were equal at 280 nm, given similar chromophores. The graph represents the average of two independent replicate experiments.
Reduction products were observed with all five of the dihydroflavonols tested, irrespective of the number of hydroxyl groups on the B ring, or the presence or absence of the 5-hydroxyl group (Fig. 6B). No reaction was observed with substrates lacking the hydroxyl group at the 3 position or with a double bond present between carbons 2 and 3 (flavone/flavonol derivatives), including quercetin, eriodictyol, kaempferol, and apigenin. LC-MS confirmed that the products of DFR acting on the dihydroflavonols had all gained exactly 2 mass units relative to the substrates (data not shown), consistent with a simple reduction using NADPH. Relative to (±)-taxifolin, (+)-taxifolin was converted more extensively (150%–160%) by both MtDFR1 and MtDFR2, confirming the previously observed stereospecificity of these enzymes. (±)-DHM (trihydroxy B ring) was converted less efficiently than taxifolin by both enzymes [10%–30% of (±)-taxifolin value]. Although (±)-DHK (monohydroxy B ring) was converted much more by MtDFR1 [250% of (±)-taxifolin value], MtDFR2 utilized this substrate much less efficiently than taxifolin [60% of (±)-taxifolin value]. (±)-Fustin, the 5-deoxy analog of (±)-taxifolin, has not been reported previously to be a substrate of DFR but was reduced 5 times faster than (±)-taxifolin by MtDFR2 and approximately 3 times faster by MtDFR1. MtDFR2 showed twice the relative activity on (-)-fustin as did MtDFR1. The reduction of (±)-fustin produced two large product peaks (retention times of 7.5 and 9.0 min), having identical UV profiles and Mrs. When (±)-fustin was used as the substrate, the two product peaks were similar in area, but when (-)-fustin [possessing the same absolute stereochemistry as (+)-taxifolin] was used as the substrate, the later eluting peak contained 90% of the product area. This suggests that the DFRs can accept both isomers of fustin and that the resulting diastereomeric products are resolved in the HPLC system. (±)-DHR, the 5-deoxy analog of (±)-DHM, was reduced as efficiently as (±)-taxifolin by both enzymes. Two product peaks were also observed for (±)-DHR. Reduction of the carbonyl group in fustin or DHR would yield the compounds commonly known as leucofisetinidin and leucorobinetinidin, known constituents of certain leguminous plants (Harborne, 1988).
Accumulation of Anthocyanins, CTs, and Leucoanthocyanidins in M. truncatula cv Jemalong Line A-17
Patterns of anthocyanin accumulation in leaf tissues vary among M. truncatula ecotypes and nearisogenic lines. In M. truncatula cv Jemalong line A-17, a central portion of the upper surface of each leaflet of the trifoliate leaves, is intensely colored by red anthocyanins when the plants are grown under certain conditions (Fig. 7A). The anthocyanin accumulation is generally higher when plants are grown under high light (greater than 200 μE) and low or moderate nitrogen levels, such as those provided by nitrogen fixation after nodulation by Rhizobium meliloti (data not shown). Anthocyanin levels are greatly reduced in leaves grown under high inorganic nitrogen (such as greater than 20 mm nitrate) and low light and approach zero in leaves of plants grown from seedlings that were vernalized at 4°C for 12 to 15 d, a treatment that is commonly used to accelerate seed set. Anthocyanin accumulation is also inducible in the epidermis of hypocotyls of vernalized seedlings by using higher light treatments, as has been demonstrated for many plant species.
Figure 7.
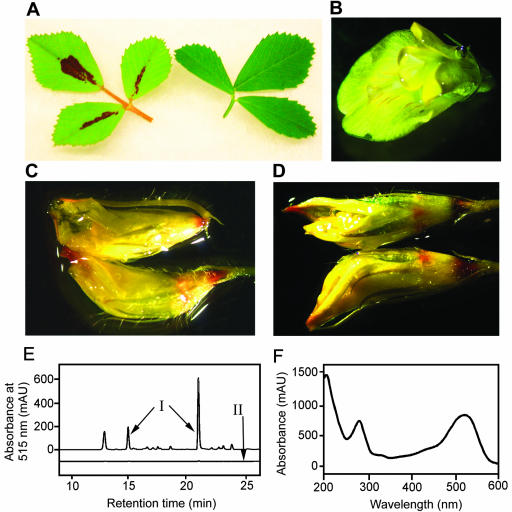
Anthocyanin and leucoanthocyanidin analysis in M. truncatula leaves and flowers. A, Trifoliate leaves from M. truncatula cv Jemalong line A-17 with (left) and without (right) leaf anthocyanin accumulation. Extended vernalization of young seedlings and high nitrate fertilizer levels reduce or eliminate leaf anthocyanin accumulation, whereas low nitrogen and high light levels promote anthocyanin accumulation. B, Open flower, submerged in water, as a negative staining control. Most of the flower petals are bright yellow, with very narrow streaks of red pigment visible on the standard (largest) petal. C, Flower buds treated with 6 m HCl for 2 to 3 min at room temperature. The very rapid formation of red color at the petal tips and bases is indicative of the conversion of colorless leucoanthocyanidins to anthocyanins. Under these mild conditions, color formation from CTs should require the presence of vanillin (Devic et al., 1999). D, Flower buds treated with 1% (w/v) vanillin in 6 m HCl. Red color is observed in the same regions of the buds with vanillin as without vanillin (C). Although CTs may be in these regions, their presence would be obscured by the other substances reacting with 6 m HCl. E, HPLC chromatograms of extracts from red and green portions of M. truncatula leaves. Elution was monitored at 515 nm to enhance the detection of and selectivity for red pigments. Three prominent peaks were present in acidic methanol extracts of red leaf sectors (line I), whereas these peaks were absent or greatly reduced in extracts of green sectors (line II). Chromatograms are offset by 25 milli-absorbance units to help differentiate the chromatograms. F, UV/visible diode array scan of the major anthocyanin component of M. truncatula leaves. The scan was captured as the peak eluted (at 21 min) from the HPLC in a mixture of phosphoric acid and acetonitrile. The scan is very similar to those of cyanidin in the same HPLC elution conditions (data not shown).
The flowers of M. truncatula contain mainly yellow pigments, most likely xanthophylls or carotenoid derivatives (Fig. 7B), although fine red veins of anthocyanins run the length of the standard petal, similar to patterns observed for alfalfa flower coloration (Hanson et al., 1988). During an investigation of the CT content of M. truncatula tissues, treating flowers with a control solution of 6 m HCl caused the formation of red pigments, consistent with the presence of leucoanthocyanidins, colorless products of DFR (Fig. 7C). Staining with 1% (w/v) vanillin in 6 m HCl to test for the presence of CTs in flowers and buds revealed no additional color compared with 6 m HCl, suggesting no CTs are present, but the presence of low levels of CTs may have been masked by the leucoanthocyanidins and other substances present (Fig. 7D). CTs were clearly present in the seed coats of young (green) seeds but are not present in other parts of the seeds (Xie et al., 2003).
A preliminary characterization of the red leaf pigments was carried out, primarily to confirm the anthocyanin nature and to determine the structure of the anthocyanidin core. Red centers of leaflets were dissected away from the surrounding green regions, and each was extracted and analyzed for anthocyanin content by HPLC. Using detection at 515 nm to selectively detect red chromophores, three major anthocyanin peaks were detected at approximately 21 (predominant peak), 15, and 13 min (Fig. 7E). Diode array scans revealed that the three peaks had almost identical UV/visible absorption patterns, with maxima at 280 and 520 to 525 nm (Fig. 7F), consistent with common anthocyanins in acidified solvents.
A small amount of the most abundant anthocyanin (21-min retention time, Fig. 7E) was partially purified and hydrolyzed by heating in acid. Two major product peaks were detected after hydrolysis. One product peak was cyanidin, based on co-elution with a commercial standard (at 24.5 min) and identical UV/visible scans. A second product peak matched the retention time and absorption spectrum of the 15-min peak. Partially purified samples of the 21- and 15-min peaks were subjected to LC/MS and LC/tandem MS analysis in positive ion mode. (The quantities of the purified 13-min peak were insufficient to allow analysis.) The parent ion observed in the 21-min peak was m/z = 667 atomic mass units (amu), with major fragments at m/z = 625, 449, and 287, whereas the 15-min peak had a parent ion of m/z = 625 amu, with major fragments at m/z = 287 and 449. The fragment masses of 287 and 449 are equal to the mass of cyanidin and possible cyanidin-3-O-glucoside ions, respectively. The 42-amu mass difference between the two parent ions is consistent with the loss of an acetyl group as ketene. Several anthocyanins and other flavonoids have been reported to be acetylated, and others that are malonylated in vivo spontaneously decarboxylate during isolation to form the acetate derivatives, especially in the presence of HCl (Harborne, 1988). The previously observed conversion of the 21-min peak to the 15-min peak during acid hydrolysis would be consistent with these structural assignments. Taken together, the data would be consistent with an acetylated and monomethylated diglucoside of cyanidin (mass of 667) for the 21-min peak and a corresponding deacetylated derivative for the 15-min peak. A conclusive structural assignment would require much larger quantities of the anthocyanins, greater purity levels, and detailed NMR analysis.
Characterization of Developmental Gene Expression Patterns of MtDFR1 and MtDFR2
The tissue specificity of the expression of the two M. truncatula DFR genes was initially examined by RNA gel-blot analysis (Fig. 8A). RNA was extracted from a variety of M. truncatula tissues, especially those accumulating CTs or anthocyanins, known products of DFR activity. A soybean 18S ribosomal RNA probe was used to confirm equal loading and transfer of the total RNA samples (Fig. 8A). The northern analysis results indicated that both MtDFR1 and MtDFR2 transcripts were highly abundant in YS tissues, the source tissue for the original cDNA library, and a known site of CT biosynthesis in alfalfa (Koupai-Abyazani et al., 1993) and M. truncatula (Xie et al., 2003). Blots probed with each 32P-labeled full-length cDNA also showed high transcripts levels in open flowers and flower buds and various stages of developing leaves with and without anthocyanin accumulation. Low transcript levels were observed in light-induced hypocotyls, accumulating low levels of anthocyanins. No transcripts were detected in dark-grown hypocotyls or roots, nodulated or nonnodulated.
Figure 8.
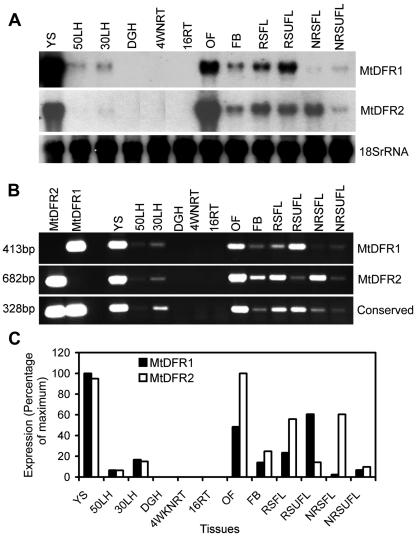
Developmental variation in DFR transcript levels in M. truncatula. A, Northern-blot hybridization analysis with different DFR probes: MtDFR1 coding region probe, MtDFR2 coding region probe, and soybean (Glycine max) 18S rRNA (loading control) probe. B, Ethidium bromide-stained agarose gel analysis of reverse transcriptase (RT)-PCR with transcript specific and general DFR primers: MtDFR1-specific primer pair, MtDFR2-specific primer pair, and conserved DFR primer pair, which amplifies both MtDFR1 and MtDFR2 transcripts. C, Normalized values of signal intensities for MtDFR1 and MtDFR2 RT-PCR products. Following quantitation of band intensities of the RT-PCR experiment, all of the numerical values for each primer set were divided by the highest value in that set, such that the tissue with highest transcript levels would be assigned 100% and the other tissue would be scored relative to this. For MtDFR1, YSs were the highest (100%), whereas for MtDFR2, open flowers were the highest, slightly higher than YSs. The analysis was repeated three times, with less than 5% variation between replicates. 50LH, 50-h Light-induced hypocotyls (with visible red epidermal anthocyanins); 30LH, 30-h light-induced hypocotyls (with visible red epidermal anthocyanins); DGH, dark-grown hypocotyls (white, with no anthocyanins); 4WNRT, 4-week-old nodulated roots (including both nodules and entire root systems); 16RT, 16-d-old roots (not inoculated with Rhizobia meliloti, and grown with 16 mm nitrate fertilizer); OF, open flowers; FB, flower buds; RSFL, red spot folded leaves; RSUFL, red spot unfolded leaves; NRSFL, non-red spot folded leaves; NRSUFL, non-red spot unfolded leaves. In M. truncatula, the younger leaflets are folded (FL, folded leaf) as they initially emerge and later unfold (UFL, unfolded leaf). If conditions allow leaf anthocyanin accumulation, the central red leaf spot will already be visible when the leaflets unfold but may darken as the leaf matures.
Because of concerns regarding possible crosshybridization of the two highly similar DFR transcripts during the above analysis, the same sets of tissue RNAs were analyzed by RT-PCR with primer pairs specific to each DFR transcript. The first two lanes of Figure 8B demonstrate the high specificity of the primer pairs for the amplification of their respective cDNA clones. RT-PCR using the MtDFR1 gene-specific primer pair produces one band 413 bp in size (Fig. 8B), whereas the MtDFR2 gene specific primer pair produces one band 682 bp in size (Fig. 8B). RT-PCR using a set of primers common to both DFR transcripts provides an estimate of the combined DFR gene expression in each tissue (Fig. 8B). Because the larger band size would automatically cause the intensity of the MtDFR2 bands to appear brighter than the MtDFR1 bands for the same amount of template, thus possibly biasing the direct visual interpretation of the results, the signals from the ethidium bromide-stained gels were also quantitated and normalized against the strongest signal observed for each primer pair (Fig. 8C) to facilitate a comparison of the RT-PCR results from the two primer pairs.
The results of these semiquantitative RT-PCR experiments paralleled those obtained by northern analysis but also revealed subtle differences in the expression patterns of the two DFR genes. For both genes, transcripts were at or near relative maximal levels in the YS mRNA pool, and no transcripts were detected with either primer pair in the two root samples or dark-grown hypocotyls. Although both MtDFR1 and MtDFR2 transcripts were detected in open flowers and flower buds, MtDFR2 had a 60% to 100% higher relative expression level than MtDFR1 in these floral samples, and MtDFR2 was slightly more highly expressed in open flowers than in the YS mRNA pool.
In younger, folded leaves, the relative levels of MtDFR2 transcripts were approximately one-half of the maximal levels observed, whereas MtDFR1 transcripts were much lower. MtDFR2 transcripts were twice as abundant as MtDFR1 transcripts in young folded leaves actively accumulating anthocyanins (red spot folded leaves), and MtDFR2 transcripts were 20 times higher than the levels of MtDFR1 transcripts in young folded leaves not accumulating anthocyanins (non-red spot folded leaves). In contrast, MtDFR1 transcripts were at about one-half of the maximal levels, approximately 4 times more abundant than MtDFR2 transcripts, in fully expanded, mature (unfolded) leaves that had accumulated anthocyanins (red spot unfolded leaves). Neither DFR gene was highly expressed in mature leaves containing no anthocyanins (non-red spot unfolded leaves). Both MtDFR1 and MtDFR2 transcript levels were higher in light-grown hypocotyls, particularly after 30 h of light exposure, compared with dark-grown hypocotyls, where no transcripts were detected.
Effect of Overexpression of MtDFR1 and MtDFR2 on Flower Color in Transgenic Tobacco
Although it was evident that the two DFR proteins exhibited different kinetic properties in vitro and had slightly different expression patterns in M. truncatula, overexpression in transgenic tobacco plants was used to assess whether or not the two DFR proteins would perform differently in plant cells. Although selected M. truncatula varieties have been reported to be transformed and regenerated, cv Jemalong A-17 is fairly recalcitrant, and the process requires several months. Binary vectors containing the coding regions of the MtDFR1 and MtDFR2 cDNA clones under the control of the CaMV35S promoter were derived from pBI121 (Jefferson et al., 1987) and were used to produce transgenic tobacco plants. Plants harboring the original pBI121 vector (CaMV35S promoter driving the beta-glucuronidase gene) were used as controls for comparison.
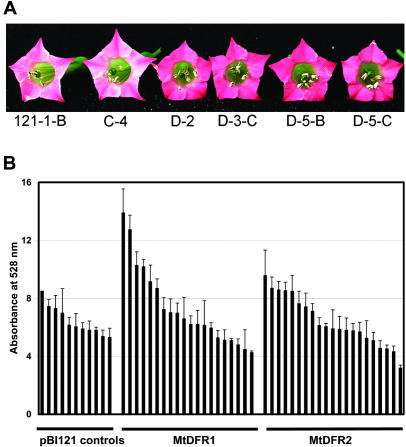
Tobacco (Nicotiana tobacum cv Xanthi), used for transformation, produces pale pink flowers under standard greenhouse conditions. When the transgenic lines were allowed to bloom, several plants harboring the MtDFR1 overexpression construct produced much darker pink flowers than were observed on untransformed plants or on plants harboring either the MtDFR2 vector or the pBI121 control vector (Fig. 9A). The anthocyanins were extracted from the corollas of flowers from each of the three types of transformants and were roughly quantitated spectrophotometrically. Although the anthocyanin content varied from flower to flower on the same plant, at least four plants harboring the MtDFR1 overexpression construct produced flowers containing significantly higher amounts of anthocyanins on a fresh weight basis (Fig. 9B). None of the MtDFR2-transformed lines showed significantly increased levels of anthocyanins relative to pBI121-transformed lines based on visual observations or spectrophotometric analysis. Northern analysis confirmed that several MtDFR2-transformed lines did accumulate MtDFR2 transcripts to the same levels as MtDFR1-transformed lines accumulated MtDFR1 transcripts (data not shown), indicating that the differences in anthocyanin accumulation were because of posttranscriptional events. Together, this indicates that the enzyme encoded by MtDFR1 successfully interacts with the endogenous tobacco anthocyanin biosynthetic pathway enzymes to increase anthocyanin accumulation in vivo, whereas the enzyme encoded by MtDFR2 is unable to increase the flux toward anthocyanin products. These data suggests that the endogenous DFR levels are rate limiting for anthocyanin biosynthesis at some stage in tobacco flower development, and MtDFR1 can increase the pathway flux to anthocyanins at that stage. A similar enhancement of anthocyanin accumulation was observed in vegetative tissues of Forsythia Intermedia cv “Spring Glory” transformed with Antirrhinum majus DFR (Rosati et al., 1997).
Figure 9.
Effect of MtDFR 1 and MtDFR2 in vivo on anthocyanin accumulation in transformed tobacco flowers. A, Overexpression of MtDFR1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter in transgenic tobacco flowers (lines D-2, D-3-C, D-5-B, and D-5-C) resulted in a visible increase in anthocyanin accumulation in the corolla, relative to untransformed lines (C-4) and lines harboring pBI121 (121-1-B). B, Spectroscopic quantitation of anthocyanin levels in transformed tobacco flowers. Corollas of three flowers from individual transformed tobacco plants were extracted in acidic methanol, and the absorbance of the extracts was measured at 528 nm to estimate relative anthocyanin levels. Error bars = sds of three measurements for each line. Four lines harboring MtDFR1 had significantly higher anthocyanin levels compared with the four highest pBI121 transformed lines (based on a Student's t test analysis limit of P ≤ 0.05; three lines passed at P ≤ 0.01). No lines overexpressing MtDFR2 showed significant increases in anthocyanins (based on a Student's t test analysis limit of P ≤ 0.05).
DISCUSSION
Two DFR cDNA clones were isolated from a YS EST library indicating the presence of a small DFR gene family in M. truncatula. Both DFR clones were functionally expressed in E. coli, confirming that they both encode active proteins and providing a means to explore substrate specificity of the proteins in vitro. Recently, three other reports of microbial expression of plant DFR clones were published (Martens et al., 2002; Peters and Constabel, 2002; Fischer et al., 2003). One reported no success with two E. coli expression systems followed by excellent expression of a G. hybrida DFR cDNA clone in yeast (Martens et al., 2002). Apple (Malus domestica) and pear (Pyrus communis) DFR cDNA clones also were expressed successfully using the same yeast system (Fischer et al., 2003). A third group (Peters and Constabel, 2002) successfully expressed DFR activity from a trembling aspen (Populus tremuloides) cDNA clone in E. coli using a system very similar to the one successfully utilized herein. The variable success in E. coli expression may be a function of subtle variations in protein sequences among the DFR clones. Although the recombinant proteins used in this study were prepared under identical conditions and levels of protein accumulation and initial activity were similar, a dramatic loss of DFR activity was observed after repeated freezing and thawing of bacterial extracts prepared using the MtDFR1 clone, whereas no loss of activity was observed in MtDFR2 extracts (data not shown).
Preliminary chemical characterization of the major foliar red pigments in M. truncatula indicated that these are derivatives of cyanidin diglucosides (3′,4′-dihydroxylated anthocyanins). Cyanidin derivatives are widespread through the plant kingdom (Harborne, 1988). Although the exact structure of the anthocyanins was not determined, the presence of a cyanidin core indicates a biosynthetic requirement for the expression of at least one DFR gene during leaf development and one that utilizes taxifolin as substrate. Pelargonidin or delphinidin chromophores were not detected. Alfalfa, an outcrossing perennial autotetraploid species, was previously reported to contain only a single DFR gene per genome equivalent (Charrier et al., 1995). The alfalfa gene sequence (GenBank accession no. X80222) is most similar (>98% identical at the amino acid level) to that of MtDFR1. It was not anticipated that a near-isogenic line of a diploid Medicago spp. like M. truncatula would have more DFR sequences than a tetraploid relative. There are striking similarities in the secondary metabolite compositions of the two legumes, such as root isoflavonoids (Baggett et al., 2002), root saponins (Huhman and Sumner, 2002), and seed coat CTs (Xie et al., 2003), although the exact profiles vary. M. truncatula cv Jemalong line A-17 does differ from alfalfa in the very intense anthocyanin accumulation on the upper, central surface of the leaves (Fig. 7A), but many alfalfa cultivars accumulate red anthocyanins in leaves and stems during stress (Hanson et al., 1988). Unlike the yellow-flowered M. truncatula, alfalfa accumulates high levels of red and blue anthocyanin pigments in flower petals, which are predominantly delphinidin and the methylated derivatives malvidin and petunidin but not cyanidin or pelargonidin derivatives (Cooper and Elliott, 1964). Future analysis of the alfalfa genome may reveal a second DFR gene similar to MtDFR2, or this may be an example of another difference between the model legume M. truncatula and cultivated alfalfa.
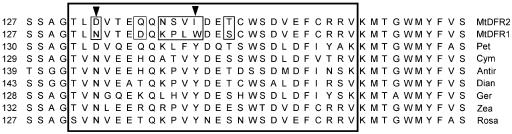
MtDFR1 and MtDFR2 provide a natural pair of dihydroflavonol reductases from the same species with slightly differing amino acid sequences to examine the active site features that determine the relative substrate preferences of these isozymes (Fig. 10). Following alignments of several DFR clones isolated from species varying in their anthocyanin hydroxylation patterns, earlier workers had postulated that a 26-amino acid region in DFR controlled the substrate specificity of the reductases, which in turn controls anthocyanin composition in some species (Beld et al., 1989). Using an elegant approach of in planta expression of cloned DFR sequences in DFR-minus mutants of petunia, Johnson et al. (2001) demonstrated that a single-amino acid change in this region could greatly alter the substrate specificity of petunia DFR. The native petunia DFR cannot accept DHK as a substrate to produce pelargonidin anthocyanins and contains an Asp residue at a position where several other DFR clones that can reduce DHK contain an Asn residue. Changing this Asp in the petunia sequence to a Leu converted the encoded enzyme from one that only accepted DHQ (dihydroxy) and DHM (trihydroxy) substrates to one that no longer accepted these substrates but that readily accepted DHK (monohydroxy). However, the authors did not report the effect of a mutation of Asp to Asn in the petunia sequence. MtDFR1 contains an Asn at the corresponding position (position 133) and reduces DHK more than twice as rapidly as DHQ (taxifolin) under the in vitro conditions used in this study (Figs. 6B and 10). MtDFR2 contains a Asp at this position, and although it still accepts DHK as a substrate, MtDFR2 reduces DHK much less readily than DHQ (taxifolin) and at only 10% to 20% of the rate of MtDFR1. The much greater activity of MtDFR1 on the monohydroxylated substrate DHK compared with that of MtDFR2 reaffirms the importance of the Asp or Asn residue in determining the rate of conversion of DHK to pelargonidin anthocyanins. However, the fact that MtDFR1 is also active on DHQ and DHM illustrates that other active site residues must also have an effect on substrate preference. Both M. truncatula DFR sequences differ from previously reported DFR sequences, including the replacement of a highly conserved Tyr at position 142 by Trp in MtDFR1 and Ile in MtDFR2 (Fig. 10).
Figure 10.
Alignment of M. truncatula MtDFR1 and MtDFR2 partial sequences with DFR sequences from seven other species, emphasizing the 26-amino acid region (boxed) proposed to determine substrate specificity (Beld et al., 1989; Johnson et al., 2001). Pet, Petunia; Cym, C. hybrida; Dian, Dianthus caryophyllus; Ger, G. hybrida; Zea, maize (Zea mays); Antir, snapdragon; Rosa, Rosa hybrida. The first arrow over residue 133 in MtDFR1 and MtDFR2 indicates the Asn or Asp residue that has a major impact on the utilization of DHK; MtDFR2 and petunia DFR both contain Asp residues at this site and process DHK poorly or not at all. The second arrow over residue 142 in MtDFR1 and MtDFR2 indicates the Ile or Trp residues that differ from the highly conserved Tyr residues in other DFR sequences.
Our in vitro assays showed that each of the recombinant M. truncatula DFR proteins can catalyze the reduction of all three of the most common dihydroflavonol substrates in nature (DHK, DHQ, and DHM) containing one hydroxyl, two hydroxyls, and three hydroxyls on the B ring, respectively. However, both MtDFR1 and MtDFR2 enzymes most efficiently reduced fustin, a 5-deoxy-dihydroflavonol analog of taxifolin, to leucofisetinidin, and reduced DHR, a 5-deoxy analog of DHM, to leucorobinetinidin. To our knowledge, this is the first report of DFR converting 5-deoxy-dihydroflavonols more efficiently than the 5-hydroxy-dihydroflavonols. DFR proteins from pear and apple were also able to reduce fustin and 5-deoxydihydrokaempferol (garbanzol), but these substrates were less efficiently processed than DHQ, and 5-deoxyflavonols do not naturally occur in these Rosaceae species (Fischer et al., 2003). Leucofisetinidin and leucorobinetinidin and their derivatives have not been identified yet in herbaceous legumes such as M. truncatula but have been isolated from woody legumes such as Acacia mearnsii, Robinia pseudacacia, and Gleditzia japonica (Harborne, 1988). Several legumes including alfalfa accumulate 5-deoxyflavonoids (such as liquiritigenin) and 5-deoxyisoflavonoids (such as daidzein, formononetin, and medicarpin) because of the activity of chalcone reductase, which reduces the polyketide intermediate produced by CHS (Guo and Paiva, 1995). Upon cyclization, a trihydroxychalcone is formed (in place of the tetrahydroxychalcone in Fig. 1), which is further processed to 5-deoxyflavonoids and 5-deoxyisoflavonoids. The high activity of MtDFR1 and MtDFR2 on 5-deoxy substrates may be related to currently unknown metabolic pathways in M. truncatula or may simply be an example of an unnatural substrate being processed more efficiently than an endogenous one.
The two MtDFRs have relative substrate preferences different from those of individual DFRs from other plant species. For example, DFR-A in petunia reduces DHM more readily than taxifolin, unlike either of the M. truncatula DFRs (Forkmann and Ruhnau, 1987). Similarly, D. caryophyllus DFR utilizes taxifolin and DHM much more readily than DHK (Stich et al., 1992), unlike the MtDFR1 enzyme. Phlobaphene pigment and 3-deoxy anthocyanin biosynthesis in monocot species such as maize and sorghum (Sorghum bicolor) requires the reduction of eriodictyol by DFR (Winkel-Shirley, 2001). The lack of activity of the two recombinant M. truncatula DFR proteins on eriodictyol is consistent with the general absence of these compounds in dicots. However, a highly purified DFR preparation from Dahlia variabilis flowers had a relative substrate preference very similar to that of MtDFR1 (DHK > DHQ > DHM) but was able to reduce naringenin and eriodictyol weakly, unlike the M. truncatula enzymes (Fischer et al., 1988). DFR proteins encoded by pear and apple cDNA clones also were able to reduce eriodictyol, but not naringenin, accounting for the accumulation of 3-deoxyflavonoids in prohexadione-treated leaves (Fischer et al., 2003).
Any differences in the respective biosynthetic roles of the two M. truncatula DFRs are unclear at this time. In many plant species, the expression of a single DFR gene accounts for the observed complex patterns of metabolite accumulation (Gerats et al., 1990; Kristiansen and Rohde, 1991; Winkel-Shirley et al., 1992; Bongue-Bartelsman et al., 1994; Sparvoli et al., 1994; Holton and Cornish, 1995; Chen et al., 1998; Elomaa et al., 1998; Nakai et al., 1998; Hoshino et al., 2001). For the two M. truncatula DFR genes, the patterns of transcript accumulation were remarkably similar in most of the tissues examined, despite the observed differences in in vitro kinetic parameters. Only in developing leaves, with and without anthocyanin accumulation, was a clear difference in relative expression levels observed. MtDFR1 expression was higher in both younger (folded) and older (unfolded) M. truncatula leaves accumulating anthocyanins (red spot folded leaves and red spot unfolded leaves) than in leaves without the red anthocyanin spot (non-red spot folded leaves and non-red spot unfolded leaves), whereas MtDFR2 transcripts levels were higher in young leaves than in older leaves, irrespective of the anthocyanin accumulation (Fig. 8). This suggests that MtDFR1 may play more of a role in leaf anthocyanin (cyanidin glucoside) biosynthesis. MtDFR1 was also slightly more induced than MtDFR2 in response to high-light treatment of dark-grown hypocotyls, in parallel with epidermal anthocyanin accumulation.
Both MtDFR1 and MtDFR2 appear to be equally expressed in seed and flower leucoanthocyanidin synthesis. Further dissection of the tissues, in parallel with a more extensive phytochemical analysis of the tissues, may reveal the association of the two enzymes with different metabolic pathways or expression in adjacent regions. The M. truncatula flowers used to isolate RNA were not dissected because of their small size and the fused architecture of the floral components, and although the majority of the mass is from petals, a number of other tissues (calyx, anthers, stigmas, and developing ovules) were present, possibly obscuring differences in DFR gene expression. Similarly, RNA was isolated from whole developing seeds, although different parts of the seed or layers of the seed coat may accumulate different metabolites. In Arabidopsis, CTs and anthocyanins accumulate in a single endothelium cell layer in the developing seed coat (Devic et al., 1999), but the sites of biosynthesis of other legume seed flavonoid metabolites has not been determined yet. High DFR transcript levels in the nearly anthocyaninless M. truncatula flowers prompted us to stain for CTs in these tissues, but we discovered instead the presence of putative leucoanthocyanidins (Fig. 7C). The phytochemical analysis described in this work emphasized the detection of known DFR-derived metabolites likely to be present in M. truncatula, but other colorless leucoanthocyanidin derivatives may be present in flowers, leaves, or seeds. Alternatively, the two DFR genes may represent “pathway redundancy” in M. truncatula or gene duplication as a possible artifact of evolution. Certain isoflavonoid pathway genes that are unique to legumes such as VR (Guo and Paiva, 1995) and isoflavone reductase (Paiva et al., 1991) are very similar to DFR sequences, and these may have arisen from DFR after gene duplication. Tools currently under development for the model legume M. truncatula may also help decipher the functions of the DFR genes via mutants lacking the expression of one functional DFR gene and the evolutionary relationship of the two DFR genes via sequencing of the M. truncatula genome (Cook, 1999; Penmetsa and Cook, 2000).
MATERIALS AND METHODS
Plant Materials
To produce immature seeds for cDNA library construction, Medicago truncatula cv Jemalong line A17 (Cook, 1999; Penmetsa and Cook, 2000) was grown in the greenhouse with supplemental light (16-h days, 20°C–30°C) until flowering and self-pollination began. Hundreds of YS pods were harvested and dissected. Immature (flat) green seeds at a variety of stages (2–5 mm in length, but before much deposition of starch and storage proteins had occurred) were immediately frozen in liquid nitrogen. Other tissues for mRNA isolation and RT-PCR analysis were prepared as described earlier (Xie et al., 2003) or in later sections herein.
Production of YS cDNA Library and ESTs
Total RNA was extracted from immature seeds using a commercial RNA isolation kit (Plant RNAeasy Midi kit, Qiagen, Valencia, CA) with the alternative guanidine hydrochloride buffer recommended for high starch tissues. mRNA was isolated from 1 mg of total RNA using immobilized oligo(dT) [Oligotex poly(A+) mRNA purification kit, Qiagen]. A primary cDNA library (YS library) containing over 6 × 106 phage was constructed following the manufacturer's instructions using the UniZapXR cDNA library kit (Stratagene, La Jolla, CA).
A portion of the YS primary cDNA library (10,000 plaque forming units) was mass excised using Stratagene's protocol. The resulting ExAssist phagemids were used to transform SOLR cells, and transformants containing pBluescript SK- plasmids with cDNA inserts were selected on Luria-Bertani agar plates supplemented with 100 μg mL-1 ampicillin. Individual colonies were randomly selected and used to inoculate 1.5 mL of Terrific Broth medium with 100 μg mL-1 ampicillin in 96-well 2-mL capacity plates. After 24 h of growth with shaking at 37°C, plasmids were isolated in a 96-well format using a modified alkaline lysis protocol (Roe et al., 1996). The 5′ end of each insert was sequenced using a lengthened M13 Reverse primer (GGAAACAGCTATGACCATG), BigDye Terminator sequencing kits, and ABI3700 DNA Analyzer (PE-Applied Biosystems, Foster City, CA). Using the data processing facilities of the Noble Medicago Genomics Initiative (Bell et al., 2001) after removal of vector sequences, EST sequences were subjected to BLASTX analysis against the National Center for Biotechnology Information nonredundant database.
Isolation of M. truncatula cDNA Clones Encoding MtDFR1 and MtDFR2 Proteins
MtDFR1
One EST sequence (NF001D05YS1F1046) with high BLASTX scores against sequences of known DFR clones was designated MtDFR1. Based on alignments of the complete sequence of the cDNA insert, this clone contained a full-length DFR ORF and 5′- and 3′-untranslated regions. The MtDFR1 ORF was subcloned from the original MtDFR1 pBluescript SK- plasmid using PCR into the Escherichia coli expression vector pSE380 (Brosius, 1989; Invitrogen). The upstream primer (001D05 primer1 5′GCGC-CCATGGGTTCTATGGCCGAAACTG3′) introduced an NcoI site at the first ATG in the ORF and the downstream primer from the vector (M13 5′GTAAAACGACGGCCAGT 3′). The temperature program for PCR was 94°C for 2 min, 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by final extension at 72°C for 10 min, using AmpliTaq (Boehringer Mannheim/Roche, Basel) and 2.5 mm MgCl2. The PCR product (1.33 kb) was gel purified and ligated into pGEM-T Easy. After resequencing, the MtDFR1 coding region and short poly(A+) tail was excised from pGEM-T Easy by NcoI and XhoI and ligated into pSE380 to produce pSE380-MtDFR1.
MtDFR2
A second EST (NF004H10YS1F1092) with high BLASTX scores against known DFR sequences was designated MtDFR2. Sequence analysis and comparison with highly similar, overlapping M. truncatula EST sequences in public databases (http://www.tigr.org) indicated that the MtDFR2 ORF was truncated. A full-length MtDFR2 sequence was obtained by PCR using 5 μL of boiled amplified YS cDNA library (1011 plaque forming units mL-1) as template, a 004H10 DFR1 primer (5′GCGCCCATGGGTTCAGTCTCAGAAACAGTTTGC3′), which introduces an NcoI site at the first ATG in the predicted ORF, and the M13 vector primer. The PCR program, purification of the 1.24-kb product, and ligation into pGEM-T Easy vector was the same as for MtDFR1. The MtDFR2 sequence from ATG to poly(A+) was cut from pGEM-T Easy by NcoI and XhoI and ligated into pSE380 to produce pSE380-MtDFR2.
Expression of MtDFR1 and MtDFR2 Proteins and in Vitro Enzyme Assay
The constructs containing MtDFR1 and MtDFR2 ORFs in the expression vector pSE380 were used to transform E. coli BL21-Gold competent cells (Stratagene). Single colonies harboring expression constructs pSE380-MtDFR1, pSE380-MtDFR2, or pSE380 (vector control) were cultured in Luria-Bertani medium with 100 μg mL-1 ampicillin, induced with 1 mm isopropyl-β-D-thio-galactoside for 3 h, harvested, and lysed as described previously (Guo and Paiva, 1995), except β-mercaptoethanol was omitted from the lysis buffer (100 mm Tris-HCl [pH 8.0], 5 mm EDTA [pH 8.0], and 100 μg mL-1 lysozyme). After 10 min of incubation at room temperature, the viscous lysate was sonicated for 15 to 20 s on ice to shear DNA and centrifuged for 15 min at ≥10,000g at 4°C.
The initial standard enzyme assay conditions included incubation at 30°C for 30 min in 0.5-mL total volume containing 370 μL of 100 mm Tris-HCl buffer (pH 7.0), 70 μL of protein extract (1.5 μg protein μL-1), 50 μL of 10 mm NADPH, and 10 μL of (±)-taxifolin (10 μg μL-1 methanol) in 1.5-mL centrifuge tubes. Enzyme reactions were stopped by adding 1 mL of ethyl acetate and vortexing. Samples were centrifuged for 2 min at 10,000g, and 0.75 mL of ethyl acetate extract was removed to a new tube and evaporated with a stream of nitrogen gas. Residues were dissolved in 150 μL of methanol and used directly for HPLC analysis (20-μL injection). The area of the leucoanthocyanidin product peaks was integrated using 32-Karat software (Beckman Instruments, Fullerton, CA).
(±)-Taxifolin (Sigma, St. Louis) was used as the initial substrate to test the effects of substrate concentration, assay pH, temperature, duration, and NADPH concentration on enzyme activity. Studies of pH dependence were conducted at 30°C for 30 min in a 0.5-mL volume consisting of 370 μL of 50 mm citrate/phosphate buffer (pH 4.6–7.0) or 100 mm Tris-HCl buffer (pH 7.0–8.8), 70 μL of protein extract, 50 μL of 10 mm NADPH, and 10 μL of 33 mm (±)-taxifolin.
Test of Different Substrates in Vitro
Dihydroflavonols including (±)-taxifolin (DHQ; Sigma), (+)-taxifolin, (-)-fustin (Dr. Daneel Ferreira), racemic (±)-fustin (Indofine, Hillsborough, NJ), DHK, DHM, and DHR (Apin, Abingdon, Oxon, UK) were dissolved in methanol at 10 mg mL-1. The flavonols, flavanones, or flavones, i.e. kaempferol, quercetin, eriodictyol, and apigenin, were dissolved in methanol at 10, 10, 2.5, and 1 mg mL-1, respectively. Reactions for MtDFR1 were conducted at 30°C for 30 min using 370 μL of 100 mm Tris-HCl buffer (pH 7.0), 70 μL of MtDFR1 enzyme extract, 10 μL of substrate, and 50 μL of 20 mm NADPH, whereas reactions for MtDFR2 substituted 370 μL of 50 mm citrate/phosphate buffer (pH 6.2), so that enzymes were assayed at their respective optimum pH values.
HPLC Analysis of Enzyme Reaction Products
Compounds were resolved on a C18-silica HPLC column (5 μm, narrow pore, 4.6 × 250 mm, Bakerbond, J.T. Baker, Phillipsburg, NJ) with detection at 280 nm, the maximum absorbance wavelength for most of the substrates and products. UV spectra were recorded with a UV diode array detector (model 168, Beckman Instruments). For routine HPLC quantitation, the solvents were 1% (v/v) H3PO4 in water (A) and methanol (B), and separation methods were developed by adapting the systems reported previously (Stafford and Lester, 1984; Dellus et al., 1997). For initial DFR assays or screening of possible substrates, the column was equilibrated at 15% (v/v) B at a flow rate of 1 mL min-1. Following injection (20 μL), a linear gradient from 15% to 60% (v/v) B in 20 min was initiated. Peaks were identified by retention time and UV spectra, compared with standards available in house, and by LC-MS analysis. For later assays using only taxifolin, the assay time was reduced by increasing the flow rate to 1.5 mL min-1 and decreasing the gradient to 8 min.
Analysis of Anthocyanins in M. truncatula and Tobacco (Nicotiana tabacum)
To prepare large quantities of anthocyanins for preliminary structural characterization, fresh leaves and stems grown under high-light conditions were extracted for 12 to 24 h with neutral acetone to remove the bulk of the chlorophyll, lipids, and other substances without extracting the majority of the anthocyanins (data not shown). The red anthocyanin sectors were still clearly visible against the decolorized portions of the leaves. The acetone was decanted and replaced with acidic methanol (1 mL HCl L MeOH-1), and anthocyanins were extracted for 1 to 2 d at room temperature with shaking in the dark. The MeOH was removed by rotary evaporation, and the red residue was redissolved in a small volume of MeOH. Anthocyanins were quantitated using HPLC (4.6- × 250-mm Bakerbond C18-silica column, as above; J.T. Baker) with detection at 515 nm and normalized against the fresh weight values. The solvents were 1% (v/v) H3PO4 in water (A) and CH3CN (B) with column equilibration at 5% (v/v) B and a flow rate of 0.8 mL min-1. Following injection (20 μL), a linear gradient from 5% to 40% (v/v) B in 35 min was initiated.
The anthocyanins were partially purified by diluting the crude solution with water to less than 10% (v/v) MeOH and adsorbing to a disposable C-18 cartridge (Discovery DSC-18 SPE, Supelco, St. Louis). After rinsing with 10% (v/v) MeOH, 30% (v/v) CH3CN was used to elute the anthocyanins. The eluate was concentrated and subjected to semipreparative HPLC (Econosil C-18, 22- × 250-mm column, Alltech, Deerfield, IL) using a gradient and solvents identical to the analytical method but with a 20 mL min-1 flow rate. Fractions containing anthocyanins were pooled and CH3CN was removed using a stream of nitrogen gas. The remaining aqueous solution was extracted with EtOAc, then the EtOAc phase back-extracted with water to remove traces of H3PO4, and EtOAc was removed by evaporation. The residue was dissolved in MeOH and subjected to LC-MS analysis. A portion of each purified anthocyanin sample was hydrolyzed by heating in 10% (v/v) HCl at 95°C for 4 h to release the anthocyanidin core.
To compare the anthocyanin content of small tissue samples from M. truncatula (100–300 mg fresh weight) or tobacco flowers, tissues were extracted with acidic MeOH (5 mL) by shaking at room temperature in the dark for 48 h. For M. truncatula tissues, extracts were dried and redissolved in 200 μL of MeOH and subjected to HPLC analysis as above. For tobacco flowers, total anthocyanins were quantitated by spectrophotometry at 528 nm (Xie et al., 2003).
LC-MS Analysis of DFR Reaction Products and Anthocyanins
For the (±)-taxifolin product, 200-fold scaled-up reactions (100-mL volume) were performed at 30°C for 3 h in 50 mm citrate/phosphate buffer (pH 6.2) with 14 mL of MtDFR2 enzyme extract, 20 mg of (±)-taxifolin, and 10 mL of 20 mm NADPH. The reaction mixture was extracted six times with 100-mL aliquots of ethyl acetate. The pooled extracts were concentrated, and the residue was dissolved in 3 mL of methanol for further purification or LC-MS analysis. For (-)-fustin, (±)-fustin, and other substrates, similar large-scale reactions (up to 6-mL reaction volume) and HPLC and LC-MS analyses were carried out.
For LC-MS analysis of the DFR products, an HP1100 series HPLC system (Agilent, Palo Alto, CA) with UV/Vis diode array detection followed by a Esquire-LC00098 mass spectrometer system (Bruker, Billerica, MA) was used. H3PO4 was omitted from the HPLC solvents without substantially altering the retention times of the DFR products. The total ion chromatograms were obtained using MS detection with negative ionization and scans of peaks were stored from m/z of 50 to 1,000 amu.
For LC-MS analysis of the extracted and hydrolyzed anthocyanins, the anthocyanin quantitation method was modified by substituting volatile 1% (v/v) acetic acid for 1% (v/v) H3PO4. Total ion chromatograms were obtained using MS detection with positive ionization as is recommended for anthocyanins, and scans of peaks were stored from m/z of 50 to 2200 amu.
Southern Hybridization
Genomic DNA of M. truncatula was extracted from young leaves using the Plant DNAeasy Maxi kit (Qiagen). DNA (10 μg) was digested overnight with HindIII and EcoRI and resolved by electrophoresis in a 0.8% (w/v) agarose gel in Tris-acetic acid-EDTA buffer. Blotting to GeneScreenPlus (DuPont, Wilmington, DE) nylon membranes and hybridization at 65°C were performed as previously described (Church and Gilbert, 1984; Hipskind and Paiva, 2000). Fragments of the cDNA clones containing the entire ORF of either MtDFR1 or MtDFR2 were labeled with α-32P-dCTP using random primer labeling (Prime-a-Gene, Promega, Madison, WI) and used as probes, although either can cross-hybridize to some extent to both DFR genes.
Gene-specific probes for MtDFR2 or MtDFR1 were designed from the DNA sequences of the 3′ portions of the ORFs. The 225-bp DNA fragment for the MtDFR1-specific probe was amplified using the PCR primer pairs of 5′GCACCAATAAGTAATGGTGTCAC3′ and 5′CGTGTAGCTCTCAATAAGG3′, whereas the 173-bp DNA fragment for the MtDFR2 specific probe was amplified using 5′CCTAAAGTTACAGAGACTCCGG3′ and 5′CAGACAACGTGACCCATAAAC3′. The PCR products were labeled with α-32P-dCTP, and membranes were hybridized and washed at 65°C as described above.
Northern Hybridization Analysis of MtDFR1 and MtDFR2 Transcript Levels
To assess MtDFR1 and MtDFR2 expression patterns in M. truncatula, total RNA was extracted from different organs including roots, hypocotyls, leaves, flower buds, open flowers, and YSs using Tri-Reagent (Molecular Research Center, Cincinnati, OH). Root samples were harvested from plants grown in perlite wetted with a nutrient solution (Hipskind and Paiva, 2000). These included 4-week-old root systems grown in 2 mm KNO3 and nodulated by Rhizobium meliloti and uninoculated 16-d-old roots grown in 16 mm KNO3. Young expanding folded leaves and fully expanded unfolded (mature) leaves containing the central red anthocyanin leaf spots were collected from un-vernalized plants grown in a growth room with a light intensity of 200 μE and low nitrogen fertilizer. Similar leaf tissues lacking the central red anthocyanin leaf spots were collected from vernalized plants (seedlings vernalized for 2 weeks during germination at 4°C). Flower buds and flowers (including the calyx) and YSs were collected from plants grown in the greenhouse. Seeds were scarified with concentrated sulfuric acid for 10 min and then washed several times with sterile MilliQ water before placing on wet filter paper in clear petri dishes for germination in the dark. Three-day-old dark-grown seedlings were transferred to light (200 μE), and red hypocotyls were harvested after 30 and 50 h. White hypocotyls lacking detectable anthocyanins were harvested from 5-d-old dark-grown seedlings.
Total RNA samples (15 μg) were electrophoresed in 1.5% (w/v) agarose MOPS-formaldehyde gels and transferred to GeneScreen Plus (NEN/DuPont, Wilmington, DE) membranes as described by Sambrook et al. (1989). The MtDFR1 and MtDFR2 coding region probes were labeled with α-32P-dCTP using a Prime-a-Gene (Promega) random primer labeling kit. Membranes were hybridized and processed as described above for Southerns. Band intensities were quantitated using ImageQuant software (Molecular Dynamics, Sunnyvale, CA) following phosphor imager analysis.
RT-PCR Analysis of Transcript Levels
One primer pair was designed from conserved regions in MtDFR1 and MtDFR2 to amplify a 329-bp PCR product from both cDNA clones. The primers were 5′CCTATGGATTTTGAGTCCAAGGACCC3′ (corresponding to bases 315–340 of MtDFR1 and 259–284 of MtDFR2) and 5′GGACCAACAACAAGAGGTGG3′ (corresponding to bases 643–624 of MtDFR1 and 587–568 of MtDFR2). MtDFR1 and MtDFR2 gene-specific primer pairs were designed based on alignments of the cDNA sequences. The MtDFR1 primers were 5′CCTCAAGGCCAAAACTGTCC3′ (bases 398–417) and 5′GATACCTCCCTTCTACTTCC3′ (bases 810–791), yielding a 413-bp PCR product. The MtDFR2 primers were 5′GACTTATGGAGCGCGGCTACACA3′ (bases 72–93) and 5′GTATCTCCCATGTGCTTTAGGG3′ (bases 753–732), yielding a 682-bp PCR product.
Total RNA (1 μg) was used for first strand cDNA synthesis, and 5 μL of the first strand cDNA was used for each PCR reaction (50 μL) using the Advantage RT-for-PCR kit (CLONTECH Laboratories, Palo Alto, CA). For the conserved primers for both MtDFR1 and MtDFR2, PCR conditions were 94°C for 2 min, 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, followed by 72°C for 10 min. Identical conditions were used for MtDFR1 gene specific primers, whereas the annealing temperature was increased to 65°C for MtDFR2 gene-specific primers. PCR products (20 μL) were resolved on 0.8% (w/v) agarose Tris-acetic acid-EDTA gels, visualized with ethidium bromide, and band intensities were quantitated using ImageQuant software (Molecular Dynamics). Band intensities were normalized for each primer pair by expressing the results relative to the highest band intensity observed in each experiment.
Tobacco Transformation
Using modified PCR primers, a BamHI site was introduced immediately upstream of the start codons in MtDFR1 and MtDFR2, and a SacI site was introduced approximately 40 bp downstream of the stop codons. These modified coding regions from MtDFR1 and MtDFR2 were subcloned into the BamHI and SacI sites of the binary vector pBI121 (Jefferson et al., 1987), replacing the GUS coding region. These pBI121-DFR derivatives and unmodified pBI121 were introduced into Agrobacterium tumefaciens LBA4404 bacteria and used to transform tobacco cv Xanthi leaf pieces. Regeneration of transformed tobacco plants was accomplished using 300 μg mL-1 kanamycin selection according to standard protocols (Horsch et al., 1988). Transformed tobacco plants harboring control (pBI121), pBI121-MtDFR1, and pBI121-MtDFR2 vectors were transferred to the greenhouse and grown with supplemental lighting (16-h days) until flowers developed. Northern and RT-PCR analysis confirmed that several plants were accumulating high levels of MtDFR1 and MtDFR2 mRNA. The corolla of newly opened flowers were harvested and immediately extracted with acidic methanol for anthocyanin analysis as described above.
Acknowledgments
We would like to thank Dr. Bettina Deavours (The Samuel Roberts Noble Foundation, Ardmore, OK) for critical reading of the manuscript. We would also like to thank David Huhman (The Samuel Roberts Noble Foundation, Ardmore, OK) for expert assistance in acquiring LC-MS data.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.030221.
This work was supported by The Samuel Roberts Noble Foundation (Ardmore, OK) and by Forage Genetics (West Salem, WI).
References
- Almeida J, Carpenter R, Robbins TP, Martin C, Coen ES (1989) Genetic interactions underlying flower color patterns in Antirrhinun majus. Genes Dev 3: 1758-1767 [DOI] [PubMed] [Google Scholar]
- Baggett BR, Cooper JD, Hogan ET, Carper J, Paiva NL, Smith JT (2002) Profiling isoflavonoids found in legume root extracts using capillary electrophoresis. Electrophoresis 23: 1642-1651 [DOI] [PubMed] [Google Scholar]
- Beld M, Martin C, Huits H, Stuitje AR, Gerats AG (1989) Flavonoid synthesis in Petunia hybrida: partial characterization of dihydroflavonol-4-reductase genes. Plant Mol Biol 13: 491-502 [DOI] [PubMed] [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW et al. (2001) The Medicago Genome Initiative: a model legume database. Nucleic Acids Res 29: 114-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongue-Bartelsman M, O'Neill SD, Tong Y, Yoder JI (1994) Characterization of the gene encoding dihydroflavonol 4-reductase in tomato. Gene 138: 153-157 [DOI] [PubMed] [Google Scholar]
- Brosius J (1989) Superpolylinkers in cloning and expression vectors. DNA 8: 759-777 [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96: 12929-12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Coronado C, Kondorosi A, Ratet P (1995) Molecular characterization and expression of alfalfa (Medicago sativa L.) flavanone-3-hydroxylase and dihydroflavonol-4-reductase encoding genes. Plant Mol Biol 29: 773-786 [DOI] [PubMed] [Google Scholar]
- Chen M, SanMiguel P, Bennetzen JL (1998) Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics 148: 435-443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991-1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR (1999) Medicago truncatula: a model in the making! Curr Opin Plant Biol 2: 301-304 [DOI] [PubMed] [Google Scholar]
- Cooper RL, Elliott FC (1964) Flower color pigments in diploid alfalfa. Crop Sci 4: 367-371 [Google Scholar]
- Dellus V, Heller W, Sandermann H, Scalbert A (1997) Dihydroflavonol 4-reductase activity in lignocellulosic tissues. Phytochemistry 45: 1415-1418 [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19: 387-398 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Steele CL (1999) Flavonoid and isoflavonoids-a gold mine for metabolic engineering. Trends Plant Sci 4: 394-400 [DOI] [PubMed] [Google Scholar]
- Elomaa P, Mehto M, Kotilainen M, Helariutta Y, Nevalainen L, Teeri TH (1998) A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J 16: 93-99 [DOI] [PubMed] [Google Scholar]
- Fischer D, Stich K, Britsch L, Grisebach H (1988) Purification and charac terization of (+)-dihydroflavonol (3-hydroxyflavanone) 4-reductase from flowers of Dahlia variabilis. Arch Biochem Biophys 264: 40-47 [DOI] [PubMed] [Google Scholar]
- Fischer TC, Halbwirth H, Meisel B, Stich K, Forkmann G (2003) Molecular cloning, substrate specificity of the functionally expressed dihydroflavonol 4-reductases from Malus domestica and Pyrus communis cultivars and the consequences for flavonoid metabolism. Arch Biochem Biophys 412: 223-230 [DOI] [PubMed] [Google Scholar]
- Forkmann G, Ruhnau B (1987) Distinct substrate specificity of dihydroflavonol 4-reductase from flowers of Petunia hybrida. Z Naturforsch C 42c: 1146-1148 [Google Scholar]
- Gerats AGM, Huits H, Vrijlandt E, Maraña C, Souer E, Beld M (1990) Molecular characterization of a nonautonomous transposable element (dTph1) of Petunia. Plant Cell 2: 1121-1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Dixon RA, Paiva NL (1994) Conversion of vestitone to medicarpin in alfalfa (Medicago sativa L.) is catalyzed by two independent enzymes. J Biol Chem 269: 22372-22378 [PubMed] [Google Scholar]
- Guo L, Paiva NL (1995) Molecular cloning and expression of alfalfa (Medicago sativa L.) vestitone reductase, the penultimate enzyme in medicarpin biosynthesis. Arch Biochem Biophys 320: 353-360 [DOI] [PubMed] [Google Scholar]
- Hahlbrock H, Grisebach H (1975) Biosynthesis of flavonoids. In JB Harborne, TJ Mabry, H Mabry, eds, The Flavonoids. Academic Press, San Diego, pp 866-915
- Hanson AA, Barnes DK, Hill RR (1988) Alfalfa and Alfalfa Improvement (Monograph No. 29). American Society of Agronomy, Madison, WI
- Harborne JB, ed (1988) Flavonoids: Advances in Research since 1980. Chapman and Hall, New York
- Helariutta Y, Elomaa P, Kotilainen M, Seppänen P, Teeri TH (1993) Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol Biol 22: 183-193 [DOI] [PubMed] [Google Scholar]
- Hipskind JD, Paiva NL (2000) Constitutive accumulation of a resveratrol glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol Plant-Microbe Interact 13: 551-562 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish E (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Joyce F, Hoffman N, Neidermeyer J, Rogers SG, Fraley RT (1988) Leaf disc transformation. In SB Gelvin, RA Schilperoot, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1-9
- Hoshino A, Johzuka-Hisatomi Y, Iida S (2001) Gene duplication and mobile genetic elements in the morning glories. 265: 1-10 [DOI] [PubMed] [Google Scholar]
- Hou DX (2003) Potential mechanisms of cancer chemoprevention by anthocyanins. Curr Mol Med 3: 149-159 [DOI] [PubMed] [Google Scholar]
- Huhman DV, Sumner LW (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347-360 [DOI] [PubMed] [Google Scholar]
- Huits HSM, Gerats AGM, Kreike MM, Mol JNM, Koes RE (1994) Genetic control of dihydroflavonol 4-reductase gene expression in Petunia hybrida. Plant J 6: 295-310 [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Johzuka-Hisatomi Y, Mori T, Takahashi S, Hayakawa Y, Peyachoknagul S, Ozeki Y, Iida S (1999) Genomic organization of the genes encoding dihydroflavonol 4-reductase for flower pigmentation in the Japanese and common morning glories. Gene 226: 181-188 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, MW Bevan (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ET, Ryu S, Yi H, Shin B, Cheong H, Choi G (2001) Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J 25: 325-333 [DOI] [PubMed] [Google Scholar]
- Johnson ET, Yi H, Shin B, Oh BJ, Cheong H, Choi G (1999) Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J 19: 81-85 [DOI] [PubMed] [Google Scholar]
- Koupai-Abyazani MR, McCallum J, Muir AD, Lees GL, Bohm BA, Towers GHN, Gruber MY (1993) Purification and characterization of a proanthocyanidin polymer from seed of alfalfa (Medicago sativa cv. Beaver). J Agric Food Chem 41: 565-569 [Google Scholar]
- Kristiansen KN, Rohde W (1991) Structure of the Hordeum vulgare gene encoding dihydroflavonol 4-reductase and molecular analysis of ant18 mutants blocked in flavonoid synthesis. Mol Gen Genet 230: 49-59 [DOI] [PubMed] [Google Scholar]
- Martens S, Teeri T, Forkman G (2002) Heterologous expression of dihydroflavonol 4-reductase from various plants. FEBS Lett 531: 453-458 [DOI] [PubMed] [Google Scholar]
- Meyer P, Heidmann I, Forkmann G, Saedler H (1987) A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 330: 677-678 [DOI] [PubMed] [Google Scholar]
- Nakajima J, Tanaka Y, Yamazaki M, Saito K (2001) Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J Biol Chem 276: 25797-25803 [DOI] [PubMed] [Google Scholar]
- Nakai K, Inagaki Y, Nagata H, Miyazaki C, Iida S (1998) Molecular characterization of the gene for dihydroflavonol 4-reductase of japonica rice varieties. Plant Biotech 15: 221-225 [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863-1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099-2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva NL (2000) An introduction to the biosynthesis of chemicals used in plant-microbe communication. J Plant Growth Regul 19: 131-143 [DOI] [PubMed] [Google Scholar]
- Paiva NL, Edwards R, Sun Y, Hrazdina G, Dixon RA (1991) Stress responses in alfalfa (Medicago sativa L.): XI. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol Biol 17: 653-667 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (2000) Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol 123: 1387-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701-712 [DOI] [PubMed] [Google Scholar]
- Roe BA, Crabtree JS, Khan AS (1996) DNA Isolation and Sequencing (Essential Techniques Series). John Wiley & Sons, New York
- Rosati C, Cadic A, Duron M, Renou JP, Simoneau P (1997) Molecular cloning and expression analysis of dihydroflavonol 4-reductase gene in flower organs of Forsythia × intermedia. Plant Mol Biol 35: 303-311 [DOI] [PubMed] [Google Scholar]
- Ross JA, Kasum CM (2002) Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu Rev Nutr 22: 19-34 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Plainview, NY
- Scalbert A, Williamson G (2000) Dietary intake and bioavailability of polyphenols. J Nutr Suppl 130: 2073S-2085S [DOI] [PubMed] [Google Scholar]
- Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C (1994) Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol Biol 24: 743-755 [DOI] [PubMed] [Google Scholar]
- Stafford HA (1990) Pathway to proanthocyanindins (condensed tannins), flavan-3-ols, and unsubstituted flavans. In: HA Stafford, ed, Flavonoid Metabolism, CRC Press, Inc., New York, pp 63-99
- Stafford HA, Lester HH (1982) Enzymic and nonenzymic reduction of (+)-dihydroquercetin to its 3,4-diol. Plant Physiol 70: 695-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford HA, Lester HH (1984) The conversion of (+)-dihydroquercetin and flavan-3,4-cis-diol (leucocyanidin) to (+)-catechin by reductase extracted from cell suspension cultures of Douglas fir. Plant Physiol 76: 184-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford HA, Lester HH (1985) The conversion of (+)-dihydromyricetin to its flavan-3,4-diol (leucodelphinidin) and to (+)-gallocatechin by reductase extracted from tissue cultures of Ginkgo biloba and Pseudotsuga menziesii. Plant Physiol 78: 791-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stich K, Eidenberger T, Wurst F, Forkmann G (1992) Enzymatic conversion of dihydroflavonols to flavan-3,4-diols using flower extracts of Dianthus caryophyllus L. (carnation). Planta 187: 103-108 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278: 31647-31656 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Kristiansen KN (1993) Synthesis of 3,4-cis-[3H]leucocyanidin and enzymatic reduction to catechin. Anal Biochem 209: 274-277 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Moore AE, Larkin PJ (1994) Proanthocyanidins inhibit hydrolysis of leaf proteins by rumen microflora in vitro. Br J Nutr 71: 947-958 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337-1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B, Kubasek W, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659-671 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396-399 [DOI] [PubMed] [Google Scholar]