Abstract
Purpose
This study addresses the species-specific and site-specific details of weight-bearing articular cartilage zone depths and chondrocyte distributions among humans and common osteoarthritis (OA) animal models using contemporary digital imaging tools. Histological analysis is the gold-standard research tool for evaluating cartilage health, OA severity, and treatment efficacy. Historically, evaluations were made by expert analysts. However, state-of-the-art tools have been developed that allow for digitization of entire histological sections for computer-aided analysis. Large volumes of common digital cartilage metrics directly complement elucidation of trends in OA inducement and concomitant potential treatments.
Materials and methods
Sixteen fresh human knees, 26 adult New Zealand rabbit stifles, and 104 bovine lateral plateaus were measured for four cartilage zones and the cell densities within each zone. Each knee was divided into four weight-bearing sites: the medial and lateral plateaus and femoral condyles.
Results
One-way analysis of variance followed by pairwise multiple comparisons (Holm–Sidak method at a significance of 0.05) clearly confirmed the variability between cartilage depths at each site, between sites in the same species, and between weight-bearing articular cartilage definitions in different species.
Conclusion
The present study clearly demonstrates multisite, multispecies differences in normal weight-bearing articular cartilage, which can be objectively quantified by a common digital histology imaging technique. The clear site-specific differences in normal cartilage must be taken into consideration when characterizing the pathoetiology of OA models. Together, these provide a path to consistently analyze the volume and variety of histologic slides necessarily generated by studies of OA progression and potential treatments in different species.
Keywords: knee, osteoarthritis, imaging, rabbit, bovine, cell density
Introduction
Joint trauma has proven to be a key factor in the development of osteoarthritis (OA),1,2 which is the most prevalent joint disease and the leading cause of disability in the adult human population.3 Posttraumatic OA caused by battlefield injuries is a primary disabling condition in warriors.4 Investigations of underlying etiology and pathophysiology of OA, and the pursuit of potential acute injury-modifying treatments to prevent OA, have utilized both in vitro explant and in vivo animal models from a variety of small (rabbits and mice) and large (cows5 and pigs6) animal species. Evaluating cartilage in these research models requires objective measures of pathological OA characteristics.
Several scoring mechanisms exist for grading human OA (Outerbridge,7 Mankin,8 Osteoarthritis Research Society International [OARSI],9 etc), and recently, the OARSI histopathology initiative achieved a consensus of scoring systems for evaluating many of the most important animal species used in OA research:10,11 dog, guinea pig, horse, mouse, rabbit, rat, sheep, and goat. To address the need for comprehensive, whole-joint histological assessment, the term “grading” was defined for microscopic/macroscopic scoring of tissues, and the term “staging” referred to overall assessment of the disease status. The histologists’ time necessary to grade and stage each slide must be weighed against the volume of specimens typically involved in a single animal study, let alone the additional demands of coordinating or comparing multicenter studies.
Until recently,12 these grading/scoring systems were susceptible to subjective application by individual observers. More objective applications of such scales exploiting semiautomated or automated digital methods are possible, but require specific quantification of normal and abnormal cartilage characteristics, such as cell density within site-specific cartilage zones. The present study introduces the use of contemporary digital image-analysis tools for objectively evaluating cartilage characteristics, useful when assigning OA scores in different anatomic sites and in multiple species. The results provide quantifiable differences in “normal” cartilage histology between four primary knee weight-bearing compartments in human knees and two common animal stifle models of OA conditions.
Materials and methods
Twenty-six stifle joints were collected from 26 normal adult New Zealand rabbits (4–5 kg males, 12–15 months old),13 killed at the end of unrelated, non-cartilage-affecting experiments that were conducted in accordance with Institutional Animal Care and Use Committee guidelines. Each knee was separated into four parts: medial and lateral femoral condyles, and medial and lateral tibial plateaus. Bovine stifle specimens (Black Angus, Angus/Simmental cross, yearling steers) were obtained from a local abattoir, and large rectangular osteochondral explants encompassing the meniscus-uncovered weight-bearing region of the lateral plateaus were harvested. Sixteen fresh human knees were garnered from surgical discard after amputations for proximal sarcomas (36 ± 17 years old, eight male and eight female), according to human subjects’ approval.
Histological preparations
Osteochondral specimens were fixed in neutral-buffered 10% formalin, and then decalcified in 5% formic acid. Decalcification was monitored by chemical end-point assay. Decalcified specimens were rinsed thoroughly in distilled water, dehydrated in graded alcohols, cleared in xylene, and embedded in paraffin (TissuePrep II; Thermo Fisher Scientific, Waltham, MA, USA). Five-micron-thick microtome slices were selected for staining with Weigert’s hematoxylin, safranin-O, and fast green. Weigert’s hematoxylin stained cell nuclei black, increasing contrast for subsequent digital identification of nuclear material. Safranin-O is a cationic dye that binds specifically to sulfated glycosaminoglycans, and the intensity of the red stain is indicative of proteoglycan content.14 Fast green provides a contrasting counterstain. Specimens were prepared according to OARSI recommendations for histological analysis,15 except for two notable modifications to the safranin-O staining protocol. The first modification was that after deparaffinizing mounted sections in xylene, slides were immersed one extra time in 100% ethyl alcohol before rehydration (as per guidelines) to water in graded alcohols for staining. The second modification was an additional decolorizing step in 95% ethyl alcohol. Sections were then fully dehydrated, cleared, and mounted as per OARSI guidelines.
Digital image capture
An essential aspect of grading cartilage is adopting a standardized slide preparation. Equally important is the need to maintain a set of standards during digital image capture that accurately retains the color and dimensions of the slide specimen. Histological sections were digitized with a QICam (QImaging, Surrey, BC, Canada) 12-bit camera mounted on an Olympus (Tokyo, Japan) BX60 microscope with 4 × objective, coupled with a stepper motor-driven stage (Prior Scientific, Rockland, MD, USA). Light intensity was set to color neutral for the camera, with a minimum amount of in-line blue filter for truer perceived color. A camera shutter speed of 2 ms was used with a default gain. Individual high-resolution image fields (1.60 × 1.21 mm field of view, 1392 × 1040 pixels, 863 pixels/mm) were concatenated using Image Pro (Media Cybernetics, Silver Spring, MD, USA) to produce stitched full-cartilage-thickness osteochondral images. Data were captured in the form of red, green, blue (RGB) 24-bit image files, with pixels encoded as integer intensities (0–255) for each of the three independent color channels.
A second imaging system was also utilized to demonstrate applicability across histological digitizing platforms. An Olympus VS110 Virtual Microscopy System consists of a 5-slot stage that is automatically controlled in x-, y-, and z-directions to move across and autofocus each area on a histological section. The controlling software (VS-ASW) integrates autofocusing, digital acquisition, background correction, and whole-slide image reconstruction. The high-resolution (eg, 322 nm/pixel for 20 × objective scans) digital histology sections were stored in a single database, which is accessible to multiple researchers’ personal computers. The images were exported (Olympus Visiomorph) at full resolution and in 4 × TIFF for digital histology analysis.
Histological site-specific cartilage definitions
Basic cartilage parameters were measured on digital images of the 5 µm-thick microscopy sections using Adobe (San Jose, CA, USA) Photoshop CS2 measure tool. For each of the nine weight-bearing cartilage types investigated, four zone depths were defined by cell size, organization, and orientation (Figure 1). The superficial zone contained small chondrocytes with flattened nuclei parallel to the articular surface. In the transitional zone, chondrocytes with round nuclei were scattered in a matrix region with intense safranin-O staining of proteoglycan. The radial zone was identified by columns of chondrocytes oriented perpendicularly to the articular surface, and the deep zone was identified by intense Safranin-O staining to the border of calcification (osteochondral junction). Each zone was recorded as a percentage of total cartilage thickness. Forty to fifty independent zone-depth measurements (by consensus) were recorded for each of the cartilage types (eg, four rabbit condyles × three groups of triplicate sagittal slices) at 1 mm spacing between groups (4 mm spacing in bovine and human) to incorporate anterior–posterior and medial–lateral variability.
Figure 1.
Zone definition within rabbit medial femoral condyle weight-bearing cartilage.
A total of 399 rabbit histology specimens were imaged and analyzed (62 medial femoral condyle, 92 lateral femoral condyle, 84 medial tibial plateau, and 161 lateral tibial plateau sections) from initial digital definitions on four rabbits subsequently applied in 22 additional rabbits’ stifles. A total of 273 human histology specimens were imaged and analyzed (57 medial femoral condyle, 87 lateral femoral condyle, 76 medial tibial plateau, and 53 lateral tibial plateau). The central weight-bearing region of 104 bovine lateral tibial plateau images was analyzed.
Three hundred ninety-seven chondrocytes were also measured for site-specific, zone-delimited size definitions in Photoshop. These cell dimensions were averaged on a zone-specific basis for each weight-bearing site in each of the species evaluated. Cell-size and cartilage-zone definitions were used in a custom cell-counting program developed in Matlab (MathWorks, Natick, MA, USA). The program identified cells in the weight-bearing cartilage by a black nucleus inside either the red-stained matrix or a white cytoplasm. Cell-size information was used to exclude identified features that were too large or too small, and the zone-depth definitions were implemented to partition whole-tissue cell counts into zonal cell counts. These counts were scaled with the known pixel-to-millimeter conversion factor to calculate the average zonal cell densities (cells/mm2). Program-generated zone boundaries and cell counts were manually verified on individual 0.5 mm-wide cartilage subimages of both 20× and 4× magnification. Continuing the rabbit-specific model, an additional 22 stifles were analyzed to produce the final site-specific, zone-specific cartilage cell-density mean and standard deviation evaluations.
Results
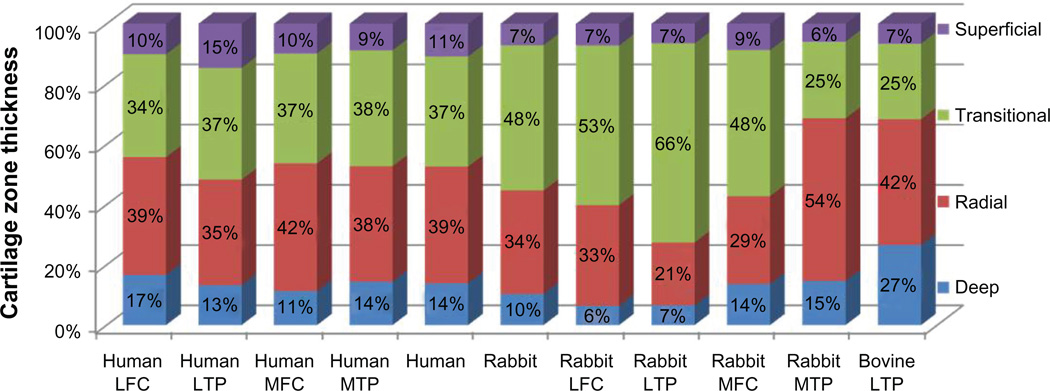
The percentage of cartilage thickness comprising each of the four cartilage zones (superficial, transitional, radial, and deep) was very similar across the four different human knee sites (Figure 2 and Table 1). The average zonal thicknesses were 11% (range 9%–15%) for the superficial, 36% (range 34%–38%) for the transitional, 40% (range 35%–42%) for the radial, and 13% (range 11%–17%) for the deep zones. In the rabbit, depths of the different cartilage zones were different from the human zonal definitions and varied from each other by site. As in humans, the superficial zone of all four rabbit sites was the thinnest, averaging 7% of total thickness (range 6%–9%). The average rabbit transitional zone thickness was thicker than the humans (48% of total thickness), but showed enormous variability by site, ranging from 25% of total cartilage thickness in the medial tibial plateau to 66% of total thickness in the lateral tibial plateau. Rabbit radial zone thicknesses were similarly variable by site, ranging from 21% to 54% of total cartilage thickness. The bovine cartilage zones were different from both the rabbits and the humans. Zone ratios of 7% (superficial), 25% (transitional), 42% (radial), and 27% (deep), make the bovine superficial and transitional zones thinner and the deep zone much thicker than the other two species’ lateral plateaus.
Figure 2.
Site-specific cartilage zone definitions.
Abbreviations: LFC, lateral femoral condyle; LTP, lateral tibial plateau; MFC, medial femoral condyle; MTP, medial tibial plateau.
Table 1.
Cartilage zone ratios, full cartilage thickness and zonal cell densities
| Site/zone |
Superficial % of depth |
Transitional % of depth |
Radial % of depth |
Deep % of depth |
Thickness mm |
|---|---|---|---|---|---|
| Zone ratios | |||||
| Human LFC (N = 37) | 10.13 ± 3.71 | 34.13 ± 5.84 | 39.13 ± 7.33 | 16.60 ± 4.94 | 2.187 ± 0.627 |
| Human LTP (N = 34) | 14.64 ± 6.01 | 37.09 ± 7.28 | 35.07 ± 7.53 | 13.19 ± 5.63 | 2.838 ± 0.959 |
| Human MFC (N = 28) | 9.83 ± 3.65 | 36.52 ± 7.17 | 42.39 ± 7.31 | 11.25 ± 2.50 | 2.155 ± 0.375 |
| Human MTP (N = 55) | 8.79 ± 3.30 | 38.49 ± 9.54 | 38.32 ± 8.01 | 14.39 ± 4.88 | 2.197 ± 0.428 |
| Human knee | 10.85 ± 4.16 | 36.56 ± 7.46 | 38.73 ± 7.55 | 13.86 ± 4.49 | |
| Rabbit stifle | 7.11 ± 2.42 | 48.23 ± 6.88 | 34.38 ± 7.27 | 10.28 ± 4.21 | |
| Rabbit LFC (N = 7) | 7.24 ± 3.74 | 52.95 ± 5.45 | 33.44 ± 8.27 | 6.37 ± 4.05 | 0.255 ± 0.087 |
| Rabbit LTP (N = 7) | 6.54 ± 1.87 | 66.05 ± 2.79 | 20.86 ± 2.67 | 6.56 ± 2.79 | 0.401 ± 0.163 |
| Rabbit MFC (N = 7) | 8.75 ± 2.18 | 48.44 ± 9.50 | 29.24 ± 9.66 | 13.56 ± 5.51 | 0.422 ± 0.182 |
| Rabbit MTP (N = 7) | 5.91 ± 1.88 | 25.47 ± 9.79 | 53.98 ± 8.48 | 14.63 ± 4.50 | 0.745 ± 0.268 |
| Bovine LTP (N = 4) | 6.57 ± 1.93 | 25.11 ± 5.25 | 41.76 ± 7.64 | 26.56 ± 7.36 | 1.844 ± 0.503 |
| Site/zone |
Superficial Cells/mm2 |
Transitional Cells/mm2 |
Radial Cells/mm2 |
Deep Cells/mm2 |
Cell diameter Microns |
| Cell density | |||||
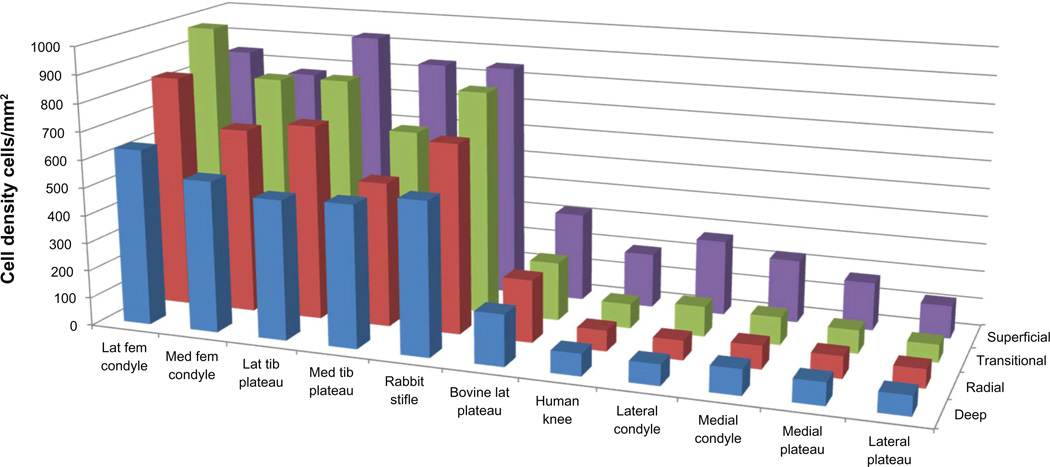
| Human LFC (N = 17) | 271 ± 101 | 108 ± 43 | 70 ± 19 | 74 ± 27 | |
| Human LTP (N = 17) | 117 ± 43 | 63 ± 31 | 66 ± 25 | 70 ± 28 | |
| Human MFC (N = 17) | 226 ± 66 | 99 ± 33 | 83 ± 37 | 91 ± 35 | |
| Human MTP (N = 17) | 172 ± 63 | 83 ± 32 | 79 ± 39 | 80 ± 30 | |
| Human knee | 197 ± 68 | 88 ± 35 | 74 ± 30 | 79 ± 30 | S = 16.9 ± 3.0 |
| D = 19.7 ± 2.6 | |||||
| Rabbit stifle | 847 ± 324 | 810 ± 152 | 682 ± 146 | 547 ± 172 | S = 13.9 ± 3.2 |
| D = 14.6 ± 3.4 | |||||
| Rabbit LFC (N = 19) | 843 ± 299 | 976 ± 132 | 838 ± 162 | 634 ± 199 | |
| Rabbit LTP (N = 19) | 929 ± 347 | 815 ± 130 | 703 ± 126 | 502 ± 178 | |
| Rabbit MFC (N = 22) | 773 ± 249 | 802 ± 197 | 668 ± 190 | 544 ± 176 | |
| Rabbit MTP (N = 22) | 843 ± 401 | 646 ± 148 | 520 ± 106 | 510 ± 136 | |
| Bovine LTP (N = 104) | 321 ± 200 | 212 ± 73 | 225 ± 61 | 184 ± 58 | |
Note: Value ± I standard deviation.
Abbreviations: S, superficial zone cells; D, deeper chondrocytes; LFC, lateral femoral condyle; LTP, lateral tibial plateau; MFC, medial femoral condyle; MTP, medial tibial plateau.
The thickness of human cartilage was similar in each of the four compartments of the knee, ranging from an average of 2.2 mm on the medial femoral condyle to 2.8 mm on the lateral tibial plateau (Table 1). As expected, the rabbit cartilage was much thinner than human, and its thickness varied by site. Rabbit cartilage was 0.3 mm thick in the lateral femoral condyle, 0.4 mm thick on the medial femoral condyle and on the lateral tibial plateau, and much thicker (0.7 mm) on the medial tibial plateau. Somewhat surprisingly, the bovine lateral plateau cartilage was thinner than human cartilage (1.9 mm), but based on the results from the other two species, it is possible that this is also one of the thinner cartilage sites in bovine stifles.
Relative dimensions of the rabbit superficial and deeper zone chondrocytes were 13.9 ± 3.2 and 14.6 ± 3.4 µm. In the larger human and bovine species, superficial and deep chondrocyte sizes were 16.9 ± 3.0 and 19.7 ± 2.6 µm, respectively. These larger measurements were due to somewhat larger chondron lacunae enclosing the cells. The wide range of zone-specific chondrocyte densities is summarized in Figure 3. The significance of pair-wise differences in multisite, multispecies zone definitions (Table 2) and cell densities (Table 3) are denoted by a Yes.
Figure 3.
Comparative digital cartilage zone cell densities.
Table 2.
Comparison of cartilage zone ratios
| Human LFC |
Human LTP |
Human MFC |
Human MTP |
Human knee |
Rabbit stifle |
Rabbit LFC |
Rabbit LTP |
Rabbit MFC |
Rabbit MTP |
Bovine LTP |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Superficial | |||||||||||
| Transitional | |||||||||||
| Human LFC | Yes | No | No | No | No | No | No | No | No | No | |
| Human LTP | No | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Human MFC | No | No | No | No | No | No | No | No | No | No | |
| Human MTP | No | No | No | No | No | No | No | No | No | No | |
| Human Knee | No | No | No | No | No | No | No | No | No | No | |
| Rabbit Stifle | Yes | Yes | Yes | Yes | Yes | No | No | No | No | No | |
| Rabbit LFC | Yes | Yes | Yes | Yes | Yes | No | No | No | No | No | |
| Rabbit LTP | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | No | No | |
| Rabbit MFC | Yes | Yes | Yes | Yes | Yes | No | No | Yes | No | No | |
| Rabbit MTP | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | |
| Bovine LTP | No | No | No | Yes | No | Yes | Yes | Yes | Yes | No | |
| Radial zones | |||||||||||
| Deep zones | |||||||||||
| Human LFC | No | No | No | No | No | No | Yes | No | Yes | No | |
| Human LTP | No | Yes | No | No | No | No | Yes | No | Yes | No | |
| Human MFC | Yes | No | No | No | Yes | No | Yes | Yes | Yes | No | |
| Human MTP | No | No | No | No | No | No | Yes | No | Yes | No | |
| Human knee | No | No | No | No | No | No | Yes | No | Yes | No | |
| Rabbit stifle | Yes | No | No | No | No | No | Yes | No | Yes | No | |
| Rabbit LFC | Yes | Yes | No | Yes | Yes | No | No | No | Yes | No | |
| Rabbit LTP | Yes | Yes | No | Yes | Yes | No | No | No | Yes | Yes | |
| Rabbit MFC | No | No | No | No | No | No | No | No | Yes | No | |
| Rabbit MTP | No | No | No | No | No | No | Yes | Yes | No | No | |
| Bovine LTP | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
Notes: The significance of pairwise differences in multisite, multispecies zone definitions is denoted by a Yes in the corresponding box, for each of the four cartilage zones. Red triangle contains the superficial zone values, green for transitional, brown for radial, and blue for deep zone significance.
Table 3.
Comparison of cell densities by cartilage zone
| Human LFC |
Human LTP |
Human MFC |
Human MTP |
Human knee |
Rabbit stifle |
Rabbit LFC |
Rabbit LTP |
Rabbit MFC |
Rabbit MTP |
Bovine LTP |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Superficial | |||||||||||
| Transitional | |||||||||||
| Human LFC | No | No | No | No | Yes | Yes | Yes | Yes | Yes | No | |
| Human LTP | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Human MFC | No | No | No | No | Yes | Yes | Yes | Yes | Yes | No | |
| Human MTP | No | No | No | No | Yes | Yes | Yes | Yes | Yes | No | |
| Human Knee | No | No | No | No | Yes | Yes | Yes | Yes | Yes | No | |
| Rabbit Stifle | Yes | Yes | Yes | Yes | Yes | No | No | No | No | Yes | |
| Rabbit LFC | Yes | Yes | Yes | Yes | Yes | Yes | No | No | No | Yes | |
| Rabbit LTP | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | Yes | |
| Rabbit MFC | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | Yes | |
| Rabbit MTP | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Bovine LTP | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Radial zones | |||||||||||
| Deep zones | |||||||||||
| Human LFC | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Human LTP | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Human MFC | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Human MTP | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Human Knee | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| Rabbit Stifle | Yes | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes | |
| Rabbit LFC | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | |
| Rabbit LTP | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Yes | Yes | |
| Rabbit MFC | Yes | Yes | Yes | Yes | Yes | No | No | No | Yes | Yes | |
| Rabbit MTP | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | Yes | |
| Bovine LTP | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | No |
Note: In the same format as Table 2, the significance of pairwise differences in multisite, multispecies cell densities are denoted by a Yes in the corresponding box, for each of the four cartilage zones.
Discussion
While common (plain radiograph) and emerging (cartilage-specific magnetic resonance imaging [MRI]) imaging tools are used to diagnose noninvasively OA and OA severity in the clinic, the definitive assessment of OA severity in a research context is often made based on histologic appearance of articular cartilage. There are a variety of scales used to evaluate histological OA severity, with the most commonly utilized index being the Mankin 14-point histological-histochemical grading scale.8 On this scale, cartilage is graded on structural compromise (0–6 points), loss of matrix staining (0–4), cellularity anomalies (0–3), and violation of tidemark integrity (0–1), with normal = 0. Mankin scorings have been dependent on human observer subjectivity, and thus have drawn criticism on the basis of the associated inter- and intraobserver variability.16
Four cartilage zone depths were defined by cell size, organization, and orientation. The deep zone was described as the cellular portion of the calcified cartilage interdigitation with the subchondral bone (Figure 1). While deep zone and radial zone are often used interchangeably in the literature, this explicit definition of deep zone provides the additional benefit of direct geometric correlation to the bone–cartilage interface of cartilage-specific quantitative MRI sequences, such as T1ρ and dGEMRIC emerging as potential noninvasive in vivo measures of cartilage condition (ie, imaging bio-markers of OA onset). Polarized light microscopy may more clearly distinguish the border between the radial and tangential zones. However, slide preparation differences would have required separate subjacent specimen microsections, which then introduce potential variability in paired-slide zone measurements.17,18 Improvement on cell-based zone definitions would likely be less than a handful of microns, and within the reported measurement variability.
Extension of such a Mankin-like grading scale to animal cartilage requires an adaptation to accommodate natural differences between normal human and normal animal cartilage. For example, a section of rabbit cartilage appears quite hypercellular relative to normal human cartilage, and could mistakenly be graded as having that particular characteristic of OA, even though the sample is normal for rabbit cartilage. At minimum, a species-specific normal definition must be established for correct application of a cartilage grading scale. Cell densities in the rabbit were 5–10 times those of human cartilage, which is consistent with previously reported measurements.19 Bovine chondrocyte density was about twice that of its human counterpart. Comparable interspecies histomorphometry parameters are reviewed in the OARSI histopathology initiative.20
This study reports comparable cartilage thickness measurements for human, but also a marked difference in rabbit medial and lateral condylar thicknesses. The thickness of human cartilage ranged from an average of 2.2 mm on the medial femoral condyle to 2.8 mm on the lateral tibial plateau. Rabbit cartilage was 0.3 mm thick in the lateral femoral condyle, 0.4 mm thick on the medial femoral condyle and on the lateral tibial plateau, and much thicker (0.7 mm) on the medial tibial plateau. These site-specific articular cartilage thicknesses are similar to reported average cartilage thicknesses of 2.2–2.5 mm in the human knee and 0.3 mm for rabbit lateral stifle compartment cartilage.21
Even within a species, if a one-size-fits-all normal cartilage definition is adopted, joint-to-joint and site-to-site biologic variation will likely produce an inaccurate assessment of cartilage condition. Stockwell alluded to this in his measurements of mammalian cartilage.19 In this study, it was found that cartilage zone depths and normal cell densities varied widely by site, even within the same joint. This difference was pronounced in the rabbit, and much less apparent in the human cartilage. The inherent site-specific variability in the composition of cartilage used to investigate OA has major implications for the acceptance or the rejection of treatments. A reference database of normal multisite, multispecies cartilage compositions measured by common digital histology methods provides an objective means to quantify trends in animal-model cartilage response to potential OA treatments, between models and laboratories. These differences must be borne in mind when interpreting animal-model studies of cartilage and their extension to the human OA condition.
Conclusion
The present study compares rabbit, human, and bovine cartilage measurements that reveal the site-dependent and interspecies variations of cartilage by using contemporary digital imaging tools. These data demonstrate multisite, multispecies differences in normal weight-bearing articular cartilage, which can be objectively quantified by a common digital histology imaging technique. The clear site-specific differences in normal cartilage must be taken into consideration when characterizing the pathoetiology of OA models. Together, these provide a path to analyze consistently the volume and variety of histologic slides necessarily generated by studies of OA progression and potential treatments in different species.
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number AR055533. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All studies are in accordance with IACCUC and IRB guidelines.
Footnotes
This is an Open Access article which permits unrestricted noncommercial use, provided the original work is properly cited.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Buckwalter JA, Brown TD. Joint injury, repair, and remodeling: roles in post-traumatic osteoarthritis. Clin Orthop Relat Res. 2004;423:7–16. [PubMed] [Google Scholar]
- 2.Felson DT, Lawrence RC, Dieppe PA, et al. Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med. 2000;133(8):635–646. doi: 10.7326/0003-4819-133-8-200010170-00016. [DOI] [PubMed] [Google Scholar]
- 3.Brown TD, Johnston RC, Saltzman CL, Marsh JL, Buckwalter JA. Posttraumatic osteoarthritis: a first estimate of incidence, prevalence, and burden of disease. J Orthop Trauma. 2006;20(10):739–744. doi: 10.1097/01.bot.0000246468.80635.ef. [DOI] [PubMed] [Google Scholar]
- 4.Rivera JC, Wenke JC, Buckwalter JA, Ficke JR, Johnson AE. Posttraumatic osteoarthritis caused by battlefield injuries: the primary source of disability in warriors. J Am Acad Orthop Surg. 2012;20(Suppl 1):S64–S69. doi: 10.5435/JAAOS-20-08-S64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin JA, McCabe D, Walter M, Buckwalter JA, McKinley TO. N-acetylcysteine inhibits post-impact chondrocyte death in osteochondral explants. J Bone Joint Surg Am. 2009;91(8):1890–1897. doi: 10.2106/JBJS.H.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakrishnan P, Hecht BA, Pedersen DR, et al. Oxidant conditioning protects cartilage from mechanically induced damage. J Orthop Res. 2010;28(7):914–920. doi: 10.1002/jor.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Outerbridge RE. The etiology of chondromalacia patellae. J Bone Joint Surg Br. 1961;43-B:752–757. doi: 10.1302/0301-620X.43B4.752. [DOI] [PubMed] [Google Scholar]
- 8.Mankin HJ, Dorfman H, Lippiello L, Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am. 1971;53(3):523–537. [PubMed] [Google Scholar]
- 9.Pritzker KP, Gay S, Jimenez SA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14(1):13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Aigner T, Cook JL, Gerwin N, et al. Histopathology atlas of animal model systems – overview of guiding principles. Osteoarthritis Cartilage. 2010;18(Suppl 3):S2–S6. doi: 10.1016/j.joca.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Berenbaum F. The OARSI histopathology initiative – the tasks and limitations. Osteoarthritis Cartilage. 2010;18(Suppl 3):S1. doi: 10.1016/j.joca.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Moussavi-Harami SF, Pedersen DR, Martin JA, Hillis SL, Brown TD. Automated objective scoring of histologically apparent cartilage degeneration using a custom image analysis program. J Orthop Res. 2009;27(4):522–528. doi: 10.1002/jor.20779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaseenon T, Tochigi Y, Heiner AD, et al. Organ-level histological and biomechanical responses from localized osteoarticular injury in the rabbit knee. J Orthop Res. 2011;29(3):340–346. doi: 10.1002/jor.21259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg L. Chemical basis for the histological use of safranin O in the study of articular cartilage. J Bone Joint Surg Am. 1971;53(1):69–82. [PubMed] [Google Scholar]
- 15.Schmitz N, Laverty S, Kraus VB, Aigner T. Basic methods in histopathology of joint tissues. Osteoarthritis Cartilage. 2010;18(Suppl 3):S113–S116. doi: 10.1016/j.joca.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 16.Ostergaard K, Petersen J, Andersen CB, Bendtzen K, Salter DM. Histologic/histochemical grading system for osteoarthritic articular cartilage: reproducibility and validity. Arthritis Rheum. 1997;40(10):1766–1771. doi: 10.1002/art.1780401007. [DOI] [PubMed] [Google Scholar]
- 17.Changoor A, Tran-Khanh N, Méthot S, et al. A polarized light microscopy method for accurate and reliable grading of collagen organization in cartilage repair. Osteoarthritis Cartilage. 2011;19(1):126–135. doi: 10.1016/j.joca.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Julkunen P, Harjula T, Iivarinen J, et al. Biomechanical, biochemical and structural correlations in immature and mature rabbit articular cartilage. Osteoarthritis Cartilage. 2009;17(12):1628–1638. doi: 10.1016/j.joca.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Stockwell RA. The interrelationship of cell density and cartilage thickness in mammalian articular cartilage. J Anat. 1971;109(Pt 3):411–421. [PMC free article] [PubMed] [Google Scholar]
- 20.Pastoureau PC, Hunziker EB, Pelletier JP. Cartilage, bone and synovial histomorphometry in animal models of osteoarthritis. Osteoarthritis Cartilage. 2010;18(Suppl 3):S106–S112. doi: 10.1016/j.joca.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Frisbie DD, Cross MW, McIlwraith CW. A comparative study of articular cartilage thickness in the stifle of animal species used in human pre-clinical studies compared to articular cartilage thickness in the human knee. Ve t Comp Orthop Traumatol. 2006;19(3):142–146. [PubMed] [Google Scholar]