Abstract
Chromosome segregation is an essential cellular function in eukaryotic and prokaryotic cells. The ParABS system is a fundamental player for a mitosis-like process in chromosome partitioning in many bacterial species. This work shows that the social bacterium Myxococcus xanthus also uses the ParABS system for chromosome segregation. Its large prokaryotic genome of 9.1 Mb contains 22 parS sequences near the origin of replication, and it is shown here that M. xanthus ParB binds preferentially to a consensus parS sequence in vitro. ParB and ParA are essential for cell viability in M. xanthus as in Caulobacter crescentus, but unlike in many other bacteria. Absence of ParB results in anucleate cells, chromosome segregation defects and loss of viability. Analysis of ParA subcellular localization shows that it clusters at the poles in all cells, and in some, in the DNA-free cell division plane between two chromosomal DNA masses. This ParA localization pattern depends on ParB but not on FtsZ. ParB inhibits the nonspecific interaction of ParA with DNA, and ParA colocalizes with chromosomal DNA only when ParB is depleted. The subcellular localization of ParB suggests a single ParB-parS complex localized at the edge of the nucleoid, next to a polar ParA cluster, with a second ParB-parS complex migrating after the replication of parS takes place to the opposite nucleoid edge, next to the other polar ParA cluster.
Introduction
Genetic information is written in long DNA molecules. A typical bacterial chromosome extends to a length over a thousand times greater than the cell in which it resides. Therefore, chromosomal DNA organization, its transcription, replication, and segregation must be highly organized in the cytoplasm and tightly coordinated in time [1], [2]. Negative DNA supercoiling is the main mechanism for bacterial chromosome compaction, generating topological domains of about 10 kb, the interwound DNA loops and their boundaries being highly dynamic [3]. In Escherichia coli, a higher-order structure of chromosomal DNA (or macrodomain) has been described, with a length between 0.8 to 1 Mb, which is organized by MatP protein and multiple matS DNA sequences [4]. In Bacillus subtilis, the nucleoid adopts an organization where the origins of chromosomal replication (oriC) are located near opposite cell poles and termini (ter) at the mid-cell [5]. In Pseudomonas aeruginosa, the oriC-ter axis is oriented from the old pole of the cell to the cell division plane or to the incipient newborn pole [6]. And in a newly divided Caulobacter crescentus cell, loci occupy specific regions in the cytoplasmic space with respect to its linear genomic position, being oriC at the old cell pole and ter at the newborn pole [7]. Unlike eukaryotic cells, chromosome segregation is coupled to chromosome replication, and loci separate progressively just after being replicated [7]–[9]. Models for chromosome segregation without mitotic-like apparatus have been proposed [10]–[13]. However, it is assumed that a chromosomal ParABS system, originally described in plasmids, acts as a mitotic-like apparatus to segregate replicated chromosomes [1], [2], [14]. ParABS systems, which have been identified in over two hundred bacterial chromosomes, consist of three components. One is the cis-acting parS site that is highly conserved among bacterial species, and is located in the oriC-proximal region of the chromosome. The majority of bacterial species have between one and four repeats of putative parS sites, although several with five to eight repeats, and even a few with twenty or more, are known [15]. The second component of ParABS systems, protein ParB, binds to the parS sites to form a large nucleoprotein complex near oriC as well as to the third component of the system, ParA. The latter is an ATPase proposed as the element that provides the force for the segregation of the “centromeric” parS sites via dynamic polymerization-depolymerization events [2], [14], [16]–[18]. Genes encoding parA and parB also are usually found in the oriC-proximal regions of the chromosome, and they have been shown to participate in proper chromosome partitioning in numerous bacteria [15], [19].
Myxococcus xanthus is a Gram-negative soil δ-proteobacterium used as a prokaryotic model for the investigation of several processes involved in multicellular development, coordinated cell movements, and cellular responses to external signals such as light [20]–[23]. M. xanthus has a single large circular chromosome of about 9.14 Mbp. It has been suggested that this enlarged genome (and those of related myxobacteria of the order myxococcales), is a consequence of extensive, but not random, gene duplications whose subsequent divergence enabled evolution of the signaling systems required for the striking multicellular lifestyle of myxobacteria [24]. For all these reasons, it is of particular interest to study the organization of the chromosome and its segregation in this bacterium. The main objective of this work was to ascertain if the DNA sequences that encode the predicted ParABS elements, taken from the M. xanthus genome annotation, have a role in chromosome segregation in M. xanthus. A second objective was to determine if these ParABS elements are physically or functionally interconnected. A conclusion of this work is that ParB binds preferentially to a parS consensus sequence in vitro. It is also shown here that ParB is essential for viability and its absence generates anucleate cells demonstrating the key role of ParB in chromosome segregation. ParA is also essential for viability and localizes in DNA-free zones such as at the cell poles and along the cell division plane, prior to cell division. However, this localization pattern is independent of FtsZ. ParA subcellular positioning depends on ParB, the absence of which causes ParA to be delocalized from DNA-free zones and colocalizes with chromosomal DNA. Therefore, ParB appears to prevent colocalization of ParA and DNA by guiding ParA to DNA-free zones. In most cells, ParB localizes at the edge of the chromosomal DNA in subpolar positions to thereby limit ParA localization to polar clusters.
Results and Discussion
parABS Loci in M. xanthus
MXAN_7477 and MXAN_7476 in the M. xanthus genome have been annotated as encoding parA and parB, respectively [24] (Fig. 1A). These two genes are co-transcribed as an operon together with MXAN_7475 (encoding the bactofilin BacM) and MXAN_7474 (encoding a putative lipoprotein of unknown function). Neither BacM nor MXAN_7474 is necessary for chromosome segregation or optimal cell growth under standard conditions [25]. Chromosomal parAB loci are usually found in the oriC-proximal region of bacterial chromosomes [15]. Consistent with this, the M. xanthus parAB locus indicated above is about 35 kb away from oriC. Bioinformatic analysis of 400 sequenced prokaryotic chromosomes indicated 1030 putative parS sites which are located, mostly, within oriC-proximal regions of their respective chromosomes [15]. From these parS sites, Livny and coworkers created a parS consensus matrix (Fig. 1B), and found that M. xanthus contains 12 putative parS sites, about 4.4 kb upstream from parA. To refine this analysis in M. xanthus, I decided to repeat the search for parS sites in the M. xanthus genome modifying the parS consensus matrix obtained previously. The search was performed using as query a putative parS sequence (16 pair of bases) that retains the highly conserved nucleotides, and in which the less conserved ones are varied (Fig. 1C). This search uncovered a 3 kb cluster of 22 putative parS sequence repeats (Fig. 1D) located about 4 kb upstream of parA, between genome positions 9108251 to 9111304 (Fig. 1A). In addition, this search also pointed out another parS site located at position 349383 to 349398, distant from the parAB locus. Two additional parS sites in the M. xanthus genome, near each end of the parS cluster, were indicated in another report using as query the 12 parS M. xanthus sequences previously identified and allowing for one mismatch [26]. The first sequence (position 9105392–9105407) has a “G” in position 7, and the second sequence (position 9111742–9111757) contains a “C” in position 3. Only two examples of each of these case exists among the 1030 predicted parS sites mentioned earlier. Positions 3 and 7 are otherwise highly conserved. The level of nucleotide conservation is important because the 1030 parS sites, found in 276 of the 400 sequenced strains, were identified using as a reference only the 15 parS sites from Streptomyces coelicolor and the 10 parS sites from B. subtilis that have been shown to bind ParB in vivo [27]–[29]. Taking the 22 putative parS sites described here (Fig. 1D), the M. xanthus parS consensus sequence can be assigned to be TGTTCCACGTGGAACG (Fig. 1E).
Figure 1. parABS locus in M. xanthus.
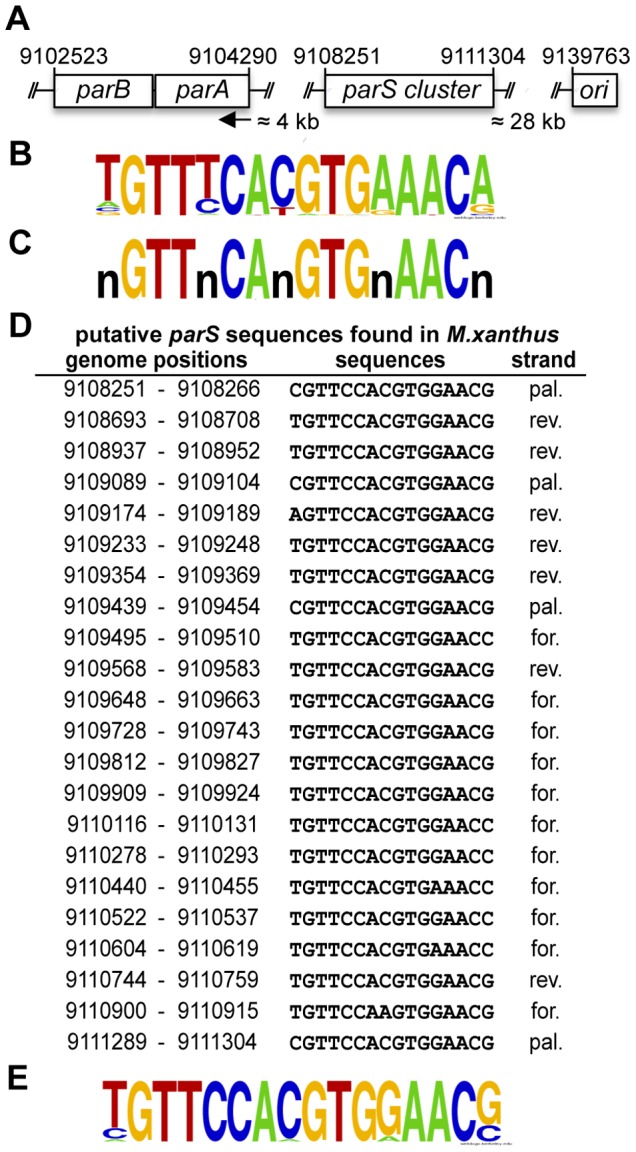
(A) Genomic organization of the parABS locus. The arrow indicates the direction of parA and parB transcription. (B) WebLogo representation [60] of the consensus of 1030 parS sites identified from 276 prokaryotic genomes [15]. (C) The 16 bp sequence used to find putative parS sites in the M. xanthus genome. (D) Putative parS sequences found in M. xanthus, for = forward, rev = reverse, pal = palindromic (E) WebLogo showing the consensus of parS sites in M. xanthus.
M. xanthus ParB Binds to a Consensus parS in vitro
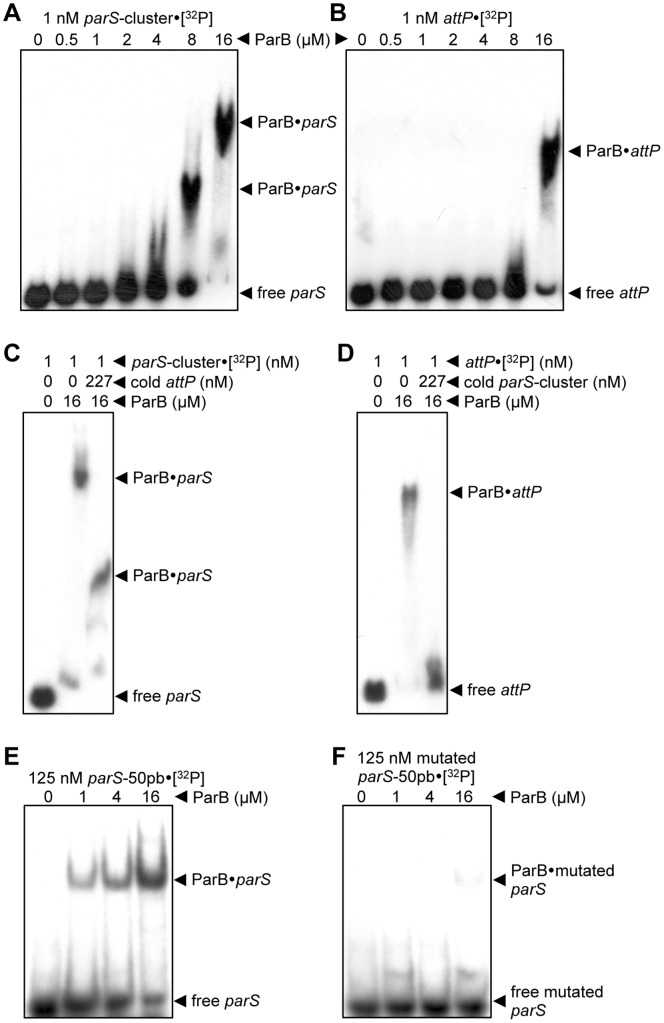
In order to determine if M. xanthus ParB is able to bind to the DNA fragment containing the 22 parS cluster described in the above section, an agarose gel electrophoretic mobility shift assay was performed. For this, a 3.12 kb 32P-radiolabeled probe, corresponding to the DNA segment from positions 9108215 to 9111334 of the M. xanthus genome and containing the 22-repeat parS stretch was incubated with increasing amounts of purified ParB, and electrophoresed in a 0.7% agarose gel. Clear mobility shifts of the parS-cluster probe appear at ParB concentrations >2 µM (Fig. 2A). This indicates that ParB interacts with the DNA fragment containing the 22 M. xanthus parS sites. It can also be observed that higher amounts of ParB resulted in slower-migrating bands. It has been previously described elsewhere [29], [30], that large DNA probe fragments show multiple slower-migrating bands as ParB concentration increase, indicating that several molecules of ParB are binding per DNA fragment, generating a large nucleoprotein complex near the origin of replication. These large structures could potentially demarcate, organize or localize the origin region of the chromosome [29], [30]. To assess its preferential binding to parS sites, ParB binding was tested with another DNA probe similar in size and G+C content but without the parS-cluster (3005 bp and 65.7% G+C versus 3120 bp and 64.9% G+C). The DNA probe chosen contained the Mx8 phage attP site involved in phage integration into the M. xanthus chromosome at the attB site [31]. ParB could bind to the attP fragment but only at ParB concentrations far higher than with the parS probe (Fig. 2B), indicative of the significantly greater of ParB for the probe bearing the parS-cluster. In order to further establish the preferential binding of ParB to the sequence with the parS-cluster, a DNA binding competition assay was performed. Prior to the 30 minutes of ParB incubation with the labeled probe with the parS-cluster, ParB was incubated with unlabeled attP probe at more than two-hundred fold excess for 1 hour. As can be seen in Fig. 2C, ParB complexes with labeled parS probe appeared even with cold attP probe present at over a 227-fold excess. By contrast, with a similar excess of cold parS probe, ParB complexes with labeled attP probe could not be detected (Fig. 2D) confirming that ParB has a greater affinity for the probe with the parS-cluster.
Figure 2. ParB binds preferentially to parS in vitro.
(A) Agarose EMSA on after incubating different amounts of ParB with a 32P-labeled 3120 bp DNA fragment containing the parS-cluster, (B) or with a 3005 bp DNA containing the Mx8 phage attP sequence. (C) Agarose EMSA of the binding of 32P-labeled parS-cluster to ParB, or to ParB previously incubated with higher amounts of unlabeled attP DNA fragment, as indicated. (D) Agarose EMSA of the binding of 32P-labeled attP fragment to ParB, or to ParB previously incubated with higher amounts of unlabeled parS-cluster, as indicated. (E) EMSA using a 6% polyacrylamide gel after incubating a 32P-labeled 50 bp DNA fragment containing the M. xanthus parS consensus sequence with increasing amounts of ParB, as indicated (F) or with this 32P-labeled 50 bp DNA fragment containing 11 point mutations of the most conserved base pairs of parS.
Next, to determine if ParB binds specifically to a single parS site, a 50 bp DNA probe containing the M. xanthus parS consensus sequence TGTTCCACGTGGAACG (Fig. 1E), which spans positions 9109710 to 9109759 of the M. xanthus genome, was used in the gel mobility assay. ParB was found to bind to this single parS site (Fig. 2E) with a single retarded band observed even at the highest ParB concentration used, in contrast to the 3 kb probe with several parS repeats which yielded a large complex at high concentrations. However, when ParB was incubated with a similar DNA fragment but containing 11-point mutations (TaccCtgCacaAggtG) of the most conserved base pairs of the parS consensus sequence (Fig. 1B), only a faint band could be barely detected even at the highest ParB concentration used (Fig. 2F). These in vitro results are consistent with specific binding of ParB to a consensus parS sequence.
ParB is Essential for Viability in M. xanthus
ParB has been shown to be important for chromosome partitioning in numerous bacteria [14], [32]. In order to determine if M. xanthus ParB participates in chromosome segregation, it was necessary to create a parB mutant. It was not possible to delete parB in M. xanthus, suggesting that ParB is essential for viability. In fact, the endogenous parB gene could be deleted only if an extra copy of the gene was also present (located at the 1.38-kb locus described in detail in reference 33). Moreover, conditional expression of parB by placing it under the control of a vanillate-inducible promoter, which is derepressed in the presence of vanillate [33], [34], resulted in viable cell growth only under permissive conditions when vanillate was present (Fig. 3A, left panel), whereas there was no growth under restrictive conditions without vanillate (Fig. 3A, right panel). Furthermore, restricting parB expression results in cells (after a 48-hour growth with no vanillate to ensure complete growth arrest) in aberrant cellular morphology when examined under a microscope, and considerable amounts of cellular debris, indicating cellular death, could be observed (Fig. 3B, right panel). Thus, these data clearly demonstrate that parB is essential for viability in M. xanthus. In most chromosomal par systems studied, mutations or deletions of the par genes did not produce lethality [14], [32], [35], a notable exception being C. crescentus [36]. For instance, in Deinococcus radiodurans and P. aeruginosa, the absence of ParB resulted in bacterial growth retardation [32], [37]. Additionally in P. aeruginosa, parB mutants were affected in swarming and swimming motility [32], and in B. subtillis, the absence of ParB ortholog Spo0J caused a sporulation defect [38]. It has been proposed that the essentiality of the par system in C. crescentus is due to a cell division defect, indicating that ParAB are required for cytokinesis [39]. Although this may also be the reason why the par system is essential in M. xanthus, the dramatic filamentous cell morphology phenotype that C. crescentus cells present in the absence of parB is not observed with M. xanthus.
Figure 3. ParB is essential in M. xanthus.
(A) Strains MR2461 (Pvan-parB) and MR2472 (Pvan-parB ΔparB) were grown in CTT media in the presence of vanillate (for parB expression) to an O.D.550 of 0.8. The cultures were serially diluted, and 5 µl of each sample was spotted onto CTT plates containing vanillate (left), and no vanillate (right). (B) Microscope DIC images from a cell culture from the strain Pvan-parB ΔparB in the presence of vanillate (left image), and 48 hours after vanillate removal (right image).
M. xanthus ParB is Involved in Chromosome Partitioning
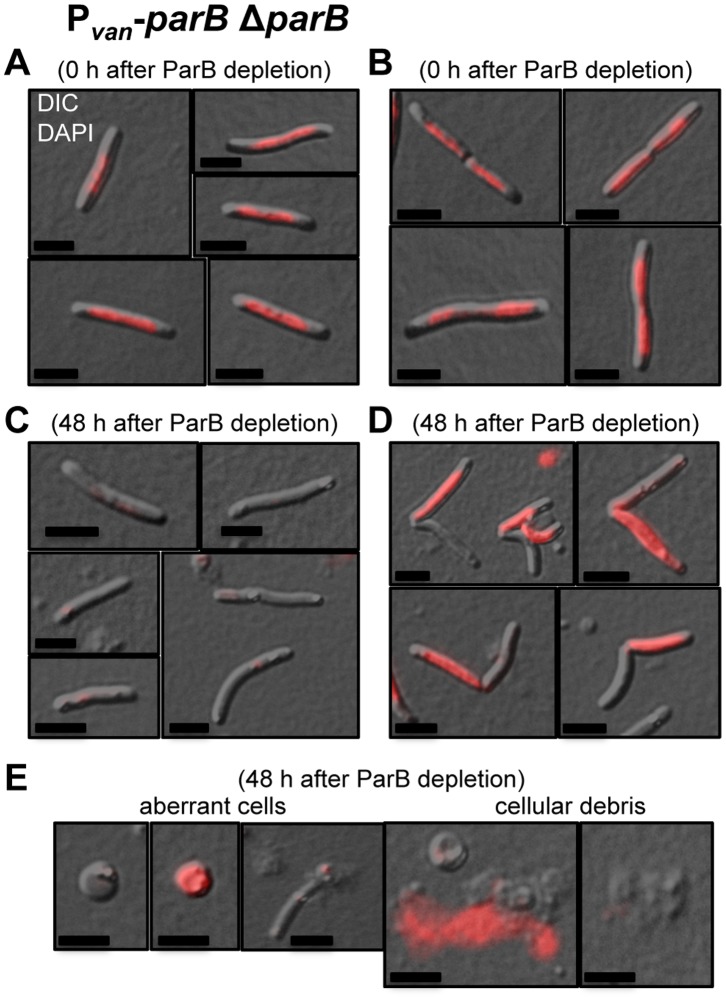
Chromosomally encoded ParB or ParA proteins have been reported to have a role in chromosome partitioning in several bacteria. It is established that a lack of Par proteins or the presence of mutant forms of theses proteins causes an increase in anucleate cells [6], [28], [36]–[38], [40]–[42]. To ascertain if the absence of ParB in M. xanthus results in anucleate cells, a culture of parB conditional strain, Pvan-parB ΔparB, was grown in the presence of vanillate. The cell culture was washed to remove vanillate from the media and the cells were examined under the microscope after 0, 24, 36, and 48 hours. Samples were incubated with DAPI 10 minutes before being placed on the agarose pad to observe chromosomal DNA by fluorescence microscopy. In the presence of vanillate, all cells had normal rod-shape morphology and contained chromosomal DNA, with dividing cells having DNA in both compartments (Table 1, Figs. 4A and B). It should be noted that the chromosomal DNA does not occupy the entire cytoplasmic space, leaving areas proximal to the cell poles free of DNA. After 24 hours of ParB depletion, anucleate cells start to appear, together with dividing cells bearing DNA only in one compartment instead of both, although the number of cells with such anomalies is small (1% of 400 cells observed; Table 1). After 36 and 48 hours of ParB depletion, the population of anucleate cells is quite significant (between 10.1 and 21.6%, n = 310 and n = 219, respectively), and the number of dividing cells with DNA in only one compartment also increases (between 14.4 and 9.4%, n = 310 and n = 219, respectively) (Table 1, Figs. 4C and D; these counts considered only cells that conserved the typical smooth rod-shape morphology). After 48 hours of ParB depletion several rounded cells (with or without DNA), significant amounts of cellular debris, and free chromosomal DNA, presumably released from dead cells into the media, can be observed (Fig. 4E). This suggests that M. xanthus ParB is involved in chromosome partitioning, and its absence provokes chromosome segregation anomalies and cellular death.
Table 1. Presence or absence of DNA in M. xanthus cells depleted of ParB.
| rod-shape cells | time after vanillate removal ⇒ | 0 h | SD | 24 h | SD | 36 h | SD | 48 h | SD |
| not dividing | with DNA | 91.0% | 2.3 | 95.1% | 4.1 | 67.9% | 7.9 | 60.9% | 5.9 |
| without DNA | 0.0% | 0.0 | 0.2% | 0.3 | 10.1% | 4.4 | 21.6% | 5.9 | |
| dividing | with DNA (in 2 compartments) | 9.0% | 2.3 | 3.9% | 4.1 | 7.6% | 3.3 | 8.1% | 1.6 |
| without DNA (in 1 compartment) | 0.0% | 0.0 | 0.8% | 0.2 | 14.4% | 7.4 | 9.4% | 2.3 | |
| total number of cells observed | 272 | 400 | 310 | 219 |
SD: standard deviation.
Figure 4. ParB is involved in chromosome partitioning in M. xanthus.
Merged DIC and fluorescence images of cells from a Pvan-parB ΔparB (MR2472) culture in the presence of vanillate (permissive conditions) (A) and (B), and after 48 hours in the absence of vanillate (restrictive conditions) (C), (D) and (E). The cultures were stained with DAPI for viewing chromosomal DNA by fluorescence, shown in red. Black scale bars represents 5 µm. (A) Non-dividing cells with DNA. (B) Dividing cells with DNA. (C) Non-dividing cells without DNA. (D) Dividing cells with DNA only in one compartment. (E) Rounded cells without DNA (first from left) and with DNA (second from left), broken cell without DNA (middle), extracytoplasmic DNA (second right) and cellular debris (first right). The mean of the results from three independent experiments and the standard deviations are shown in Table 1.
Subcellular Localization of M. xanthus ParA
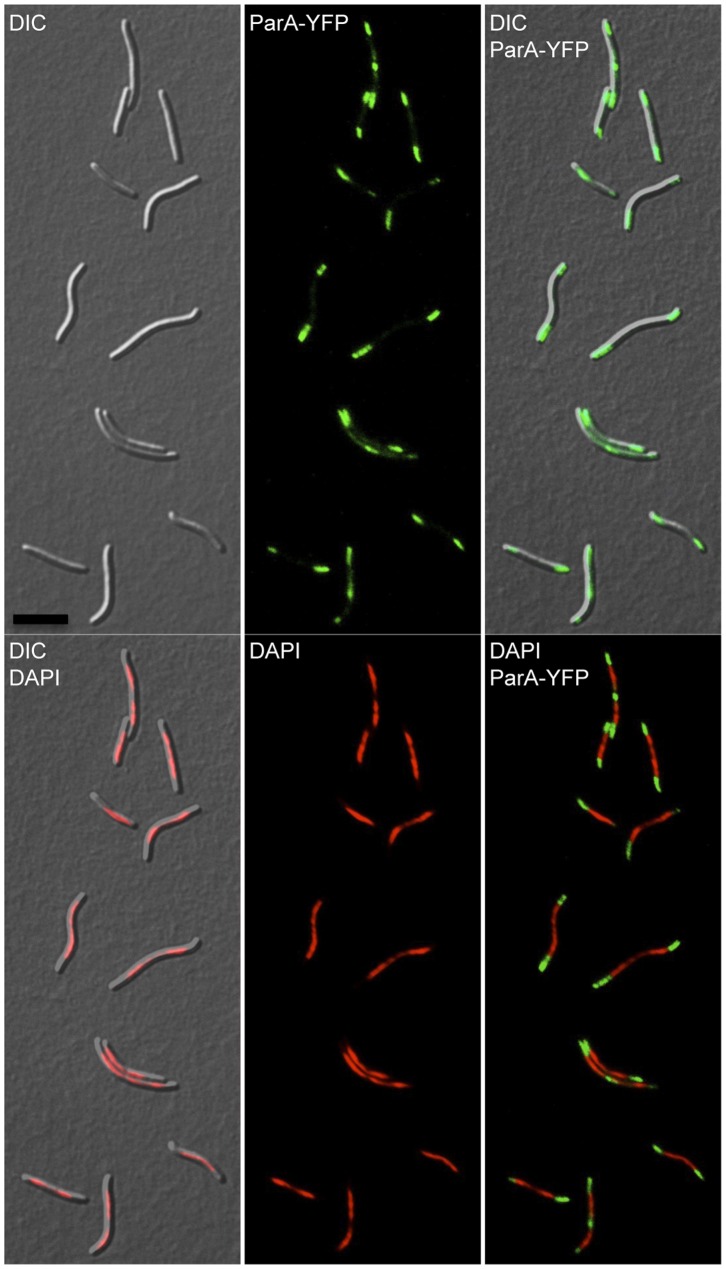
All attempts to delete chromosomal parA from M. xanthus were unsuccessful. The chromosomal parA gene was only deleted when the strain harbored another copy of parA in trans under the control of the Pvan promoter, indicating that, as with parB, parA is essential for viability in M. xanthus (data not shown). In this case, basal levels of expression from the Pvan promoter without the addition of vanillate, sufficed to allow the cells to live. To determine the subcellular localization of ParA, a strain was created harboring Pvan-parA-yfp, bearing a copy of parA fused to yfp, whose expression was under the control of the inducible vanillate promoter and inserted at the 1.38-kb locus. After growing this strain, cells were examined under the microscope and as fluorescent ParA-YFP could be observed even in the absence of vanillate, it was not included in these analyses. ParA-YFP fluorescence was found to cluster at both poles, and in some cells at the cell division plane (Fig. 5, upper panels). Merging the images obtained for ParA-YFP with that of the chromosomal DNA visualized using DAPI staining, it is clearly apparent that ParA-YFP localizes at the DNA-free regions. As mentioned in a previous section, the chromosome in M. xanthus does not occupy all regions of the cytoplasmic space, with DNA-free regions at the poles and, in some cells at the cell division plane (Fig. 5, bottom panels). Approximately 84% out of the 615 cells had two ParA-YFP clusters only at the two poles (Figs. 6A and B), and in the remaining cells an additional ParA-YFP cluster was observed in the cell division plane as well (Figs. 6C–E). Even after separation of the dividing cells, ParA remains localized at the newborn pole, which earlier was part of the cell division plane in the mother cell (Fig. 6E). Therefore, the polar localization of ParA appears to be a consequence of a prior location at the midcell and raises the question of how ParA is being recruited to the cell division site. In C. crescentus, ParA forms a cloud-like structure extending from the new pole towards the old one. The duplicated ParB-parS complex associates with the ParA-cloud and is pushed apart towards the new pole, by a ParB-dependent ParA-ATPase activity [16], [43]. A similar mechanism has been proposed for V. cholerae chromosome I [44]. It has also been suggested that the nucleoid forms a structural matrix for the assembly of a track-like structure of ParA that guides the ParB-parS complex movement [16], [45]. In the multigenomic aerial hyphae of S. coelicolor, ParA accumulates at the tip of the hyphae and it extends from the tip towards the rest of the hyphae as helical filaments providing a scaffold for a regular distribution of several ParB-parS complexes [46]. In B. subtillis, the ParA ortholog Soj localizes to the septa and as relatively faint punctuate foci within the cytoplasm [18]. In M. xanthus, the symmetrical ParA localization at DNA-free poles, observed in this study, seems to be incompatible with ParB-parS transport by ParA through a nucleoid structural matrix. This, however, should not be discarded since a faint ParA fluorescence can be detected throughout the cytoplasm even in the regions with DNA.
Figure 5. Subcellular localization of ParA in M. xanthus.
Fluorescence microscope images of cells from the strain Pvan-parA-yfp (MR2504) grown without vanillate. DIC (top left), ParA-YFP fluorescence (top middle, in green), merged DIC with ParA-YFP fluorescence (top right), merged DIC with DAPI fluorescence (bottom left), DAPI fluorescence (bottom middle, in red), and merged DAPI (in red) with ParA-YFP (in green). Black scale bar represents 10 µm.
Figure 6. Distribution of cells according to its ParA localization.
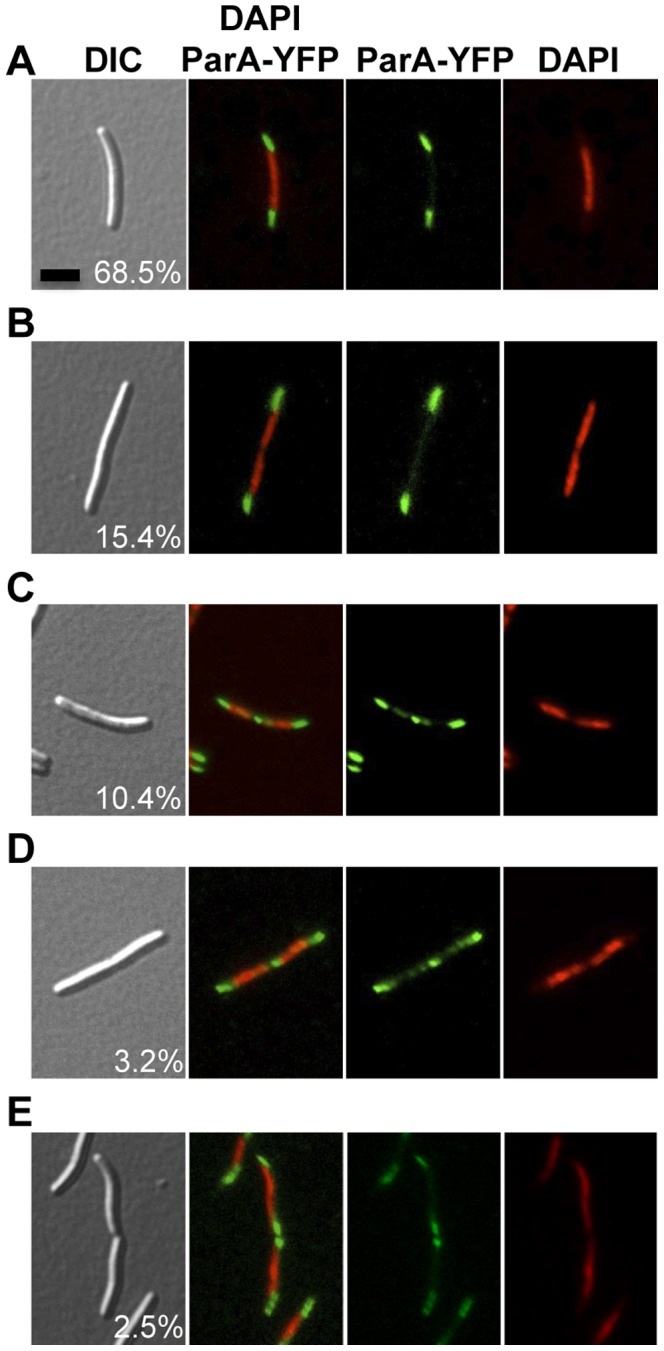
DIC, ParA-YFP (in green), and DAPI (in red) microscope fluorescence images of cells from the strain Pvan-parA-yfp (MR2504) grown without vanillate. Black scale bar represents 5 µm. A total of 615 cells from three independent experiments were examined and the mean and the standard deviation are reported. (A) A representative cell with two polar clusters of ParA-YFP and one chromosomal mass (68.5±10.2%). (B) Cell having two polar clusters of ParA-YFP and two distinct chromosomal masses (15.4±8.7%). (C) Representative cell with two polar clusters of ParA-YFP, two distinct chromosomal masses, and an additional cluster of ParA-YFP in the cell division plane but with no pinch in its cellular morphology (10.4±2.3%). (D) Same as in (C) but with incipient constriction along the cell division plane (3.2±0.7%). (E) Cells recently divided, showing two polar clusters of ParA-YFP and one chromosomal mass, in each of the two cells (2.5±1.4%).
The Localization of ParA is not Dependent on FtsZ
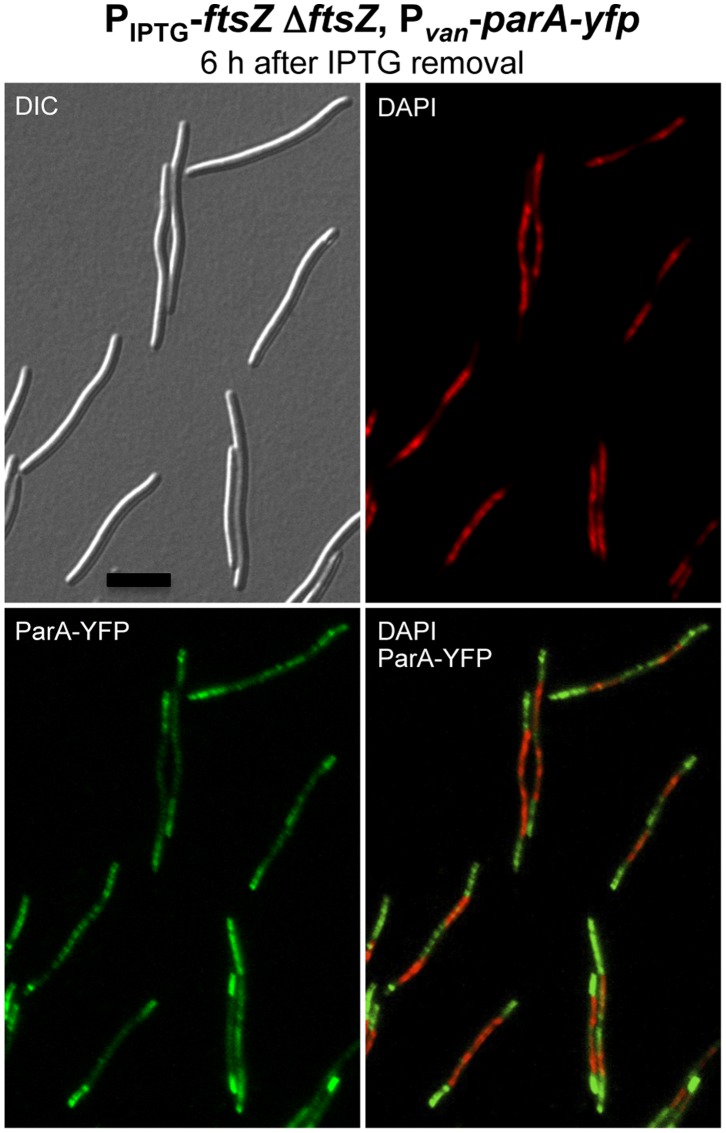
To obtain more insight into ParA cellular localization, this was examined in the absence of FtsZ, the bacterial tubulin homolog that forms a ring in the midcell region whose constriction culminates in cell division [47]–[49]. In M. xanthus, FtsZ localizes at the cell division plane in most cells, and its absence results in filamentous morphology and, eventually, cell death [33]. Whether ParA-YFP localization at the midcell depends on FtsZ, was studied by inserting a copy of Pvan-parA-yfp at the Mxan_18–19 locus (described in detail in reference 33) in strain MR2196 to generate strain MR2536. In MR2196 the only copy of the ftsZ gene is under the control of the IPTG-inducible promoter, and its growth and viability depends on the presence of IPTG [33]. MR2536 was grown in the presence of IPTG, and in the absence of vanillate. Then the cells were washed repeatedly to remove IPTG and grown during 6 hours. This depletion of FtsZ produces elongated cells in which ParA-YFP continues to localize at the cell poles, and in the space between chromosomes in many cells (Fig. 7). Although in the absence of FtsZ ParA-YFP fluorescence appears to be more dispersed throughout the cytoplasm than when FtsZ is present, the overall ParA-YFP localization pattern seems to persist. Thus, the midcell localization of ParA-YFP does not appear to be correlated with that of FtsZ.
Figure 7. ParA localization is not dependent on FtsZ.
DIC, DAPI (in red), and ParA-YFP (in green) microscope fluorescence images of cells from the strain MR2536 (PIPTG-ftsZ ΔftsZ, Pvan-parA-yfp), after 6 hours of IPTG removal (FtsZ depletion). Scale bar represents 10 µm.
ParB Controls ParA Localization
Since ParB has been shown to influence ParA localization in several bacteria [16], [18], [50], [51], this was tested in M. xanthus by examining ParA-YFP in ParB-depleted cells. For this, a copy of parA-yfp under the control of the IPTG promoter was inserted at the Mxan_18–19 locus in the strain (MR2472) described earlier, which contains the only parB copy under the control of the vanillate-inducible promoter. The resulting strain MR2538 is thus viable in presence of vanillate in the medium. After 1 hour of IPTG-induced parA-yfp expression, ParA-YFP is observed as a bright signal at the DNA-free zones at the poles and, in some cells, at the cell division plane (Fig. 8A). A faint ParA-YFP fluorescence also appears throughout the rest of the cytoplasm. Therefore, localization of ParA-YFP in this strain under permissive conditions resembles that in the wild-type strain. ParA-YFP (again after 1 hour of IPTG-induction) was still observed at the poles but also overlapped with DAPI-stained DNA, 24 hours after vanillate (and hence ParB) depletion compared to when vanillate was present (Fig. 8B). Aberrant cell morphology and abnormal distribution of DNA, generating the presence of anucleate cells, were evident after 48 hours of ParB depletion, with the ParA-YFP signal (IPTG-induced for 1 hr) markedly coincident with chromosomal DNA, although it can be seen in other DNA-free zones (Fig. 8C). In anucleate cells or cellular compartments without DNA, ParA-YFP was not detected. This result indicates that, in the absence of ParB, ParA may bind to chromosomal DNA. Indeed, various studies have reported that ParA from other bacteria can bind to DNA in a nonspecific ATP-dependent manner [16], [52], [53]. Also, as observed in M. xanthus in this study, association of the B. subtilis ParA ortholog Soj to the nucleoid has been observed to occur when the ParB ortholog Spo0J is absent [18], [50], [51], and C. crescentus ParA-YFP heterologously expressed in E. coli, which lacks a Par system [15], was found to localize on the nucleoid [16].
Figure 8. ParB inhibits ParA to localize with the nucleoid.
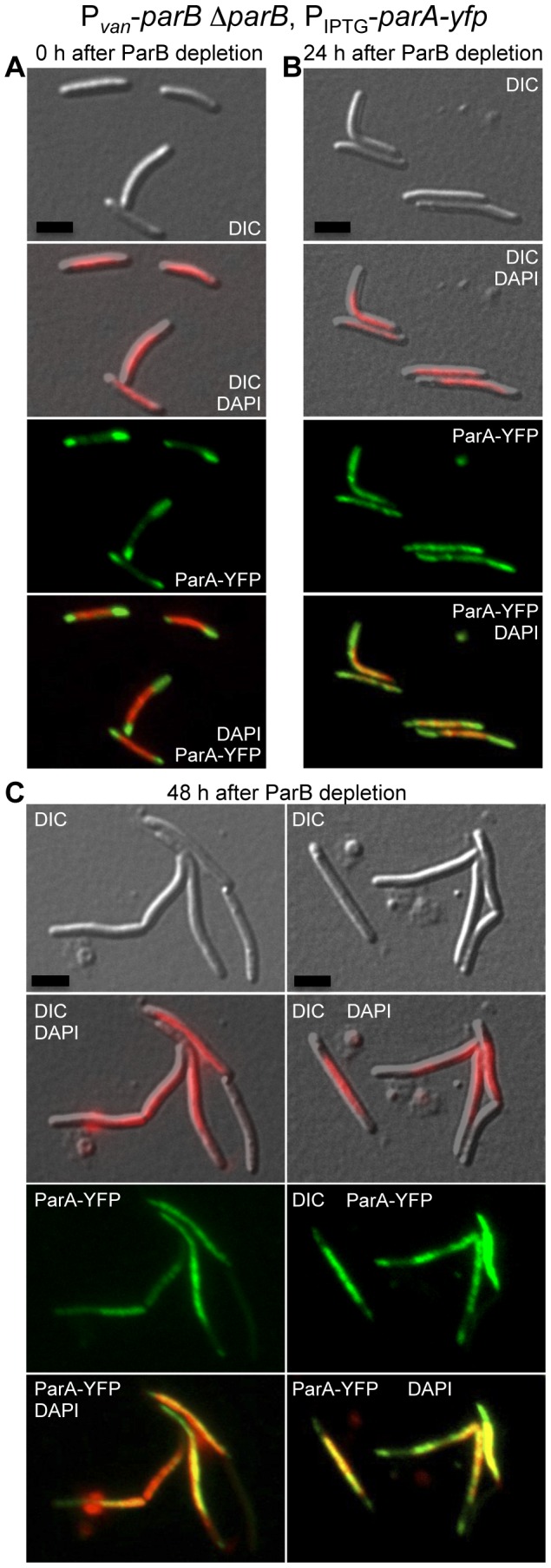
DIC, DAPI (in red), ParA-YFP (in green), and DAPI with ParA-YFP merged (in yellow) microscope fluorescence images of cells from the strain MR2538 (Pvan-parB ΔparB, PIPTG-parA-yfp) grown in presence of vanillate (A), 24 hours (B), and 48 hours after vanillate removal (C). All images were taken 1 hour after IPTG incubation for parA-yfp expression. Scale bar represents 5 µm.
Subcellular Localization of ParB
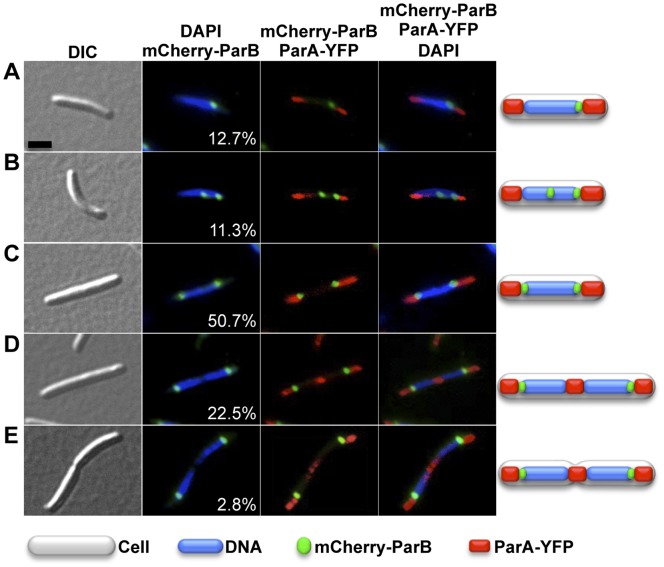
To determine intracellular localization of ParB, plasmid pMR3828 encoding an mCherry-ParB fusion controlled by an IPTG-inducible promoter [33] was integrated at the M. xanthus 1.38-kb locus to generate strain MR2526. Additionally, strain MR2526 also has the plasmid pMR3826 with the vanillate-inducible parA-yfp construct integrated at the Mxan_18–19 locus. MR2526 was grown in CTT media to exponential phase. After a 3-hours of IPTG (1 mM) induction of mCherry-parB expression, samples were taken and stained with DAPI for microscopy. In 12.7% of the cells observed (n = 550), a single focus of mCherry-ParB was seen just at the edge of the nucleoid (Fig. 9A). Most of the cells presented two mCherry-ParB foci at both edges of a single nucleoid (50.7% of the cells; Fig. 9C), or two foci at both subpolar edges of the two separated nucleoids (25.3% of the cells; Figs. 9D and E). The remaining cells had one focus localized at the edge of the nucleoid and another in an intermediate position (11.3% of the cells; Fig. 9B). The localization pattern of ParB in M. xanthus thus resembles those previously described in other bacteria, and where ParB localization was linked to its ability to bind parS [16], [44], [54]. Since, as shown in this study, M. xanthus ParB binds preferentially to a consensus parS sequence in vitro (Fig. 2E), the single mCherry-ParB focus seen in 12.7% of the cells may correspond to ParB bound to a not as yet replicated or segregated, parS (Fig. 9A). Then, the presence of cells having one focus at the edge of the nucleoid and other in an intermediate position (Fig. 9B) could indicate that parS has replicated and is being moved to the other edge of the nucleoid, resulting in cells with two ParB-parS clusters at both edges of the nucleoid (as seen in 50.7% of the cells). This final location of both ParB-parS complexes persists even after the two chromosomes have been segregated (Figs. 9D and E). Thereby, the division of the cell would provide two daughter cells with a single ParB-parS complex, completing the cell cycle.
Figure 9. Distribution of cells according to its ParB localization.
DIC, mCherry-ParB (in green), DAPI (in blue), and ParA-YFP (in red) microscope fluorescence images of cells from the strain MR2526 (Pvan-parA-yfp, PIPTG -mCherry-parB) grown without vanillate, and with IPTG (1 mM) during 3 hours for mCherry-parB expression. Black scale bar represents 5 µm. A total of 550 cells from three independent experiments were examined and the mean and standard deviation are reported. (A) Cell having one chromosomal mass and a single mCherry-ParB focus at the edge of the nucleoid (12.7±1.8%). (B) Cell having one chromosomal mass, a mCherry-ParB focus at the edge of the nucleoid and another mCherry-ParB focus in an intermediate nucleoid position (11.3±0.3%). (C) Cell presenting one chromosomal mass, and two mCherry-ParB foci at both edges of a single nucleoid (50.7±3.7%). (D) Cell having two chromosomal masses, and two mCherry-ParB foci at the subpolar edges of both nucleoids (22.5±1.7%). (E) The same as in (D) but with some sign of cellular pinch at the cell division plane (2.8±1.5%).
Simultaneous observation of mCherry-ParB and ParA-YFP shows that ParB is in close proximity to polar ParA in 76% of the cells (Figs. 9C–E), which correspond to cells with the two ParB-parS complexes fully segregated. While ParB may inhibit the presence of ATP-bound ParA within the nucleoid, it may not affect ParA polymerization at the poles, where no DNA- ParB complex is present. When the two newly replicated chromosomes separate, a DNA-free space is created at the cell division plane where, presumably, the dispersed ParA could polymerize to create a new midcell ParA cluster. Therefore, the localization of ParB appears to be consistent with its role controlling ParA localization.
Materials and Methods
Bacterial Strains and Growth Conditions
E. coli strain DH5α was used for plasmid constructions and was grown at 37°C in Luria broth medium (LB) supplemented with the appropriate antibiotics. M. xanthus was grown at 33°C in rich Casitone-Tris (CTT) medium [55]. Media were supplemented with inducer (0.5 mM vanillate or 1 mM isopropyl β-D-thiogalactoside (IPTG)) or antibiotic (40 µg/ml kanamycin (KanR), 10 µg/ml oxytetracycline (TetR) for solid media, and 2,5 µg/ml oxytetracycline for liquid media), as required.
Construction of Strains and Plasmids
M. xanthus strains and plasmids used in this study are listed in Table 2 and Table 3. Standard protocols and commercially available kits were used in the preparation and manipulation of chromosomal and plasmid DNA. All constructs were verified by DNA sequencing. Plasmids were introduced into M. xanthus by electroporation, and integration of the plasmids by homologous recombination was selected on CTT plates containing the appropriate antibiotic and/or by negative selection via a galK gene that confers sensitivity to galactose (GalS).
Table 2. Relevant strainsa.
| strain | integrated plasmid(s) | relevant genotype or description | source |
| DK1050 | M. xanthus wild type | [61] | |
| DK1622 | M. xanthus wild type | [62] | |
| MR2461 | pMR3594 | M. xanthus DK1050 1.38-kb::Pvan-parB | This study |
| MR2472 | pMR3594 | M. xanthus DK1050 1.38-kb::Pvan-parB, ΔparB | This study |
| MR2504 | pMR3785 | M. xanthus DK1050 1.38-kb::Pvan-parA-yfp | This study |
| MR2526 | pMR3826, pMR3828 | M. xanthus DK1050 Mxan_18–19::Pvan-parA-yfp, 1.38-kb::PIPTG -mCherry-parB | This study |
| MR2536 | pMR3636, pMR3826 | M. xanthus DK1622 1.38-kb::PIPTG -ftsZ, ΔftsZ, Mxan_18–19::Pvan-parA-yfp | This study |
| MR2538 | pMR3594, pMR4051 | M. xanthus DK1050 1.38-kb::Pvan-parB, ΔparB, Mxan_18–19::PIPTG -parA-yfp | This study |
Other strains, precursors to those listed here, are described in the text.
Table 3. Relevant plasmidsa.
| plasmid | relevant genotype or description | source |
| pMR3594 | M. xanthus 1.38-kb::Pvan-parB, TetR | This study |
| pMR3636 | M. xanthus 1.38-kb::PIPTG -ftsZ, TetR | [33] |
| pMR3684 | Intein_tag-M. xanthus-parB | This study |
| pMR3785 | M. xanthus 1.38-kb::Pvan-parA-yfp, KanR | This study |
| pMR3826 | M. xanthus Mxan_18–19::Pvan-parA-yfp, KanR | This study |
| pMR3828 | M. xanthus 1.38-kb::PIPTG -mCherry-parB, TetR | This study |
| pMR4051 | M. xanthus Mxan_18–19::PIPTG -parA-yfp, KanR | This study |
Other plasmids, precursors to those listed here, are described in the text.
M. xanthus parB coding sequence was PCR-amplified using genomic DNA from wild type DK1050, as DNA template, and primers 14_ParB.for (5′-gctggagtcaccatatggtgaaagcagaca-3′) and 15_ParB.rev (5′-aaaagaattcctactccttcctgagaagct-3′). This PCR product and the plasmid pMR3553, which bears the 1.38-kb sequence for chromosome integration and the Pvan promoter [33], were digested with NdeI and EcoRI and ligated, obtaining the plasmid pMR3594. To generate the plasmid pMR3620 for deleting chromosomal parB, two PCR products were generated. The first PCR product contains about 0.92 kb of parB upstream sequence, and it was obtained using DK1050 genomic DNA as a template and the primers 26_UpParB-for (5′-aaaaaagcttagcagcgtggatcagcgcgc-3′) and 27_UpParB.rev (5′-aaaaatcgatcacgtcgtgactccagccag-3′). The second PCR product has around 0.94 kb of parB downstream sequence, and it was obtained using the primers 28_DownParB.for (5′-aaaaatcgattaggacgtggcgctccttgg-3′) and 29_DownParB.rev (5′-aaaatctagatggcacagaggaacaagtcg-3′), and DK1050 genomic DNA as a template. Both PCR products were digested with HindIII-ClaI and ClaI-XbaI, respectively, and cloned into HindIII-XbaI-digested pBJ114 plasmid, which contains the galK gene [56]. To generate plasmid pMR3785, the parA gene was PCR-amplified to be translationally fused to yfp, using the primers 56_ParA.for (5′-aaaaaacatatggtgcactgcatcacgcgc-3′) and 57_ParA.rev (5′-aaagaattcccagccacgcgcctgcgagggct-3′), and DK1050 genomic DNA as a template. After NdeI-EcoRI digestion, parA from this PCR product was cloned into a NdeI-EcoRI-digested pMR3653 plasmid [33], exchanging ftsZ gene for parA. The plasmid pMR3826 was made cloning the parA-yfp sequence, by digesting pMR3785 with NdeI and NheI, into the plasmid pMR3690 [33] previously digested with the same restriction enzymes. To make plasmid pMR3828, the M. xanthus parB coding sequence was PCR-amplified using M. xanthus DK1050 genomic DNA as a template and the primers 18_ParB.for (5′-aaagaattccgtggtgaaagcagacatgca-3′) and 15_ParB.rev (5′-aaaagaattcctactccttcctgagaagct-3′), digested by EcoRI and cloned into the EcoRI-digested vector pVCHYN-2 [34], resulting in the plasmid pMR3733 encoding an mCherry-parB fusion. Then, mCherry-parB fragment was amplified by PCR using plasmid pMR3733 as DNA template, and the primers 49_mCherry.for (5′-aaaatctagaatggtgagcaagggcgagga-3′) and 42_ParB.rev (5′-aaaatctagactactccttcctgagaagct-3′). This PCR product was digested by XbaI and cloned into XbaI-digested plasmid pMR3487 [33], resulting in the plasmid pMR3828 which has the mCherry-parB fusion under the control of a IPTG-inducible promoter, and the 1.38-kb sequence for chromosomal integration. Plasmid pMR3684 used for ParB purification was obtained isolating the parB coding sequence fragment after the digestion of plasmid pMR3594 by NdeI and EcoRI, and cloning into these sites in pTYB12 (New England Biolabs). In order to create plasmid pMR4051, it was necessary to generate two precursor plasmids. First, a DNA fragment of 1.861 kb containing the IPTG inducible promoter, a multicloning site, and the lacI gene repressor was obtained digesting pMR3487 [33] with PstI and NdeI. This fragment was cloned into a PstI-NdeI-digested pMR2700 plasmid [57], generating the plasmid pMR4046. The plasmid pMR4046 was digested with HindIII, releasing the M. xanthus 1.38-kb sequence for chromosomal integration, and ligated with M. xanthus Mxan_18–19 sequence, used for a chromosomal integration in a previous work [33], obtaining the second precursor plasmid pMR4048. The 1.319 kb Mxan_18–19 sequence was isolated after HindIII digestion of plasmid pMR3691 [33]. Finally, a PCR-amplified parA-yfp sequence, obtained using pMR3785 as DNA template and the primers 97_parA.for (5′-aaaaaatctagaatggtgcactgcatcacgcg-3′) and 98_yfp.rev (5′-aaaaaaggtaccttacttgtacagctcgtcca-3′), was digested with KpnI and XbaI, and cloned into a KpnI-XbaI-digested pMR4048, producing the plasmid pMR4051.
To generate the M. xanthus parB conditional mutant strain, MR2472, the wild type DK1050 strain was electroporated with plasmid pMR3595, obtaining the strain MR2461. This strain contains the Pvan-parB sequence integrated at the M. xanthus 1.38-kb locus. Then, MR2461 was electroporated with plasmid pMR3620, which contains sequences upstream and downstream of parB in the genome to generate a parB deletion and the galK gene, creating strain MR2462. MR2462 was grown for several generations with 0.5 mM of vanillate and no Kan and plated on CTT plates supplemented with 2% galactose and 0.5 mM of vanillate to select for the loss of the GalS marker. This evicts vector DNA bearing either wild type parB or the ΔparB allele by intramolecular recombination events. GalR KanS colonies were diagnosed by PCR to isolate a strain harboring the inducible Pvan-parB construct and the ΔparB allele (MR2472). The strain MR2504 was obtained electroporating plasmid pMR3785 into the wild type DK1050 strain. The strain MR2526 was obtained electroporating the strain MR2520 with plasmid pMR3826, and MR2520 by electroporating DK1050 with plasmid pMR3828. The strain MR2536 was generated by electroporating MR2916, the strain that conditionally expresses ftsZ from an IPTG-inducible promoter [33], with plasmid pMR3826 in presence of 1 mM IPTG. The strain MR2538 was obtained by electroporating the parB conditional mutant strain MR2472 with plasmid pMR4051, in presence of 0.5 mM vanillate.
ParB Expression and Purification
To overexpress intein-tagged M. xanthus ParB, 10 ml starter culture of freshly transformed E. coli BL21(DE3) containing plasmid pMR3684 was grown at 37°C in LB medium with 100 µg/ml of ampicillin (Amp) to an OD600 of 0.6. It was added to 1 l of fresh LB/Amp, grown at 37°C to an OD600 of 0.55, and after 30 min incubation at 18°C, overexpression of intein-tagged ParB was induced overnight at 18°C with 1 mM IPTG. After overnight induction with IPTG, cells were harvested by centrifugation (15 min at 5000×g) and the pellet was stored at –70°C until further use. Intein-tagged ParB was purified using chitin resin and the intein was removed by on-column intramolecular cleavage in the presence of 50 mM dithiothreitol using the IMPACT kit protocols (New England Biolabs). The cleaved protein was passed through a small amount of chitin resin a second time to remove residual intein and dialyzed extensively against 25 mM Tris pH 8, 50 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol and 2 mM β-mercaptoethanol.
Mobility Shift Assays
Electrophoretic mobility shift assays (EMSA) in agarose gels: The 3120-bp DNA probe containing the 22-repeat parS-cluster was obtained by PCR using primers 58_parS.for (5′-ccgttcgctttcgtgacgggtccaggttcc-3′) and 59_parS.rev (5′-agtaacgcagcgtcagcaccacttcgacgt-3′) 32P end-labeled with [γ-32P]ATP and T4 polynucleotide kinase, and DK1050 genomic DNA as a template. For the 3005-bp attP probe, the primers used were 78_attP.for (5′-aaaaaaaagcttggggatggagccagacgg-3′) and 79_attP.rev (5′-aaaaaaaagcttgggatgcggtggaccatg-3′), and pMAT4 [58] as DNA template. 15 µl samples with DNA at 1 nM, was incubated with M. xanthus ParB protein for 30 minutes at 30°C in binding buffer (40 mM Na-phosphate pH 8, 20 mM NaCl, 7% glycerol, 20 µg/ml BSA, and 100 µg/ml sheared salmon sperm DNA), and loaded onto an 0.7% agarose gel and run at 100 V at 4°C in 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). Gels were dried and analyzed by autoradiography. In competitive binding assays, 227 nM of unlabeled DNA probe was incubated with ParB protein for 1 hour at 30°C, before the inclusion of the 1 nM 32P-labeled DNA sample.
EMSA in polyacrylamide gels: A 50 bp DNA duplex that contains a parS site was generated diluting oligonucleotides 60_parS.hib (5′-tgctcgagtcatccttcgttccacgtggaacacggaggccatgagtgagt-3′) and 61_parS.hib for parS (5′- actcactcatggcctccgtgttccacgtggaacgaaggatgactcgagca-3′) to a final concentration of 5 µM each. A 50 bp DNA duplex that contains a mutated parS site was generated diluting oligonucleotides 89_parS.hib (5′- tgctcgagtcatccttcaccttgtgcagggtacggaggccatgagtgagt-3′) and 90_parS.hib (5′- actcactcatggcctccgtaccctgcacaaggtgaaggatgactcgagca-3′) to a final concentration of 5 µM each. Each mixture was heated to 95°C for 10 min and slowly cooled to room temperature, and then 32P end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. Labeled DNA sample was incubated for 1 hour at 30°C with M. xanthus ParB protein in binding buffer, and 15 µl samples were loaded on a 6% polyacrylamide gels (37.5∶1 acrylamide:bis-acrylamide), and run at 150 V at 4°C. Gels were dried and analyzed by autoradiography.
Microscopy
Samples (100 µl) of M. xanthus cultures taken at an optical density of 0.1 at 550 nm were incubated, when appropriate, with the fluorescence dye 4′-6-diamino-2-phenylindole (DAPI) to achieve a final concentration of 2 ng/µl for 10 minutes. A 1 µl drop of this mixture was immobilized on 1% agarose (Pronadisa) slices prepared in TPM medium (10 mM Tris-hydrochloride pH 7.6, 1 mM KH2PO4-K2HPO4 pH 7.6, and 8 mM MgSO4). Cells were visualized with Nikon Eclipse 80i microscope equipped with a Nikon Plan Apo VC 100×/1.4 differential interference contrast (DIC) objective and a Hamamatsu ORCA-AG charge-coupled-device camera. Images were processed with Metamorph version 4.5 (Universal Imaging Group) and Photoshop CS3 10.0 (Adobe Systems). Each reported image is representative and was verified in at least three separate experiments.
Addendum in Proof
While this paper was under review, similar findings were reported by Harms et al. (2013) [59], who also showed that ParB and ParA are essential proteins, examined their subcellular localization patterns, and confirmed the in vitro binding of ParB to a consensus parS sequence and ParB participation in chromosome partitioning. The present work suggests that, in addition, ParB helps in correct chromosome segregation by inhibiting the nonspecific interaction between ParA and DNA and thereby prevents ParA colocalization with chromosomal DNA. It is also shown here that the polar and mid-cell localization pattern of ParA does not depend on the presence of FtsZ, the critical element for bacterial cell division.
Acknowledgments
I specially thank Professors Monserrat Elías-Arnanz and Francisco Murillo for the use of fluorescence microscopy and other laboratory facilities, Dr. Francisco García-Heras and Prof. Monserrat Elías-Arnanz for providing M. xanthus ParB protein, Drs. Esteban Toro, Monserrat Elías-Arnanz and S. Padmanabhan for critical reading of the manuscript, and J. A. Madrid for technical support.
Funding Statement
Source of funding: Ministerio de Economía y Competitividad, http://www.mineco.gob.es/portal/site/mineco/. Government of Spain Grants BFU2011-255422 and RYC-2009-04190. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Toro E, Shapiro L (2010) Bacterial Chromosome Organization and Segregation. Cold Spring Harbor Perspectives in Biology 2: a000349–a000349 10.1101/cshperspect.a000349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Montero Llopis P, Rudner DZ (2013) Organization and segregation of bacterial chromosomes. Nat Rev Genet 14: 191–203 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Postow L, Hardy CD, Arsuaga J, Cozzarelli NR (2004) Topological domain structure of the Escherichia coli chromosome. Genes Dev 18: 1766–1779 10.1101/gad.1207504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mercier R, Petit M-A, Schbath S, Robin S, Karoui El M, et al. (2008) The MatP/matS Site-Specific System Organizes the Terminus Region of the E. coli Chromosome into a Macrodomain. Cell 135: 475–485 10.1016/j.cell.2008.08.031 [DOI] [PubMed] [Google Scholar]
- 5. Teleman AA, Graumann PL, Lin DC-H, Grossman AD, Losick R (1998) Chromosome arrangement within a bacterium. Current Biology 8: 1102–1109 10.1016/S09609822(98)704646 [DOI] [PubMed] [Google Scholar]
- 6. Vallet-Gely I, Boccard F (2013) Chromosomal Organization and Segregation in Pseudomonas aeruginosa . PLoS Genet 9: e1003492 10.1371/journal.pgen.1003492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, et al. (2004) Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA 101: 9257–9262 10.1073/pnas.0402606101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S (2006) Progressive segregation of the Escherichia coli chromosome. Mol Microbiol 61: 383–393 10.1111/j.13652958.2006.05245.x [DOI] [PubMed] [Google Scholar]
- 9. Lesterlin C, Gigant E, Boccard F, Espéli O (2012) Sister chromatid interactions in bacteria revealed by a site-specific recombination assay. EMBO J 31: 3468–3479 10.1038/emboj.2012.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lemon KP, Grossman AD (2001) The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev 15: 2031–2041 10.1101/gad.913301 [DOI] [PubMed] [Google Scholar]
- 11. Dworkin J, Losick R (2002) Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA 99: 14089–14094 10.1073/pnas.182539899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woldringh CL (2002) The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol Microbiol 45: 17–29 10.1046/j.13652958.2002.02993.x [DOI] [PubMed] [Google Scholar]
- 13. Jun S, Mulder B (2006) Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA 103: 12388–12393 10.1073/pnas.0605305103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mierzejewska J, Jagura-Burdzy G (2012) Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67: 1–14 10.1016/j.plasmid.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 15. Livny J, Yamaichi Y, Waldor MK (2007) Distribution of Centromere-Like parS Sites in Bacteria: Insights from Comparative Genomics. J Bacteriol 189: 8693–8703 10.1128/JB.0123907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, et al. (2010) A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12: 791–798 10.1038/ncb2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toro E, Hong S-H, McAdams HH, Shapiro L (2008) Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA 105: 15435–15440 10.1073/pnas.0807448105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murray H, Errington J (2008) Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135: 74–84 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- 19. Gerdes K, Howard M, Szardenings F (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141: 927–942 10.1016/j.cell.2010.05.033 [DOI] [PubMed] [Google Scholar]
- 20. Kaiser D, Robinson M, Kroos L (2010) Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harbor Perspectives in Biology 2: a000380 10.1101/cshperspect.a000380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Ducret A, Shaevitz J, Mignot T (2012) From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus . FEMS Microbiol Rev 36: 149–164 10.1111/j.15746976.2011.00307.x [DOI] [PubMed] [Google Scholar]
- 22. Velicer GJ, Vos M (2009) Sociobiology of the myxobacteria. Annu Rev Microbiol 63: 599–623 10.1146/annurev.micro.091208.073158 [DOI] [PubMed] [Google Scholar]
- 23. Elías-Arnanz M, Padmanabhan S, Murillo FJ (2011) Light-dependent gene regulation in nonphototrophic bacteria. Curr Opin in Microbiol 14: 128–135 10.1016/j.mib.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 24. Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, et al. (2006) Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci USA 103: 15200–15205 10.1073/pnas.0607335103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koch MK, McHugh CA, Hoiczyk E (2011) BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol 80: 1031–1051 10.1111/j.13652958.2011.07629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treuner-Lange A, Aguiluz K, van der Does C, Gómez-Santos N, Harms A, et al. (2012) PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus Mol Microbiol. 10.1111/mmi.12094 [DOI] [PubMed]
- 27. Breier AM, Grossman AD (2007) Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol 64: 703–718 10.1111/j.13652958.2007.05690.x [DOI] [PubMed] [Google Scholar]
- 28. Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF (2000) Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J Bacteriol 182: 1313–1320 10.1128/JB.182.5.13131320.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin DC, Grossman AD (1998) Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685 10.1016/S00928674(00)811356 [DOI] [PubMed] [Google Scholar]
- 30. Murray H, Ferreira H, Errington J (2006) The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol 61: 1352–1361 10.1111/j.13652958.2006.05316.x [DOI] [PubMed] [Google Scholar]
- 31. Orndorff P, Stellwag E, Starich T, Dworkin M, Zissler J (1983) Genetic and physical characterization of lysogeny by bacteriophage MX8 in Myxococcus xanthus . J Bacteriol 154: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G (2009) ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology 155: 1080–1092 10.1099/mic.0.0246610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iniesta AA, García-Heras F, Abellón-Ruiz J, Gallego-García A, Elías-Arnanz M (2012) Two systems for conditional gene expression in Myxococcus xanthus inducible by isopropyl-β-D-thiogalactopyranoside or vanillate. J Bacteriol 194: 5875–5885 10.1128/JB.0111012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thanbichler M, Iniesta AA, Shapiro L (2007) A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus . Nucleic Acids Res 35: e137–e137 10.1093/nar/gkm818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charaka VK, Misra HS (2012) Functional characterization of the role of the chromosome I partitioning system in genome segregation in Deinococcus radiodurans . J Bacteriol 194: 5739–5748 10.1128/JB.0061012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mohl DA, Gober JW (1997) Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus . Cell 88: 675–684 10.1016/S0092-B674(00)819108 [DOI] [PubMed] [Google Scholar]
- 37. Charaka VK, Misra HS (2012) Functional characterization of the role of the chromosome I partitioning system in genome segregation in Deinococcus radiodurans . J Bacteriol 194: 5739–5748 10.1128/JB.0061012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ireton K, Gunther NW, Grossman AD (1994) spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis . J Bacteriol 176: 5320–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohl DA, Easter J, Gober JW (2001) The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus . Mol Microbiol 42: 741–755 10.1046/j.13652958.2001.02643.x [DOI] [PubMed] [Google Scholar]
- 40. Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G (2004) ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J Bacteriol 186: 6983–6998 10.1128/JB.186.20.69836998.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dubarry N, Pasta F, Lane D (2006) ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J Bacteriol 188: 1489–1496 10.1128/JB.188.4.14891496.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saint-Dic D, Frushour BP, Kehrl JH, Kahng LS (2006) A parA homolog selectively influences positioning of the large chromosome origin in Vibrio cholerae . J Bacteriol 188: 5626–5631 10.1128/JB.0025006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schofield WB, Lim HC, Jacobs-Wagner C (2010) Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J 29: 3068–3081 10.1038/emboj.2010.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fogel MA, Waldor MK (2006) A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20: 3269–3282 10.1101/gad.1496506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ringgaard S, van Zon J, Howard M, Gerdes K (2009) Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA 106: 19369–19374 10.1073/pnas.0908347106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwińska J, Chater KF (2007) Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae . Mol Microbiol 65: 625–641 10.1111/j.13652958.2007.05815.x [DOI] [PubMed] [Google Scholar]
- 47. Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 10.1038/nrmicro2198 [DOI] [PubMed] [Google Scholar]
- 48. Erickson HP, Anderson DE, Osawa M (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74: 504–528 10.1128/MMBR.0002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mingorance J, Rivas G, Vélez M, Gómez-Puertas P, Vicente M (2010) Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol 18: 348–356 10.1016/j.tim.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 50. Marston AL, Errington J (1999) Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol Cell 4: 673–682 10.1016/S10972765(00)803780 [DOI] [PubMed] [Google Scholar]
- 51. Quisel JD, Lin DC, Grossman AD (1999) Control of development by altered localization of a transcription factor in B. subtilis . Mol Cell 4: 665–672 10.1016/S10972765(00)803779 [DOI] [PubMed] [Google Scholar]
- 52. Leonard TA, Butler PJ, Löwe J (2005) Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J 24: 270–282 10.1038/sj.emboj.7600530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Easter J, Gober JW (2002) ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell 10: 427–434 10.1016/S10972765(02)005944 [DOI] [PubMed] [Google Scholar]
- 54. Donovan C, Schwaiger A, Krämer R, Bramkamp M (2010) Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum . J Bacteriol 192: 3441–3451 10.1128/JB.0021410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bretscher AP, Kaiser D (1978) Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol 133: 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ueki T, Inouye S, Inouye M (1996) Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183: 153–157 10.1016/S03781119(96)00546-X [DOI] [PubMed] [Google Scholar]
- 57. Pérez-Marín MC, López-Rubio JJ, Murillo FJ, Elías-Arnanz M, Padmanabhan S (2004) The N terminus of Myxococcus xanthus CarA repressor is an autonomously folding domain that mediates physical and functional interactions with both operator DNA and antirepressor protein. J Biol Chem 279: 33093–33103 10.1074/jbc.M405225200 [DOI] [PubMed] [Google Scholar]
- 58. Gomez-Santos N, Treuner-Lange A, Moraleda-Munoz A, Garcia-Bravo E, Garcia-Hernandez R, et al. (2012) A comprehensive set of integrative plasmid vectors for copper inducible gene expression in Myxococcus xanthus . Appl Environ Microbiol 78: 2515–2521 10.1128/AEM.0750211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harms A, Treuner-Lange A, Schumacher D, Søgaard-Andersen L (2013) Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet 9: e1003802 10.1371/journal.pgen.1003802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crooks GE, Hon G, Chandonia J-M, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ruiz-Vázquez R, Murillo FJ (1984) Abnormal motility and fruiting behavior of Myxococcus xanthus bacteriophage-resistant strains induced by a clear-plaque mutant of bacteriophage Mx8. J Bacteriol 160: 818–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kaiser D (1979) Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA 76: 5952–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]