Abstract
The availability of nitrogen is a limiting factor for plant growth in most soils. Allantoin and its degradation derivatives are a group of soil heterocyclic nitrogen compounds that play an essential role in the assimilation, metabolism, transport, and storage of nitrogen in plants. Allantoinase is a key enzyme for biogenesis and degradation of these ureide compounds. Here, we describe the isolation of two functional allantoinase genes, AtALN (Arabidopsis allantoinase) and RpALN (Robinia pseudoacacia allantoinase), from Arabidopsis and black locust (Robinia pseudoacacia). The proteins encoded by those genes were predicted to have a signal peptide for the secretory pathway, which is consistent with earlier biochemical work that localized allantoinase activity to microbodies and endoplasmic reticulum (Hanks et al., 1981). Their functions were confirmed by genetic complementation of a yeast mutant (dal1) deficient in allantoin hydrolysis. The absence of nitrogen in the medium increased the expression of the genes. In Arabidopsis, the addition of allantoin to the medium as a sole source of nitrogen resulted in the up-regulation of the AtALN gene. The black locust gene (RpALN) was differentially regulated in cotyledons, axis, and hypocotyls during seed germination and seedling growth, but was not expressed in root tissues. In the trunk wood of a mature black locust tree, the RpALN gene was highly expressed in the bark/cambial region, but had no detectable expression in the sapwood or sapwood-heartwood transition zone. In addition, the gene expression in the bark/cambial region was up-regulated in spring and fall when compared with summer, suggesting its involvement in nitrogen mobilization.
Nitrogen is a key component of plant metabolism and its availability is often a limiting factor for plant growth in most soils. Heterocyclic nitrogen compounds (i.e. purines, pyrimidines, and their degradation products) represent major sources of soil organic nitrogen (Schulten and Schnitzer, 1998). Among these, allantoin (ALN) and its degradation product allantoic acid (ALA) are nitrogen-rich organic compounds with a C:N ratio of 1:1, and they play an essential role in the assimilation, metabolism, transport, and storage of nitrogen in plants (Schubert and Boland, 1990). In addition, they serve as effective carriers of the biologically fixed nitrogen in ureide-type legumes, and provide nitrogen storage with minimal expense of reduced carbon. For example, these compounds constitute 70% to 80% (w/v) of the organic nitrogen in the xylem sap of nodulated soybean (Glycine max; McClure and Israel, 1979). In a tree legume (black locust, Robinia pseudoacacia), about 3% (w/w) of xylem nitrogen was ureide (Atkins et al., 1991). Furthermore, ALN and ALA are central metabolites in the seasonal nitrogen cycle of perennial species such as maple (Acer spp.) and comfrey (Symphytum officinale; for review, see Schubert and Boland, 1990), indicating their key role in nitrogen storage and cycling. In legumes, the organic nitrogen fixed by soil bacteria (e.g. Rhizobium sp.) can be transported to the other parts of the plant as amides (Asn and Gln; amide-type legume) or as ureides (ALN and ALA; ureide-type legume).
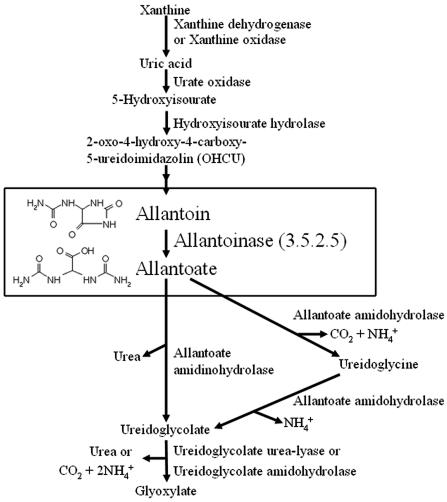
The major route for ALN biogenesis is the purine oxidation pathway (also called ureide pathway and ALN degradation pathway; Fig. 1). The first step in this pathway is the degradation of nucleic acid purine moieties (adenine and guanine) to uric acid. After two consecutive oxidation reactions by urate oxidase and hydroxyisourate hydrolase, the uric acid is converted to ALN (Raychaudhuri and Tipton, 2002). Allantoinase (ALN amidohydrolase, EC 3.5.2.5) catalyzes the hydrolysis of ALN to form allantoic acid, which is a key reaction for biogenesis and the degradation of ureides (Vogels et al., 1966; Noguchi et al., 1986). The resulting ALA is then further metabolized to ammonium, urea, and glyoxylate (Muñoz et al., 2001). Allantoate degradation can be catalyzed by allantoate amidohydrolase (EC 3.5.3.9) or allantoate amidinohydrolase (EC 3.5.3.4) to produce ureidoglycolate, which is metabolized to glyoxylate by ureidoglycolate urea-lyase (EC 4.3.2.3) or ureidoglycolate amidohydrolase (EC 3.5.3.19). Allantoate amidohydrolases and ureidoglycolate amidohydrolases generate ammonium, whereas allantoate amidinohydrolase and ureidoglycolate urea-lyase release urea. This pathway serves different roles and is evolutionarily distinct in plants, animals, and microorganisms. It is primarily used for salvage or excretion of nitrogen from purines in animals (Campbell and Bishop, 1970; Stryer, 1988). On the other hand, microorganisms use it to extract nitrogen from a variety of sources in the external environment (Cooper, 1980). Although its main function in animals is nitrogen excretion, many leguminous plant species use this pathway to store and recycle nitrogen. The pathway for ureide degradation in nonlegume plants has not been well documented. Recently, Desimone et al. (2002) demonstrated that Arabidopsis, a nonlegume model species, could take up and use ALN as a sole nitrogen source. This requires enzymatic degradation of the mobilized ALN inside the cells. The first step of the catabolism is catalyzed by allantoinase.
Figure 1.
Catabolism of ureide in plants.
Allantoinase is present in a wide variety of organisms, including animals, bacteria, fungi, and plants (for review, see Schubert and Boland, 1990). It is important for ureide biogenesis and degradation. Despite the long history of allantoinase study in plants (mainly legumes), our understanding of the physiological significance of the enzyme in plant growth and development is still limited. Molecular analysis of allantoinase genes is the first step in our approach to elucidate the relevance of ALN for plant nutrition. The genes encoding allantoinase have been cloned from yeast (Saccharomyces cerevisiae; Buckholz and Cooper, 1991) and bullfrog (Rana catesbeiana; Hayashi et al., 1994), but no plant allantoinase genes have been characterized yet. In this report, we isolated and characterized functional plant allantoinase genes from a tree legume (black locust, Robinia pseudoacacia) and Arabidopsis (nonlegume model species). The black locust allantoinase gene was discovered as a part of our trunk wood expressed sequence tag (EST) project, and the Arabidopsis gene was identified by a homologous search with the black locust gene. Their functions were confirmed by genetic complementation of a yeast mutant defective in allantoinase function. Differential expression of the genes was investigated with regard to the stage of development, seasonal variation, presence or absence of nitrogen, and different tissues.
RESULTS
Allantoinase Genes from Black Locust and Arabidopsis
In our previous EST analysis from the trunk wood of black locust (Yang et al., 2003), we found an EST that was homologous to other reported allantoinase genes such as those found in Escherichia coli (Kim et al., 2000), yeast (Buckholz and Cooper, 1991), and bullfrog (Hayashi et al., 1994). To obtain a full-length cDNA clone, we used 4- to 6-d-old seedlings to construct a cDNA library, and then screened them using the EST as a probe. The full-length cDNA, named RpALN, was comprised of 1,820 bp with an open reading frame of 1,536 bp, which encodes a 56,375-D protein with 512 amino acid residues. The protein was predicted by TargetP V1.0 (http://www.cbs.dtu.dk/services/TargetP/; Nielsen et al., 1997; Emanuelsson et al., 2000) to contain a signal peptide for the secretory pathway and a cleavage site at the 30th amino acid. Southern-blot analysis confirmed that the gene has a single copy in the black locust genome (data not shown).
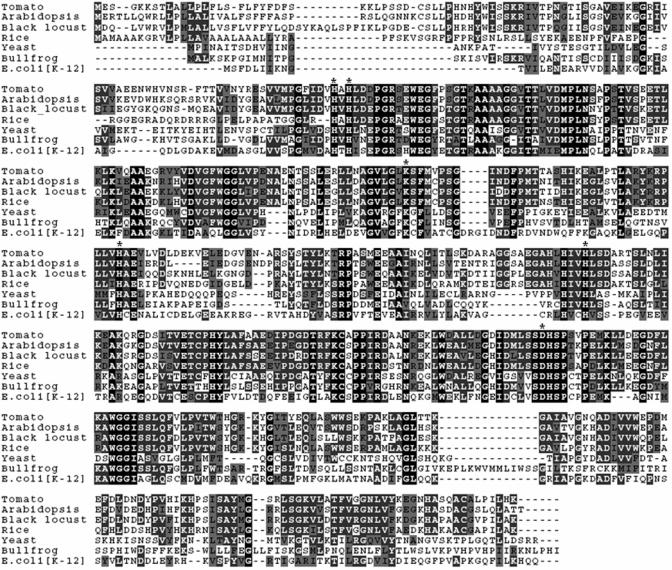
The RpALN sequence information was used to carry out BLASTN searches against public databases (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov and The Institute for Genomic Research [TIGR], http://www.tigr.org). We found an Arabidopsis homologous gene at the locus (accession no. AT4G04955). The BLASTX analysis with the RpALN amino acid sequence further confirmed its homology with an E value of 0.0 (BLASTX score = 670). We obtained a full-length cDNA clone (The Arabidopsis Resource [TAIR] clone no. 3989737) for this gene from TAIR (http://www.Arabidopsis.org). The size of the inserted gene was 1,552 bp with an open reading frame of 1,518 bp, encoding a protein with 506 amino acid residues and a calculated molecular mass of 55,438 D. Again, this protein was predicted to have a signal peptide for the secretory pathway and a cleavage site at the 26th amino acid. The Arabidopsis genome has only one copy of this gene. The deduced amino acid sequences of the RpALN and AtALN share highly conserved residues that are responsible for metal binding that were found in other reported ALN proteins (Fig. 2).
Figure 2.
Alignment of allantoinase amino acid sequences. Black locust putative allantoinase, Arabidopsis putative allantoinase, and tomato (Lycopersicon esculentum) putative allantoinase were compared with published allantoinases from E. coli, yeast, and bullfrog. Completely conserved amino acids in all species are marked in black; identical amino acids in dark gray; similar amino acids in light gray; and different amino acids in white. The conserved residues responsible for metal biding are indicated with asterisks.
Function of RpALN and AtALN
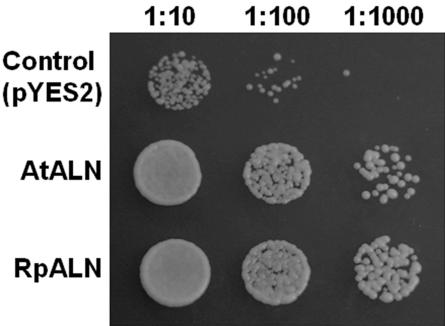
After cloning the RpALN and AtALN, we confirmed their functions via genetic complementation of a yeast mutant (dal1) deficient in ALN hydrolysis. This dal1 mutant grows slowly with ALN as a sole nitrogen source because no enzymatic degradation of ALN to urea can occur (Winkler et al., 1988). However, both transformants carrying RpALN or AtALN cDNA gene grew well (Fig. 3), suggesting the enzymes encoded by the plant genes could catalyze the hydrolysis of ALN to allantoic acid. Recently, Desimone et al. (2002) have shown that an Arabidopsis ureide permease (AtUPS1) could genetically complement an ALN uptake-deficient yeast mutant.
Figure 3.
Functional complementation of a yeast allantoinase mutant. The dal1 mutant was transformed with the empty vector (pYES2), with pYES2 carrying AtALN, or with pYES2 carrying RpALN. Three cell dilutions (1:10, 1:100, and 1:1,000) were plated on yeast minimal medium containing 0.1% (w/v) ALN as sole nitrogen sources and were incubated for 5 d at room temperature.
Arabidopsis Mutant ataln
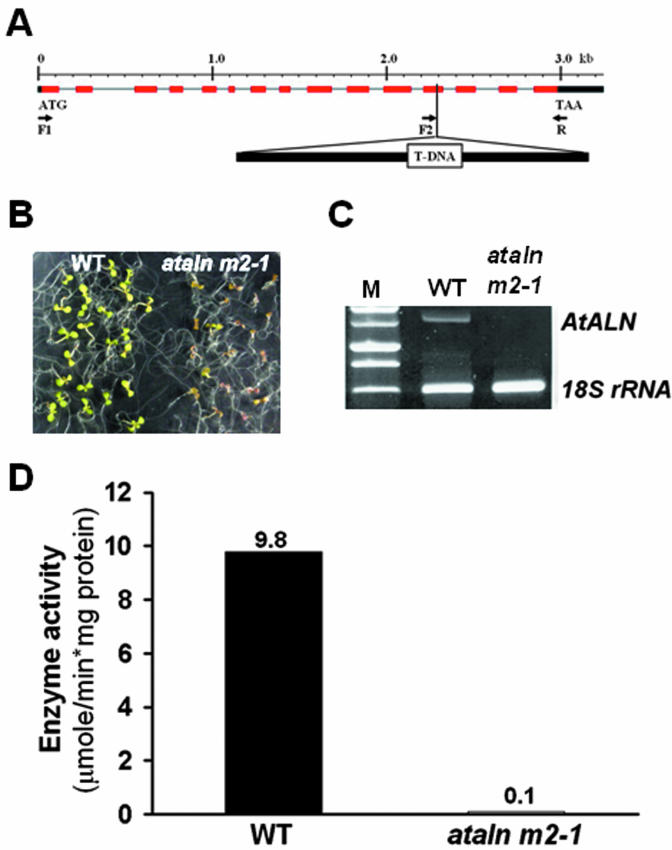
Consisting of 15 exons and 14 introns, the AtALN gene is located on chromosome 4 of the Arabidopsis genome (Fig. 4A). To examine the phenotypic consequence of the AtALN gene mutation, we screened T-DNA insertion mutants received from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/). First, single-copy T-DNA insertion was confirmed by 3:1 (resistant:sensitive) segregation in kanamycin selection. From the SALK-000327 line, we identified one mutant line (ataln m2-1) that did not grow well on Murashige and Skoog medium (Murashige and Skoog, 1962), which was composed of 10 mm ALN, instead of ammonium nitrate (Fig. 4B). We confirmed the T-DNA insertion in the Arabidopsis genome using PCR analysis with the gene-specific primer sets, which failed to detect stable AtALN transcripts in the mutant plants when compared with wild-type plants (Fig. 4, A and C). As expected, the progenies of the homozygous mutant line showed the mutant phenotype and kanamycin resistance with no segregation. No allantoinase activity was detected in the mutant plants. However, wild-type seedlings had an allantoinase activity of about 10 μmol min-1 mg-1 protein when cultured on ALN media (Fig. 4D). This level was higher than the activity of wild-type seedlings (1 μmol min-1 mg-1 protein) and the black locust seedlings (5 μmol min-1 mg-1 protein) cultured on Murashige and Skoog media, suggesting that the AtALN gene may be regulated by exogenous nitrogen conditions. In the presence of 10 mm ALN, the mutant seedlings were smaller when compared with the wild-type seedlings. Furthermore, the leaf of the mutant was colored pale green, and the mutant cotyledons showed very little red. Notably, wild-type seedlings and mutant seedlings that were subjected to poor nitrogen conditions (less than 10 μm ammonium nitrate) showed similar phenotypes. Regardless of the nitrogen levels in the media, the ataln mutant growth patterns did not differ from those of the wild-type seedlings previously reported by Desimone et al. (2002). However, when the mutant seedlings were transferred from the ALN media onto Murashige and Skoog media containing 10 mm ammonium nitrate, the seedlings recovered, leaf color returned to dark green, and the plants grew normally and produced flowers.
Figure 4.
Identification of T-DNA insertional mutants of Arabidopsis allantoinase. A, Diagram of AtALN genomic structure and site of T-DNA insertion in ataln as well as the position of gene-specific primers. Positions of introns (black line), exons (red box), and untranslated regions (black box) as defined in the wild-type gene from the Colombia ecotype are shown. The actual size of the T-DNA insertions is not known. Positions of primers used to confirm T-DNA insertion and/or to perform RT-PCR analysis are shown as arrows. B, Phenotype of ataln mutants grown with 10 mm ALN as a sole nitrogen source. Wild-type plants (WT) and ataln mutants (ataln m2-1) are shown at the age of 8 d after germination (DAG). C, Reverse transcriptase (RT)-PCR analysis of allantoinase gene expression. Total RNAs were prepared from 8-d-old wild-type plants (WT) and ataln mutants (ataln m2-1) grown with 10 mm ALN as a sole nitrogen source, and gene-specific primers (F1 and R) were used for amplifying AtALN mRNAs. The PCR product size of AtALN was 1.5 kb and the size of 18S RNA was 315 bp. D, Enzyme assay of allantoinase for wild-type and ataln mutant plants. Total proteins were extracted from the same samples shown as Figure 4B. Results represent means of two independent experiments.
Phylogenetic Analysis
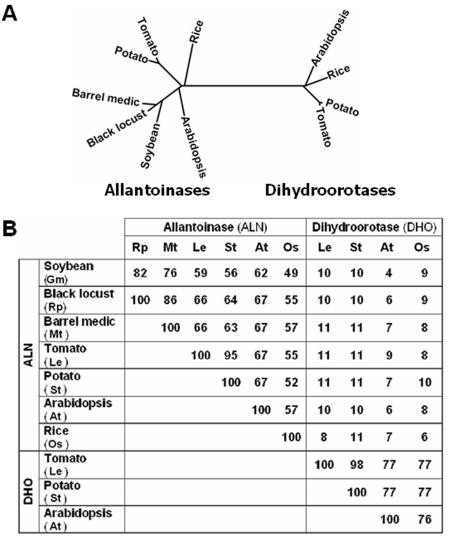
This Arabidopsis ALN (AtALN) has been annotated as a putative dihydroorotase (DHO) or DHO-like protein. Because ALN and DHO, along with hydantoinase and dihydropyrimidinase, belong to the same superfamily of cyclic amidohydrolases (EC 3.5.2.), this annotation is not surprising. Furthermore, many plant ALN gene sequences were not correctly annotated (mainly as “unknown” or “expressed protein”). Amino acid sequence alignment of RpALN identifies seven plant ALNs (Arabidopsis [Arabidopsis thaliana], barrel medic [Medicago truncatula], potato [Solanum tuberosum], rice [Oryza sativa], soybean [Glycine max], and tomato [Lycopersicon esculentum]) with high levels of amino acid sequence similarity (Fig. 5). To examine the structural relationship of ALNs and DHOs, we constructed an unrooted phylogenetic tree with the seven plant ALNs and four plant DHOs (Fig. 5). First, the analysis clearly showed that the two enzymes were unmistakably different at the amino acid level. The two enzyme groups (ALNs and DHOs) were distinctly separated in the phylogenetic tree, ALNs having very low similarity to DHOs (less than 12% similarity at the amino acid level). Within the same enzyme group, it was obvious that the proteins from the same family had the most structural similarity to each other. For instance, black locust ALN was most similar to that of soybean (GmALN; 82% similar) and barrel medic (MtALN; 86% similar), just as potato ALN was almost identical to the tomato ALN (95% similar). Furthermore, potato DHO (StDHO) was the most similar to tomato DHO (LeDHO; 98% similar).
Figure 5.
Dendrogram and similarity comparing the amino acid sequences of allantoinases and dihydroorotases. A, PHYLIP unrooted phylogenetic tree. Plant putative allantoinase are grouped at the left, and plant reported DHO are grouped at the right. B, Similarity between allantoinases and DHOs. The similarities of amino acid sequences were calculated by CLUSTALW using default parameters, and the phylogenetic tree was created by DRAWTREE (PHYLIP unrooted phylogenetic tree) using the entire sequence of each accession number. GenBank accession numbers: black locust ALN (AY466437), potato ALN (AB061260), Arabidopsis ALN (NM 116734; locus, AT4G04955), rice ALN (CAE05739), Arabidopsis DHO (NM 118422; locus, AT4G22930), rice DHO (NM 191198), and potato DHO (AX088927). TIGR tentative consensus sequences (TCs) number: soybean ALN (TC175849), barrel medic ALN (TC77627), tomato ALN (TC116684), and potato DHO (TC119412).
In this report, we confirmed the function of putative ALNs from two species (black locust and Arabidopsis) and we suggest that the six plant sequences, which are currently annotated as “unknown” proteins, are ALNs based on sequence homology and phylogenetic relationship. Allantoinase belongs to a superfamily of metal-containing amidohydrolases and is predicted to have a common three-dimensional fold (Holm and Sander, 1997). Especially, the conserved residues (Fig. 5, asterisks) responsible for metal biding were found in all reported ALNs. Recently, the recombinant E. coli ALN was shown to have the highest activity levels when observed in cell extracts from cultures supplemented with zinc and cobalt (Mulrooney and Hausinger, 2003).
RpALN Gene Expression
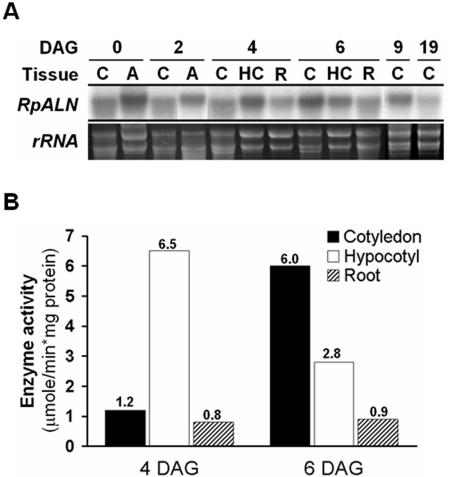
During seed germination and seedling growth, nitrogen metabolism is a critical factor for the survival of the tree in a natural environment. To determine the RpALN expression pattern during early development of black locust, northern-blot analysis was carried out using total RNAs isolated from cotyledons, axis, hypocotyls, and roots in different seed germination stages. In black locust, an axis emerges from the seed coat at 1 d postimbibition. The axis continues to grow and shows the line of root and shoot junction at 4 DAG. At 6 DAG, cotyledons open, become wrinkled, and true leaves sprout. In the axis and hypocotyls, the RpALN gene expression level peaked at 2 to 4 DAG, and then dropped at 6 DAG. On the other hand, its expression in cotyledon was the highest at 6 to 9 DAG (Fig. 6A) and reduced to the basal level at 19 DAG. No significant signal was detected in root tissues (4-6 DAG). The differential expression of the gene during the seed germination and seedling growth was also observed at the level of enzyme activity. The allantoinase activity was the highest in axis/hypocotyledon at 4 DAG and in cotyledon at 6 DAG (Fig. 6B).
Figure 6.
Differential expression of black locust allantoinase during the seed germination and seedling growth. A, Northern-blot analyses of RpALN in black locust seedlings. RpALN mRNA is differentially expressed in seedling tissues. According to the number of DAG, total RNA (10 μg) from the indicated tissues was electrophoresed and blotted. The RNA was then probed with [α32P]dCTP-labeled RpALN cDNA. Each bottom panel presents the corresponding agarose gel stained with ethidium bromide, showing ribosomal RNAs. B, Enzyme assay of allantoinase of seedling tissues. Total proteins were extracted from three tissues of 4- or 6-d-old or seedlings: cotyledon (black bar), hypocotyl (white bar), and root (hatched bar). Results represent means of two independent experiments.
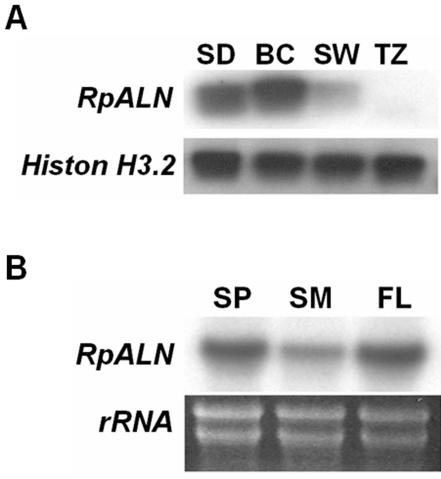
Trunk wood serves as a conduit for long-distance transport of nutrient and represents one of the largest sink tissues in trees. The expression pattern of RpALN gene was examined in the bark/cambial zone, sapwood, and sapwood-heartwood transition zones of mature black locust trees (Fig. 7). The bark/cambial zone is the outer most region, the sapwood is the bright-colored middle region, and the heartwood is the dark, inner region. The transition zone, the interphase between sapwood and heartwood, is where the carbohydrates are converted to heartwood extractives. Because most of the cells in trunk wood are dead, the quantity and quality of mRNA that can be extracted from such tissue is limited. To overcome this problem, we used antisense northern-blot analysis, which included an in vitro transcription process to amplify mRNAs (Yang et al., 2003). Our results showed that the bark/cambial region had the highest expression level (Fig. 7A). We then examined the seasonal regulation of RpALN gene expression using bark/cambial zone tissues collected in spring, summer, and fall. In temperate deciduous trees, leaf nitrogen is translocated to perennial tissues (e.g. trunk wood) during the fall and the stored nitrogen is remobilized for spring shoot growth (Taylor and May, 1967; Ryan and Bormann, 1982). We found that the expression level of RpALN gene was higher in the spring and fall samples compared with that of the summer sample (Fig. 7B), suggesting that the RpALN gene may be involved in nitrogen resorption and storage during the fall and resupply during the spring growth.
Figure 7.
RpALN gene expression in trunk wood of black locust. A, Antisense northern-blot analysis of RpALN mRNA in seedling (SD), bark/cambial zone (BC), sapwood (SW), and transition zone (TZ) by antisense northern-blot analysis. Histon H 3.2 transcript levels were indicated as a control. Each sample lane contained an equal amount of 200 ng of aRNAs. B, Northern-blot analysis of RpALN mRNA using three different seasons of trunk wood. RpALN mRNA is differentially expressed in bark/cambial zones according to spring (SP), summer (SM), and fall (FL). Total RNA (10 μg) from the indicated season samples was separated and blotted, and the mRNA was then detected with [α32P]dCTP-labeled RpALN cDNA. Bottom panel presents the corresponding agarose gel stained with ethidium bromide, showing ribosomal RNAs.
Effects of Nitrogen on RpALN and AtALN Gene Expression
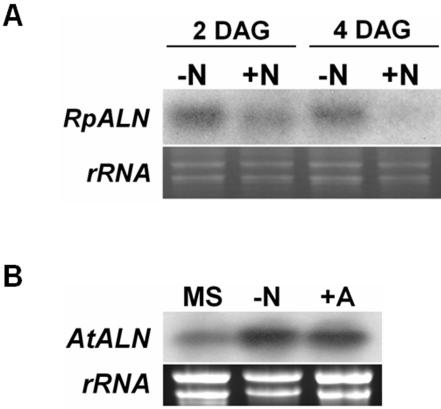
To test whether nitrogen affects allantoinase gene expression, we carried out northern-blot analysis using black locust and Arabidopsis seedlings that were cultured on Murashige and Skoog medium supplemented with or without nitrogen. The absence of nitrogen in the medium appears to induce the expression of RpALN gene (Fig. 8A) during seedling germination and growth (2 and 4 DAG). In Arabidopsis seedlings, the absence of nitrogen or the presence of ALN as a sole nitrogen source dramatically increased AtALN gene expression (Fig. 8B).
Figure 8.
Nitrogen influence on the expression of allantoinase genes. A, Expression of RpALN mRNA under nitrogen conditions. Total RNAs were isolated from 2- or 4-d-old seedlings grown under nitrogen absent condition (-N) or supplied with 10 mm ammonium nitrate (+N). B, Northern-blot analysis of AtALN mRNA in different nitrogen conditions. Total RNAs were isolated from 8-d-old seedlings cultured on Murashige and Skoog medium supplemented with nitrogen (MS), without nitrogen (-N), or with 10 mm ALN instead of the nitrogen sources (+A). Total RNA (10 μg) from the indicated season samples was separated and blotted, and the mRNA was then detected with [α32P]dCTP-labeled RpALN or AtALN cDNA. Each bottom panel presents the corresponding agarose gel stained with ethidium bromide, showing ribosomal RNAs.
DISCUSSION
Biochemical analyses have shown that ureides such as ALN and ALA are present in higher plants, including ureide-type legume species such as soybean (Tracey, 1961; Atkins et al., 1991; Cheng et al., 2000). Desimone et al. (2002) demonstrated that even nonlegume Arabidopsis can take up and use ALN as a nitrogen source. Here, we cloned and characterized two full-length cDNA clones (AtALN and RpALN) encoding allantoinase, further confirming that the ALN degradation pathway exists in Arabidopsis as well as in amide-type legumes (e.g. black locust). To our best knowledge, this is the first report on the cloning of functional allantoinase genes in plants. Using these two ALN gene sequences in a BLAST search against public databases, we identified additional ALN genes from various plant species including barrel medic, potato, soybean, potato, rice, and tomato. Multiple sequence alignment showed that all of the ALNs share high sequence similarity with known ALNs from other nonplant organisms (bullfrog, E. coli, and yeast), and identified characteristic conserved residues for metal binding. On the other hand, their sequence similarity with plant DHOs was very low (≤11%). Plant ALN genes appear to be expressed abundantly in various tissues, suggesting that the ureide pathway is universal among plant species. In fact, Desimone et al. (2002) demonstrated that Arabidopsis could uptake and use ALN as a sole nitrogen source. The BLAST search against EST databases and the rice genome sequence database on the TIGR identified 15 ESTs from developing stems, developing leaves, drought-treated plants of barrel medic, 12 ESTs from dormant tuber, mature tuber, leaves/petioles of potato, and 31 ESTs from most tomato organs. Other plant species for which ALN genes were identified include rice, cotton (Gossypium hirsutum), soybean, and barley (Hordeum vulgare).
Until now, plant allantoinase genes have been mis-annotated as putative, unknown, or DHO. Particularly, the current Arabidopsis genome annotation calls AtALN a putative DHO or DHO-like protein. At least three pieces of evidence confirm that the RpALN and AtALN genes encode allantoinase. First, both of the genes were able to genetically complement a yeast mutant defective in ALN hydrolysis. Second, an Arabidopsis mutant (ataln m2-1) with T-DNA insertion in the 12th exon of AtALN was not able to use ALN as sole source of nitrogen, whereas wild-type plants grew on ALN medium. Third, a phylogenetic tree constructed with seven plant ALNs and four plant DHOs showed that the two enzyme groups were unmistakably different at the amino acid level.
Seed germination and seedling growth are the developmental stages that can affect the survival of the plant in its natural ecosystem. Developmental and metabolic programs are accomplished during the early stage of plant life (Howell, 1998; Holdsworth et al., 1999; Eastmond et al., 2000). Different metabolic pathways are carried out in each tissue of the seedling during its growth and development. For example, a nutrient source tissue (e.g. cotyledon) performs metabolisms that generate nutrients transported to sink tissues, whereas a sink source tissue (e.g. axis) conducts metabolisms involved in cell growth. Thus, an axis cell likely demands more nitrogen and carbon than does a cotyledon cell. It is necessary even for the same metabolism to be differentially controlled depending on the tissues involved. Enzyme activities required for the production of ureides were observed in young soybean seedlings (Polayes and Schubert, 1984). Our results showed that black locust seedlings differentially express allantoinase genes depending on tissue type and developmental stages. Although these findings suggest that allantoinase may be differentially regulated according to the nitrogen demand, it is not known whether the allantoinase degradation pathway plays any nutritional role in seedling development and growth.
Many plant purine biosynthetic enzymes appear to be localized in organelles (Smith and Atkins, 2002). Previous reports indicate that allantoinase is localized in microbodies of plants and animals (for review, see Schubert and Borland, 1990). Hanks et al. (1981) reported that allantoinase was localized in the endoplasmic reticulum (ER). It has been suggested that this ER localization may play a role in the export of ALA from the cell. However, the ER localization of allantoinase has not been independently confirmed. In this regard, our finding that both of the two plant allantoinases have putative transit sequences is interesting. However, further protein localization studies, using green fluorescent protein (GFP) fusion proteins in transgenic plants, can provide direct evidence for the intracellular localization of allantoinase. The cloning of the two functional ALN genes can facilitate such studies.
Signals derived from nitrogen treatment or nitrogen shortage are certainly involved in triggering widespread changes in gene expression and metabolic pathways. Cooper et al. (1990) have shown that in yeast, the ureide pathway is transcriptionally up-regulated under the nitrogen-deficient conditions. It appears that allantoinase genes are down-regulated in nitrogen-rich conditions, but up-regulated in nitrogen-limiting conditions. For example, the yeast allantoinase gene (dal1) was up-regulated by nitrogen depletion as well as amino acid starvation (Gasch et al., 2000; also web supplement at http://genome-www.stanford.edu/yeaststress/index.shtml). In Arabidopsis, cDNA microarray data available from the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD/) showed that the ALN gene is down-regulated by nitrogen treatment, potassium nitrate versus no treatment (experiment ID nos. 3787 and 3789), the potassium chloride versus ammonium chloride (experiment ID nos. 12308 and 12309), and the potassium chloride versus potassium nitrate (experiment ID nos. 10849 and 10851). In addition, previous studies showed that nitrate treatment decreased the nodule mass and nitrogen-fixing activity, which subsequently resulted in the reduction of ureide content in the xylem sap in soybean (Israel and McClure, 1980; McNeil and LaRue, 1984) and cowpea (Vigna unguiculata; Pate et al., 1980) plants. Our data showing nitrogen-induced down-regulation of ALN gene expression confirm these earlier physiological and biochemical observations.
In addition to the developmental regulation of ALN, a differential gene expression among different tissue types and seasonal variation might offer some insight on nitrogen metabolism in perennial woody plant species. In the current study, we found that black locust ALN genes had the highest expression levels in bark/cambium tissue compared with sapwood, sapwood-heartwood transition zone, or seedlings. In the fall, nitrogen in the leaves is mobilized and transported to storage tissues such as bark and xylem ray cells (Kang and Titus, 1980; Chapin and Kedrowswki, 1983). The stored nitrogen is then remobilized to support the spring growth (Taylor and May, 1967; Ryan and Bormann, 1982). It is notable that the RpALN gene expression was up-regulated in spring and fall compared with summer, again supporting the previous physiological observations. This seasonal differential expression of RpALN gene strongly suggests that RpALN may be involved in the seasonal cycle of nitrogen resorption, storage, and recycling in temperate deciduous trees. Furthermore, the cloning of two plant ALN genes will facilitate further studies on the molecular and biochemical basis for nitrogen uptake, metabolism, and recycling in plants.
MATERIALS AND METHODS
Plant Materials and Culture Conditions
Arabidopsis (Colombia ecotypes) and black locust (Robinia pseudoacacia) seedlings were grown in petri dishes under light conditions (16 h of light day-1) at 25°C. For black locust seed germination and seedling growth, seeds were heated in boiling water for 1 min, soaked in sterile water for 1 h, and transferred into glass jars containing 0.7% (w/v) agar (without nitrogen) or 0.7% (w/v) agar plus 10 mm ammonium nitrate (with nitrogen). For Arabidopsis plants, seeds were subjected to cold treatments for 3 d in sterile water, and then were transferred onto Murashige and Skoog (1962) medium with 2% (w/v) Glc and 10 mm ammonium nitrate or 10 mm ALN (ALN medium) as a nitrogen source. All procedures were performed under sterile conditions.
Cloning, Multiple Alignment, and Phylogenetic Analysis
We used the cDNA library prepared from the 4- to 6-d-old seedlings of black locust to obtain the full-length cDNA clone of black locust allantoinase. About 100,000 plaques were screened with the partial cDNA clone (GenBank accession no. BI643000) and were labeled with [α-32P]dCTP. Five positive phages were randomly selected and inserts were recovered as pBluescript SK phagemids according to the manufacture's instruction (Stratagene, Cedar Creek, TX). All of them were sequenced and shown to be identical clones. We used the sequenced black locust full-length cDNA clone to search for other plant homologous clones on the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the TIGR (http://www.tigr.org) sequence databases. The full-length cDNA clone of Arabidopsis allantoinase gene (TAIR clone no. 3989737) was obtained from TAIR. The multiple alignment of amino acid sequences was carried out by CLUSTALW (Thompson et al., 1994) using default parameters, and the phylogenetic tree was created by DRAWTREE (PHYLIP unrooted phylogenetic tree, Felsenstein, 1989).
Identification of ataln T-DNA Insertional Mutant
Seven T-DNA insertion mutants (SALK-000325, SALK-000327, SALK-000282, SALK-013427, SALK-013985, SALK-013986, and SALK-142607) of the AtALN gene generated by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/) were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The seeds were planted on kanamycin plates and the resulting kanamycin-resistant plants were transferred to soil and seeds were harvested separately from individual plants. Subsequently, the seeds were screened on Murashige and Skoog media supplied with 10 mm ALN as a sole nitrogen source. After an additional phenotypic selection, we randomly chose one mutant line (ataln m2-1) that did not grow well on ALN media for further analyses. To confirm the mutant line as a homozygote, PCR was performed with the genomic DNA of ataln m2-1 mutants using gene-specific oligonucleotides (forward1, 5′-AGAGATATGGAGAGAACTTTGCTT-3′; forward2, 5′-GGAAGGAGACATTGATATGCTGAG; reverse, 5′-AAGTTAAGTAGTTGCAAGTTGCAG-3′) or one gene-specific primer and one left-border-specific primer. As expected, the progenies of the homozygous mutant line showed the mutant phenotype and kanamycin resistance with no segregation. The mutant line did not have any noticeable morphological alterations or growth defects under the normal condition.
RNA Isolation
Using the Trizol reagent (Invitrogen, Carlsbad, CA), total RNAs were isolated from seedlings of Arabidopsis and black locust. RNA extraction from inner wood was performed as previously described (Yang et al., 2003). Briefly, mature trees (20 cm diameter at breast height) were felled using a chain saw and were made into 25-cm-long logs. The logs were immediately placed on ice and brought back to a wood shop, where thin cookies (approximately 1 cm thick) were made using a table saw. The cookies were immediately submerged in RNA extraction buffer (20 mm EDTA, pH 8.0, 50 mm Tris-HCl, pH 8.0, 0.2% [w/v] SDS, and 10 mm β-mercaptoethanol). While submerged, the trunk wood sections (bark/cambial region, sapwood, and transition zone) were carved out by using a chisel and hammer, and were washed with RNase Away solution (Invitrogen). The isolated sections (approximately 1-cm3 cubicles) were frozen in liquid nitrogen and stored at -80°C until needed. For RNA isolation, the frozen samples were first ground in a blender and then further ground to a fine powder using a mortar and pestle. The grounded sample was first passed through the shredder column of DNeasy Maxi kit (Qiagen, Hilden, Germany), and was then subjected to total RNA isolation using the RNeasy Maxi kit (Qiagen) and cleaned up by RNeasy Mini kit (Qiagen).
Northern Blots
Total RNA (5 or 10 μg) was separated on a 1.2% (w/v) agarose gel containing 1× MOPS buffer with 2.2 m formaldehyde at 60 V for 3.5 h. The RNA was then washed twice in water and transferred by capillary action to a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Piscataway, NJ). Membranes were prehybridized for 2 h at 42°C in 50% (v/v) formamide, 6× SSC, 5× Denhardt's solution, 0.5% (w/v) SDS, and 20 μg mL-1 sonicated salmon sperm DNA. Hybridizations were performed overnight at 65°C after adding the randomly primed 32P-labeled cDNA probe in the prehybridization buffer. Membranes were washed at 65°C for 10 min each with washing solution I (2.0× SSC and 0.1% [w/v] SDS), washing solution II (0.5× SSC and 0.1% [w/v] SDS), or washing solution III (0.1× SSC and 0.1% [w/v] SDS). Random-labeled probes were made with a kit (Rediprime II; Amersham Pharmacia Biotech) using an internal fragment from the full-length cDNA of AtALN and RpALN1.
RT-PCR
One microgram of total RNA, isolated from wild-type and mutant plants, was reverse transcribed using a random primer in a 20-μL reverse transcription reaction as recommended by the manufacturer (SuperScript II reverse transcriptase; Invitrogen). One microliter of the first-strand cDNA reaction was used as a template for PCR amplification with specific oligo-nucleotide primers for AtALN (forward, 5′-AGAGATATGGAGAGAACTTTGCTT-3′; reverse, 5′-AAGTTAAGTAGTTGCAAGTTGCAG-3′). As an internal control, plant 18s primers (Ambion, Austin, TX) were used. The PCR products were loaded on ethidium bromide-stained agarose gels.
Antisense Northern Blot
Antisense northern-blot analysis was performed as previously described (Yang et al., 2003). For the generation of antisense RNAs, total RNAs (2 μg) were amplified using Message AmpTM aRNA kit (Ambion). We began by synthesizing first stand cDNAs from the total RNAs of seedling, bark/cambial region, sapwood, or transition zone, and the first cDNAs were used as templates for the synthesis of second cDNAs. Finally, aRNAs from the second cDNAs were generated by in vitro transcription (amplification). About 200 ng of aRNAs was separated in a formamide agarose gel and transferred onto the nylon membrane using the capillary transfer method. The membrane was then hybridized with an isotope-labeled probe. The signal was exposed and detected on an x-ray film. Histon 3.2, which showed constitutive expression in black locust tissues (Yang et al., 2003), was used as a control.
Genetic Complementation of Yeast (Saccharomyces cerevisiae) Mutant with RpALN and AtALN Genes
To conduct the functional complementation test of RpALN and AtALN in yeast, we found the yeast allantoinase (DAL1) gene deletion mutant on the Saccharomyces Genome Deletion Project web page (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html), and bought the homozygous diploid strain of dal1 deletion mutant (item no. 4035962) from American Type Culture Collection (Manassas, VA). The homozygous diploid of the mutant is in the background of BY4743 (BY4741: MATa his3δ1 leu2δ0 met15δ0 ura3δ0/BY4742: MATα his3δ1 leu2δ0 lys2δ0 ura3δ0). The deficiency of the mutant, using ALN as a sole nitrogen source, was determined on solid plates containing different concentrations of ALN as sole nitrogen sources. Optimal selective conditions were found at 2 g L-1. The dal1 mutant was transformed with an expression vector pYES2 (Invitrogen) harboring a full-length cDNA of RpALN or AtALN gene. PCR products generated from the full-length cDNA of RpALN or AtALN gene were digested by BamHI and XbaI and ligated into the BamHI-XbaI site of plasmid pYES2. At the first, transformants were selected on solid plates that were composed of a yeast nitrogen base, 2% (w/v) dextrose, 0.5% (w/v) ammonium sulfate, and a synthetic complete supplement mixture minus uracil (Qbiogene, Carlsbad, CA). RpALN or AtALN transformants were then confirmed by PCR analysis using gene-specific primers. After uracil selection, each transformant was reselected on a solid plate containing yeast nitrogen base, 2% (w/v) Gal, 1% (w/v) raffinose, and 0.2% (w/v) ALN (Sigma, St. Louis) as a nitrogen source, and 1/100-fold reduced synthetic complete supplement mixture minus uracil to supply Leu and His. The colonies capable of growing were reselected under the same conditions. As a negative control, the mutant was transformed with the empty vector. After a single colony from each transformed mutant was chosen, growth complementation tests were performed using ALN as a sole nitrogen source (Desimone et al., 2002).
Protein Extraction and Enzyme Assay
Crude extracts for ALN assays of RpALN and AtALN were prepared according to the method described by Bell and Webb (1995) with slight modification. Plant samples (0.5-1.0 g) were ground with 4× volumes of 20 mm Tris-HCl and 500 mm NaCl, pH 7.5, 100 μm dithiothreitol, 10 mm EDTA, 0.1% (w/v) polyvinyl-polypyrrolidone, and a mixture of protease inhibitors, including phenylmethylsulfonyl fluoride (2 mm), aprotini (10 μg mL-1), leupeptin (10 μg mL-1), and pepstatin (10 μg mL-1) in a chilled mortar, with a pestle. Extracts were centrifuged at 30,000g for 30 min at 4°C and were filtered through two layers of Miracloth (Calbiochem, San Diego), and the supernatant was collected. Then, the supernatant was filtered through 0.2-μm filters (Millipore, Bedford, MA) to remove particulate matter. Extracts were kept on ice or -20°C until they were used.
The assay for allantoinase activity was based on the procedure reported by Schubert (1981) and Romanov et al. (1999). The enzyme extract was incubated for 20 min at room temperature with 10 mm ALN in 50 mm Tris-HCl buffer. The pH was set to 7.5. Reaction solution (0.5 mL) was then treated with 0.125 mL of 0.2 n HCl, and was heated in a boiling water bath for 4 min to degrade ALA to glyoxylate and urea. After cooling for 30 s, 0.4 mL of concentrated HCl and 0.125 mL of potassium ferricyanide solution (16 mg mL-1) were added and incubated at 37°C for 30 min. The total reaction solution was immediately applied to measure the A520 arising from the final product of this reaction, glyoxylate diphenylformazan. All assays were replicated at least twice, and controls were included to account for endogenous ALA in the enzyme extract and nonenzymatic degradation of ALN.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034637.
This work was supported by the U.S. Department of Agriculture (grants to the Eastern Hardwood Utilization Program at Michigan State University, nos. 98-34158-5995, 00-34158-9236, and 01-34158-11222).
References
- Atkins CA, Storer PJ, Young EB (1991) Translocation of nitrogen and expression of nodule-specific uricase (nodulin-35) in Robinia pseudoacacia. Physiol Plant 83: 483-491 [Google Scholar]
- Bell JA, Webb MA (1995) Immunoaffinity purification and comparison of allantoinases from soybean root nodules and cotyledons. Plant Physiol 107: 435-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholz RG, Cooper TG (1991) The allantoinase (DAL1) gene of Saccharomyces cerevisiae. Yeast 7: 913-923 [DOI] [PubMed] [Google Scholar]
- Campbell JW, Bishop SH (1970) Nitrogen metabolism in molluscs. In JW Campbell, ed, Comparative Biochemistry of Nitrogen Metabolism, Vol 1. Academic Press, London, pp 103-206 [Google Scholar]
- Chapin FS, Kedrowski RA (1983) Seasonal changes in nitrogen and phosphorus fractions and autumn retranslocation in evergreen and deciduous taiga trees. Ecology 64: 376-391 [Google Scholar]
- Cheng XG, Nomura M, Takane K, Kouchi H, Tajima S (2000) Expression of two uricase (Nodulin-35) genes in a non-ureide type legume, Medicago sativa. Plant Cell Physiol 41: 104-109 [DOI] [PubMed] [Google Scholar]
- Cooper TG (1980) Selective gene expression and intracellular compartment: two means of regulating nitrogen metabolism in yeast. Trends Biochem Sci 5: 332-334 [Google Scholar]
- Cooper TG, Ferguson D, Rai R, Bysani N (1990) The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol 172: 1014-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimone M, Catoni E, Ludewig U, Hilpert M, Schneider A, Kunze R, Tegeder M, Frommer WB, Schumacher K (2002) A novel superfamily of transporters for allantoin and other oxo derivatives of nitrogen heterocyclic compounds in Arabidopsis. Plant Cell 14: 847-856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA (2000) Post-germinative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 10: 5669-5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005-1016 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5: 164-166 [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241-4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks JF, Tolbert NE, Schubert KR (1981) Localization of enzymes of ureide biosynthesis in peroxisomes and microsomes of nodules. Plant Physiol 68: 65-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Jain S, Chu R, Alvares K, Xu B, Erfurth F, Usuda N, Rao MS, Reddy SK, Noguchi T et al. (1994) Amphibian allantoinase: molecular cloning, tissue distribution, and functional expression. J Biol Chem 269: 12269-12276 [PubMed] [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R (1999) Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Pharmacol Sci 4: 275-280 [Google Scholar]
- Holm L, Sander C (1997) An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28: 72-82 [PubMed] [Google Scholar]
- Howell SH (1998) Seedling development. In Molecular Genetics of Plant Development. Cambridge University Press, Cambridge, UK, pp 83-102
- Israel DW, McClure PR (1980) Nitrogen translocation in the xylem of soybeans. In FT Corbin, ed, World Soybean Research Conference II: Proceedings. Westview Press, Boulder, CO, pp 111-127
- Kang SM, Titus JS (1980) Qualitative and quantitative changes in nitrogenous compounds in senescing leaf and bark tissues of apple. Physiol Plant 50: 285-290 [Google Scholar]
- Kim GJ, Lee DE, Kim HS (2000) Functional expression and characterization of the two cyclic amidohydrolase enzymes, allantoinase and a novel phenylhydantoinase, from Escherichia coli. J Bacteriol 182: 7021-7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure PR, Israel DW (1979) Transport of nitrogen in the xylem of soybean plants. Plant Physiol 64: 411-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil DL, LaRue TA (1984) Effect of nitrogen source on ureides in soybean. Plant Physiol 74: 227-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney SB, Hausinger RP (2003) Metal ion dependence of recombinant Escherichia coli allantoinase. J Bacteriol 185: 126-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz A, Piedras P, Aguilar M, Pineda M (2001) Urea is a product of ureidoglycolate degradation in chickpea: purification and characterization of the ureidoglycolate urea-lyase. Plant Physiol 125: 828-834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473-479 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1-6 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujiwara S, Hayashi S (1986) Evolution of allantoinase and allantoicase involved in urate degradation in liver peroxisomes: a rapid purification of amphibian allantoinase and allantoicase complex, its subunit locations of the 2 enzymes, and its comparison with fish allantoinase and allantoicase. J Biol Chem 261: 4221-4223 [PubMed] [Google Scholar]
- Pate JS, Atkins CA, White ST, Rainbird RM, Woo KC (1980) Nitrogen nutrition and xylem transport of nitrogen in ureide-producing grain legumes. Plant Physiol 65: 961-965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polayes DA, Schubert KR (1984) Purine synthesis and catabolism in soybean seedlings: the biogenesis of ureides. Plant Physiol 75: 1104-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri A, Tipton PT (2002) Cloning and expression of the gene for soybean hydroxyisourate hydrolase: localization and implications for function and mechanism. Plant Physiol 130: 2061-2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov V, Merski MT, Hausinger RP (1999) Assays for allantoinase. Anal Biochem 268: 49-53 [DOI] [PubMed] [Google Scholar]
- Ryan DF, Bormann FH (1982) Nutrient resorption in northern hardwood forests. BioScience 32: 29-32 [Google Scholar]
- Schubert KR (1981) Enzymes of purine biosynthesis and catabolism in Glycine max. Plant Physiol 68: 1115-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert KR, Boland MJ (1990) The ureides. In BJ Miflin, PJ Lea, eds, The Biochemistry of Plants A Comprehensive Treatise, Vol 16: Intermediary Nitrogen Metabolism. Academic Press, San Diego, pp 197-282
- Schulten HR, Schnitzer M (1998) The chemistry of soil organic nitrogen: a review. Biol Fertil Soils 26: 1-15 [Google Scholar]
- Smith PMC, Atkins CA (2002) Purine biosynthesis: big in cell division, even bigger in nitrogen assimilation. Plant Physiol 128: 793-802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L (1988) Biochemistry, Ed 3. WH Freeman, New York, pp 618-623
- Taylor BK, May LH (1967) Nitrogen metabolism, translocation and recycling in apple trees. Hort Rev 4: 204-246 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey MV (1961) Urea and ureides. In K Peach, M Tracey, eds, Modern Methods of Plant Analysis, Vol 4. Springer-Verlag, Heidelberg, pp 119-141 [Google Scholar]
- Vogels GD, Trijbels F, Uffink A (1966) Allantoinases from bacterial, plant and animal sources: purification and enzymatic properties. Biochim Biophys Acta 122: 482-496 [Google Scholar]
- Winkler RG, Blevins DG, Polacco JC, Randall DD (1988) Ureide catabolism in nitrogen-fixing legumes. Trends Biochem Sci 13: 97-100 [DOI] [PubMed] [Google Scholar]
- Yang J, Park S, Kamdem DP, Keathley DE, Retzel E, Paule C, Kapur V, Han K-H (2003) Novel gene expression profiles define the metabolic and physiological processes characteristic of wood and its extractive formation in a hardwood tree species, Robinia pseudoacacia. Plant Mol Biol 52: 935-956 [DOI] [PubMed] [Google Scholar]