Abstract
The gas phase dissociation behavior of peptides containing acyl-arginine residues is investigated. These acylations are generated via a combination of ion/ion reactions between arginine-containing peptides and N-hydroxysuccinimide (NHS) esters and subsequent tandem mass spectrometry (MS/MS). Three main dissociation pathways of acylated arginine, labeled Paths 1-3, have been identified and are dependent on the acyl groups. Path 1 involves the acyl-arginine undergoing deguanidination, resulting in the loss of the acyl group and dissociation of the guanidine to generate an ornithine residue. This pathway generates selective cleavage sites based on the recently discussed “ornithine effect”. Path 2 involves the coordinated losses of H2O and NH3 from the acyl-arginine side chain while maintaining the acylation. We propose that Path 2 is initiated via cyclization of the δ-nitrogen of arginine and the C-terminal carbonyl carbon, resulting in rapid rearrangement from the acyl-arginine side chain and the neutral losses. Path 3 occurs when the acyl group contains α-hydrogens and is observed as a rearrangement to regenerate unmodified arginine while the acylation is lost as a ketene.
Keywords: Ion/ion, gas phase covalent modification, acyl-arginine, dissociation behavior, CID
1. Introduction
Chemical modification of specific functional groups of proteins or peptides has long played a role in bioanalysis. Traditionally, solution-based experiments have largely involved covalent modifications to specific functionalities, such as the modification of primary amines of lysine or N-termini using N-hydroxysuccinimide (NHS) esters or the modification of sulfhydryls of cysteines using maleimides. Covalent modifications in mass spectrometry applications have been implemented in order to facilitate ionization,1 elucidate structural information via cross-linking,2,3,4,5,6,7,8 enhance or alter fragmentation behavior,9,10,11 label with chromophores,12 or prepare reactive surfaces for gas phase reactions.13 Conjugation approaches with strong analogies to those used in solution have recently been pursued in the gas phase in order to characterize the analytes of interest using tandem mass spectrometry (MS/MS) experiments. We have recently shown, for example, the reactivity of primary amines,7,11,14,15,16,17,18,19 such as the N-terminus and the ε-amine of lysine, and of the guanidine of arginine20 with sulfo-NHS esters in the gas phase via ion/ion reactions. The ability to make selective covalent modifications to peptides and proteins in the gas phase is attractive from the standpoint of speed, selectivity, and flexibility in using covalent modifications in biological mass spectrometry. These gas phase reactions often involve the interaction of a multiply protonated peptide with a singly deprotonated bi-functional anionic reagent to form a long-lived electrostatic complex. The bi-functionality of the anionic reagent includes a “sticky” charge site, often a sulfonate, capable of maintaining a long-lived electrostatic interaction with a protonated site of a peptide. The second functionality of the reagent is a reactive site, such as an (sulfo-)NHS ester. Upon activation of the electrostatic complex, nucleophilic attack of an unprotonated amine or guanidine on the carbonyl carbon of the ester results in the formation of an amide bond and loss of (sulfo-)NHS, which is a signature for the modification in the gas phase.7,8,15,16,20
Condensed phase reactions with primary amines are well known and have been used extensively. This has allowed the thorough study of fragmentation behavior of both acyl-lysine and acyl-N-terminal amines. Gas phase activation of peptides containing acyl-lysine residues exhibit predictable fragmentation patterns, wherein the most favorable cleavages occur at the backbone amide bonds. In contrast, due to the high basicity of arginine (pKA>12)21 and the propensity for esters to hydrolyze under highly basic aqueous conditions, condensed phase reactions with arginine are restricted to very specific reaction conditions in anhydrous solvents with reaction times requiring several days.22 For this reason, there has been little motivation for exploring the dissociation behavior of acyl-arginine containing peptides in the condensed phase. The gas phase, however, provides an environment in which both unprotonated arginine and esters of (sulfo-)NHS are able to readily interact, allowing for the acylation of arginine to be performed. This report explores the dissociation behavior of acyl-arginine in the gas phase. Unlike the acyl-modified lysines, the identity of the acyl substituent group has a strong effect on the preferred fragmentation channels of acyl-arginine, which is relevant to the selection of reagents for gas phase covalent modification of arginine-containing peptide and protein ions.
2. Experimental
2.1 Materials
Benzoic acid, phenylacetic acid, deuterated acetic acid-d4, hydrazine hydrate and coumarin-3-carboxylic acid were purchased from Sigma Aldrich (St. Louis, MO). N-hydroxysuccinimide (NHS), N-hydroxysulfosuccinimide (sulfo-NHS) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) were purchased from PierceNet (Rockford, IL). Acetic anhydride, methanol and N,N-dimethylformamide (DMF) were purchcased from Macron Chemicals (Nashville, TN). The peptide RARARAA was synthesized by CHI Scientific (Maynard, MA) and the peptides KAKAKAA and ARAAARA were synthesized by NeoBioSci (Cambridge, MA).
2.2 Peptide acetylation
N-terminal acetylation was performed by dissolving approximately 1mg of solid peptide in water with a 10mM pH 9 sodium borate buffer. To this, 20-50μL of acetic anhydride was added and the mixture was allowed to react to completion. The peptide solutions were ionized via nanoelectrospray ionization (nESI).
2.3 (Sulfo-)NHS ester reagent preparation
NHS and sulfo-NHS esters were prepared in similar fashions. The carboxylic acid derivative of the acylating reagents was activated using an equimolar amount of EDC in a mixture of DMF and H2O. To this, equimolar amounts of either NHS in DMF or sulfo-NHS in H2O were added, which resulted in the formation of an ester between the carboxyl group and the N-hydroxy of the succinimide. Final concentrations of NHS and sulfo-NHS reagents were prepared to between 25 and 50mM.
2.4 Mass spectrometry
All mass spectrometry experiments were performed on QqQ hybrid triple quadrupole/linear ion trap (LIT) mass spectrometers (QTRAP 2000 and QTRAP 4000, AB Sciex, Concord, ON, Canada), modified with home-built, alternately pulsed nanoelectrospray ionization (nESI) sources.23 Analyte and reagent ions were sequentially mass-selected in the Q1 mass filter, and then subsequently transferred to the q2 collision cell for mutual storage ion/ion reactions24 up to 500ms in length. The reaction product ions were then transferred to the Q3 LIT, where they were mass-selected and collisionally activated using ion trap collision-induced dissociation (CID). For all subsequent fragmentation steps, additional RF-DC isolations followed by ion-trap CID were performed. The ions were then mass analyzed using mass-selective axial ejection (MSAE).25
3. Results and Discussion
3.1 Gas phase acylation of nucleophilic functionalities
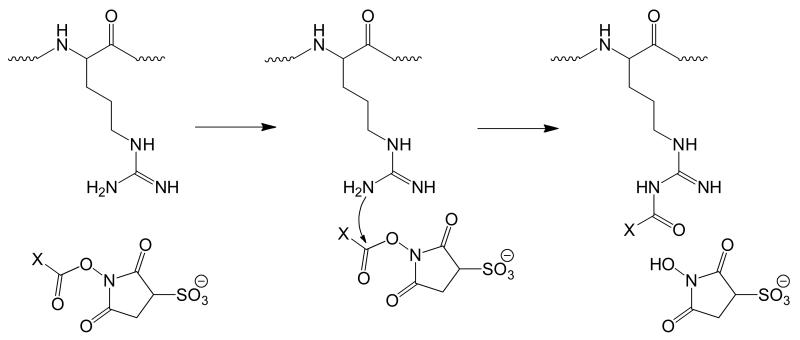
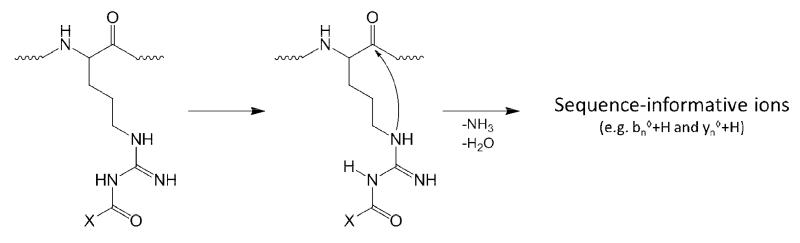
Gas phase ion/ion reactions between a multiply protonated peptide and a singly deprotonated anionic reagent were performed. With a suitable acidic group on the reagent, such as a sulfonate group, a strong electrostatic interaction with a protonated site can give rise to a long-lived complex. Subsequent collisional activation of the complex generally results in one of two observations. The first involves loss of the intact reagent as a neutral species, which reflects the transfer of a proton from the peptide to the reagent. This process indicates that no covalent bond formation occurred. The second observation is the loss of a neutral sulfo-NHS group from the complex. This observation indicates that a nucleophilic attack on the carbonyl carbon of the sulfo-NHS ester by a nucleophilic nitrogen of the peptide, being either from the unprotonated guanidine of arginine or a primary amine, has occurred, resulting in the formation of an amide bond with the peptide and cleavage of the ester bond of the reagent. The extent to which collisional activation is necessary to drive the covalent reaction prior to disruption of the electrostatic interaction that retains the sulfo-NHS leaving group is not clear; however, the extent to which this activation facilitates covalent bond formation may well be case dependent (see below). A gas phase mechanism for covalent modification is illustrated in Scheme 1 for unprotonated arginine.
Scheme 1.
General mechanism for arginine reacting with a sulfo-NHS ester.
This process results in the formation of a peptide bond between the attacking η-nitrogen of the guanidine and the carbonyl carbon of the ester. The mechanisms by which primary amines or the η-nitrogens of arginine are acylated in the gas phase are essentially identical, with the loss of (sulfo-)NHS being a signature for covalent modification. Subsequent CID of the acylated products results in quite differing fragmentation behavior between acyl-guanidines of arginine residues and acyl-amines of the N-termini and lysine residues. Differences in both the nature of the guanidine compared to an amine and the lengths of the side chains between arginine and lysine are presumed to underlie this behavior. Figure 1 illustrates these differences in the fragmentation behavior between peptides containing either acyl-arginine or acyl-lysine.
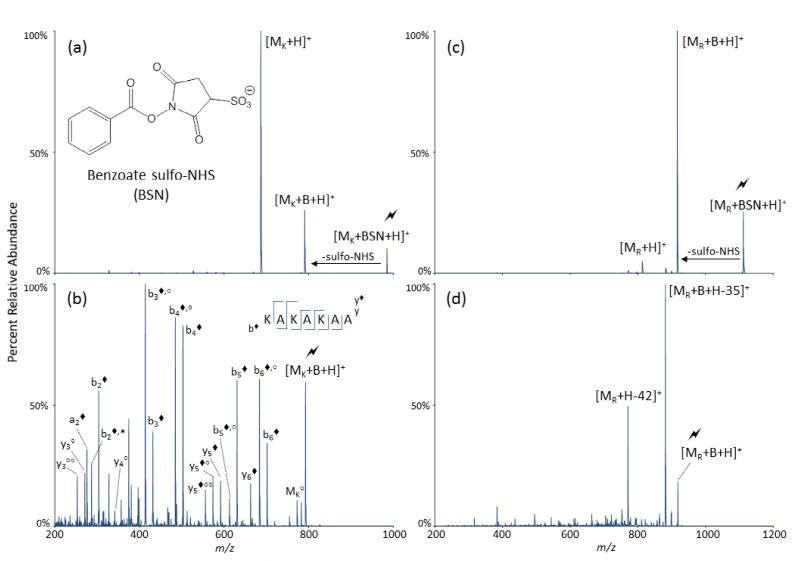
Figure 1.
CID mass spectra of benzoate sulfo-NHS (BSN) in reactions with guanidine or primary amines. (a) CID of the electrostatic complex of [MK+BSN+H)]+, where MK is the peptide KAKAKAA, to produce proton transfer or a covalent modification. (b) CID of [MK+B+H]+. (c) CID of the electrostatic complex of [MR+BSN+H]+, where MR is the peptide Ac-RARARAA. (d) CID of [MR+B+H]+. The activated peaks are labeled with a lightning bolt ( ) and covalently-modified peaks are labeled with a solid diamond (◆). Water losses are labeled with degree signs (°) and ammonia losses are labeled with astericks (*).
) and covalently-modified peaks are labeled with a solid diamond (◆). Water losses are labeled with degree signs (°) and ammonia losses are labeled with astericks (*).
The model peptides used, KAKAKAA and RARARAA, where A is alanine, K is lysine and R is arginine, and the charge states used ensure that at least one basic site be unprotonated. The specific residues which are protonated or not will be governed by Coulombic interactions; however, during ion/ion reactions it is likely that the peptide ion population consists of a mixture of peptide ions with different combinations of protonation sites. Therefore, within an ion population, there should be a mixture of sites that become acylated via the ion/ion reactions. A slight preference toward protonation of the terminal basic sites has been observed, such that, for the model peptides used, either K3 or R3 are unprotonated more frequently. This phenomenon is attributed to Coulombic repulsion. Furthermore, as there is a slight preference toward protonation sites for these model peptides, there is also a slight preference toward site-specific acylation of the unprotonated site. Subsequent fragmentation pathways, which are discussed in further detail below, may then be observed.
We have found that the acylation of unprotonated arginine is at least as facile, and likely more so, than the acylation of unprotonated lysine. Evidence for this conclusion is provided in comparing Figure 1a with Figure 1c. Figure 1a shows the CID of the electrostatic complex [MK+BSN+H]+, where MK is KAKAKAA and BSN is benzoate sulfo-NHS (Figure 1a, inset), while Figure 1c shows the CID of the analogous arginine complex [MR+BSN+H]+, where MR is N-terminally acetylated RARARAA. In each case, there is nominally one unprotonated basic residue in the peptide that can provide a reactive site. Activation of the lysine-containing complex results in both covalent modification, as reflected by the loss of sulfo-NHS, and the rupturing of the electrostatic interaction with a proton transfer, as reflected by the loss of the intact reagent, with the latter pathway being much more favorable. Conversely, activation of the arginine-containing complex results almost entirely in covalent modification, with proton transfer occurring as a minor pathway. This might be taken as evidence that the arginine side chain is inherently more reactive than that of lysine. However, somewhat higher collisional activation amplitudes are required to fragment the arginine-containing complex than the lysine-containing complex due to the higher strength of the arginine-sulfonate interaction.26 In any case, we have found that acylation of arginine residues is facile when they are unprotonated. Figure 1b shows the CID spectrum of the covalently-modified lysine-containing peptide, which results almost entirely in backbone bond cleavage to produce b- and y-type ions that have been covalently modified, indicated by a solid diamond (◆). The only unmodified ions identified are of singly and doubly dehydrated y3 and y4 ions. These ions likely arise from the K5 residue being protonated, and are therefore unreactive towards modification. Figure 1d shows the CID spectrum of the covalently-modified arginine-containing peptide. The fragmentation behavior of Figure 1d differs significantly from that of the lysine-analogue in Figure 1b. CID of the acyl-arginine peptide produces two main fragments, coordinated losses of H2O and NH3 from the modified peptide and loss of the modified guanidine from the peptide, resulting in a mass 42 Da lower than that of the unmodified peptide. These processes are described in more detail below.
3.2 Activation behavior of acyl-arginine with no alpha hydrogens
The two fragmentation pathways of Figure 1d resulting in either deguanidination or the concerted losses of H2O and NH3 are the most common fragmentation pathways observed for peptides containing acyl-arginine. In order to distinguish the different fragmentation pathways of acyl-arginine, the path that leads to deguanidination (loss of 42 Da from the original peptide mass) has been labeled Path 1. It should be noted that the mass of 42 Da also corresponds to the mass of the N-terminal acetylation. Previous work20 compared gas phase acylation of arginine for both N-terminal acetylated (addition of 42 Da, labeled Ac-) and N-terminal propionylated (addition of 56 Da, labeled Pr-) peptides. Upon CID of covalently-modified Pr-RARARAA, the loss of 42 Da from the peptide corresponded to deguanidination and the loss of 56 Da was not observed. This is to be expected as the N-terminal acylation is incorporated as a non-reactive site on the peptide. The second main fragmentation path, which leads to the concerted losses of H2O and NH3, has been labeled Path 2.
3.2.1 Path 1: Deguanidination
A proposed mechanism resulting in the loss of the acyl group and dissociation of the guanidine, referred to as Path 1, is shown in Scheme 2. Deguanidination results from loss of the acyl group bound to the –N=C=NH moiety of guanidine. This product is usually formed as a neutral species but has been observed as an anion from the reaction of doubly deprotonated sulfobenzoate sulfo-NHS, a reagent with a sulfonate on the acyl group and on the NHS group, with singly protonated Ac-ARAAARA (see Figure S1). The loss of the –N=C=NH moiety from the arginine side chain generates the side chain of ornithine. It has recently been shown that the presence of ornithine in peptide residues introduces a highly selective cleavage, termed the ornithine effect.27 This process is effected by the formation of a 6-membered lactam via a nucleophilic attack of the unprotonated δ-amine on its C-terminal carbonyl carbon, cleaving the amide bond C-terminal to the ornithine. Figure 2a shows the CID spectrum of the [MR+H-42]+ peak from Figure 1d. The product ion spectrum shows evidence for the conversion of each of the arginine residues in MR to an ornithine residue. For example, the y6-ion is consistent with the ornithine effect for conversion of R1 to O1, where O is ornithine, the b3-42 and y4-ions are consistent with the ornithine effect for the conversion of R3 to O3, and the b5-42 ion is consistent with the ornithine effect for conversion of R5 to O5. The b5-2(42) ion likely occurs through a second deguanidination event to form ornithine, as is described in more detail below. Path 1, therefore, provides a means for converting arginine to ornithine in the gas phase.
Scheme 2.
Proposed mechanism for fragmentation Path 1 of acylated arginine. Upon activation, the acyl-arginine undergoes deguanidination, resulting in the loss of an acyl carbodiimide (shown) or cyanamide (not shown), leaving an ornithine residue.
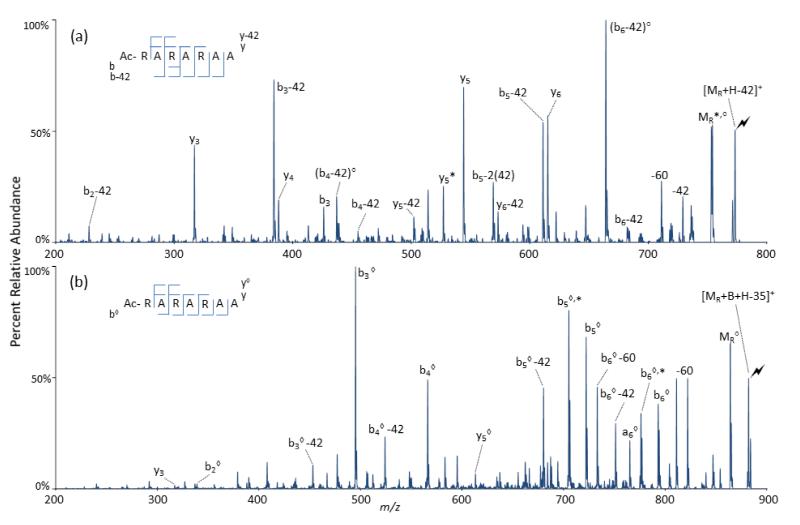
Figure 2.
CID of (a) Path 1 formed by deguanidination and (b) Path 2 formed by concerted losses of H2O and NH3, both formed from CID of [MR+B+H]+, where MR is the peptide Ac-RARARAA. The activated peaks are labeled with a lightning bolt ( ) and covalently-modified peaks that have lost both H2O and NH3 are labeled with a hollow diamond (◇). Independent water losses are labeled with degree signs (°) and independent ammonia losses are labeled with astericks (*).
) and covalently-modified peaks that have lost both H2O and NH3 are labeled with a hollow diamond (◇). Independent water losses are labeled with degree signs (°) and independent ammonia losses are labeled with astericks (*).
The presence of the b5-2(42) ion appears from an additional fragmentation event. For the formation of this ion, it is plausible that R5 was unprotonated while the electrostatic interaction between a protonated arginine and anionic sulfonate occurred at one of the two arginine residues of R1 and R3. The unprotonated arginine residue, R5, would then initiate a nucleophilic attack on the reagent ester to form an amide bond. Subsequently, the electrostatic interaction between the protonated arginine and the sulfonate would be ruptured via a proton transfer from arginine to the sulfonate, neutralizing both functionalities. By this point, R5 has been acylated and one of the arginines of R1 and R3 has been neutralized. Activation of the acylated peptide then results in deguanidination of the acyl arginine via Path 1, as is shown in Figure 1d as [MR+H-42]+. Activation of this ion likely directly produces the b5-42 ion with subsequent deguanidination of the unprotonated arginine, which is commonly observed for unprotonated arginine residues (data not shown). The second deguanidination event would result in the observation of the b5-2(42) ion.
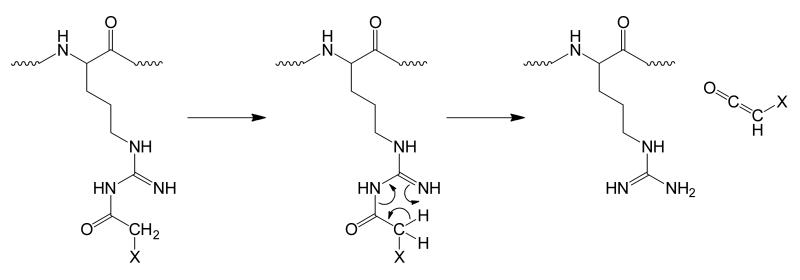
3.2.2 Path 2: Cooperative losses of H2O and NH3
The loss of both H2O and NH3 in a cooperative fashion is likely an inherent quality of the gas phase, although this is not certain. We propose that this process occurs initially by the formation of a cycle, via nucleophilic attack of the δ-nitrogen of arginine on its carbonyl carbon, similar to the cyclization observed in the ornithine effect. Following the formation of the cycle, rearrangements occur that involve losses of both H2O and NH3 (Scheme 3). Details of this mechanism remain unclear and are the subject of continued investigation.
Scheme 3.
Initial proposal for the behavior of fragmentation Path 2 of acylated arginine. Activation of acyl-arginine results in concerted losses of water and ammonia, presumably at the acyl-arginine site. Fragmentation of the Path 2 product produces sequence-informative ions that have a mass loss of 35Da.
The hypothesis of cycle formation has been supported by experimental and theoretical work. First, homoarginine, which contains an additional methylene in the side chain of arginine, was acylated in the gas phase in an analogous experiment to that of arginine. We would expect the formation of a 7-membered ring from homoarginine to be much less favorable than that of the 6-membered ring formed by arginine. It was found that activation of acyl-homoarginine did not result in a large loss of 35 Da (data not shown). Furthermore, theoretical work has shown the formation of a lactam from arginine to be likely and, in some cases, favorable.28
Figure 2b shows the CID spectrum of the [MR+B+H-35]+ ion from Figure 1d. The hollow diamond (◇) is used to represent ions that contain the covalent modification but that have also lost both H2O and NH3. All of the ions that could be modified are present except for the b1 ◇ ion, which is likely due to the low basicity of the b1 ◇ fragment. In the absence of α-hydrogens on the acyl group, Path 2 has been observed to be a prominent process in the dissociation of peptides with modified arginine side chains. In the limited number of peptides examined to date, however, modified ions generated via Path 2 have not shown fragmentation that is particularly distinct from that of the unmodified ions. Nevertheless, this pathway leaves open the possibility for use of tailored acyl groups for influencing peptide fragmentation behavior.
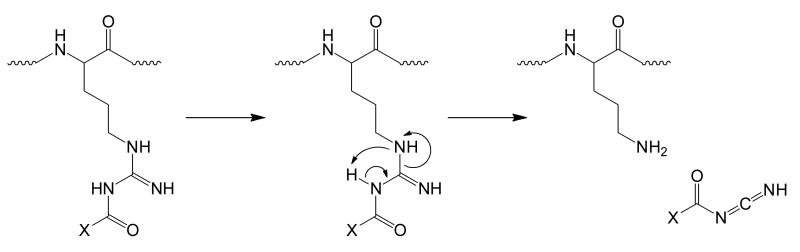
3.3 Path 3: Activation behavior of acyl-arginine with alpha hydrogens
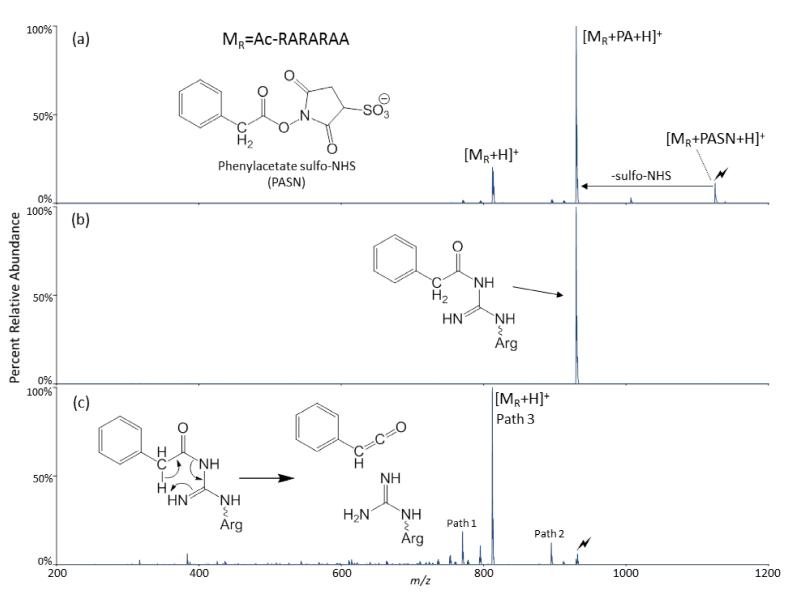
Acylation of arginine using an acyl group that contains an α-hydrogen has been found to show interesting behavior. Although the process by which the guanidine is acylated does not change based on whether the α-carbon of the acyl group contains hydrogens or not, the subsequent activation of the acyl-arginine species differs significantly. Including both the α-carbon and the hydrogen bonded to it, this form of acyl-arginine is able to form a 6-membered ring that includes the carbonyl carbon and nitrogen of the side chain amide bond, the guanidino carbon, and the unmodified guanidino η-nitrogen, as illustrated in Scheme 4. The ring formation in Scheme 4 shows the keto-form of the carbonyl, although it may be as likely that the enol-form is present during the initial ring formation. The reorganization of electrons in the cycle is characteristic of a pericyclic reaction, which, in this case, produces two separate species. In this reaction there is a net loss of one σ bond and a net gain of one π bond. Figure 3 shows the reaction processes involving [MR+PASN+H]+, where PASN is phenyl acetate sulfo-NHS (Figure 3a, inset). Activation of the electrostatic complex produces both a covalently-modified peptide [MR+PA+H]+ and proton transfer [MR+H]+, shown in Figure 3a. Figure 3b shows the isolation of only the covalently-modified peptide, [MR+PA+H]+. Figure 3c shows that CID of [MR+PA+H]+ produces almost entirely [MR+H]+. This process results in the total loss of the acylation to regenerate unmodified arginine. This rearrangement, shown in the inset of Figure 3c, has been further tested using acyl groups containing α-deuterons, the result of which is the formation of [MR+D]+, with no [MR+H]+ observed (Figure S2).
Scheme 4.
Proposed rearrangement mechanism of acyl arginine when the acyl group contains at least one α-hydrogen. Path 3 involves the rearrangement of acyl-arginine to regenerate unmodified arginine, while the acyl group is lost as a ketene.
Figure 3.
The reaction and activation of [MR+2H]2+, where MR is the peptide Ac-RARARAA, with [phenylacetate sulfo-NHS-H]− (PASN). (a) CID of the electrostatic complex between [MR +2H]2+ and [PASN-H]−. (b) RF-DC isolation of the modified peptide [MR+PA+H]+. (c) CID of the modified peptide. The activated peaks are labeled with a lightning bolt ( ).
).
4 Conclusions
Acylation of arginine residues in polypeptides in solution using sulfo-NHS esters is challenging due to incompatible solution conditions for generating unprotonated arginine residues and for maintaining the stability of the reagent. Conditions can be generated in the gas phase, however, to enable efficient acylation of arginine residues via ion/ion reactions. CID of peptides acylated at arginine residues is dominated by side chain cleavages, in contrast with the behavior of peptides acylated at lysine residues. Three main pathways have been noted with their relative importance determined by the nature of the acyl group. Acyl groups that contain an α-hydrogen in relation to the carbonyl group regenerate the arginine residues. In this pathway, activation of the modified peptide results in the formation of a 6-membered cyclic intermediate and, through a pericyclic-like rearrangement, regenerates the unmodified guanidine and forms a ketene. When no α-hydrogens are present on the acyl group, two competitive dissociation pathways dominate. One involves an acyl-arginine side chain rearrangement that eventually results in the coordinated losses of water and ammonia while maintaining the acyl-modification. The other process involves a cleavage that leads to deguanidination of the arginine residue. This pathway provides a means for converting arginine residues to ornithine residues in the gas phase.
Supplementary Material
Acknowledgements
Dr. Jorden Kass is acknowledged for many helpful discussions on the topic. This work was supported by the National Institutes of Health under Grant GM 45372.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yang WC, Mirzaei H, Liu X, Regnier FE. Enhancement of Amino Acid Detection and Quantification by Electrospray Ionization Mass Spectrometry. Anal. Chem. 2006;78:4702–4708. doi: 10.1021/ac0600510. [DOI] [PubMed] [Google Scholar]
- [2].Beardsley RL, Sharon LA, Reilly JP. Peptide de Novo Sequencing Facilitated by a Dual-Labeling Strategy. Anal. Chem. 2005;77:6300–6309. doi: 10.1021/ac050540k. [DOI] [PubMed] [Google Scholar]
- [3].Tubb MR, Gangani RA, Silva RAGD, Fang J, Tso P, Davidson WS. A Three-dimensional Homology Model of Lipid-free Apolipoprotein A-IV Using Cross-linking and Mass Spectrometry. J. Biol. Chem. 2008;283:17314–17323. doi: 10.1074/jbc.M800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mädler S, Bich C, Touboul D, Zenobi R. Chemical Cross-Linking with NHS Esters: a Systematic Study on Amino Acid Reactivities. J. Mass Spectrom. 2009;44:694–706. doi: 10.1002/jms.1544. [DOI] [PubMed] [Google Scholar]
- [5].Singh P, Panchaud A, Goodlett DR. Chemical Cross-Linking and Mass Spectrometry as a Low-Resolution Protein Structure Determination Technique. Anal. Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- [6].Chen Z, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA Polymerase II-TFIIF Complex Revealed by Cross-Linking and Mass Spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mentinova M, McLuckey SA. Intra- and Inter-Molecular Cross-Linking of Peptide Ions in the Gas-Phase: Reagents and Conditions. Rapid Commun. Mass Spectrom. 2011;22:912–921. doi: 10.1007/s13361-011-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Webb IK, Mentinova M, McGee WM, McLuckey SA. Gas-Phase Intramolecular Protein Crosslinking via Ion/Ion Reactions: Ubiquitin and a Homobifunctional sulfo-NHS Ester. J. Am. Soc. Mass Spectrom. doi: 10.1007/s13361-013-0590-4. DOI: 10.1007/s13361-013-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tsumoto H, Murata C, Miyata N, Kohda K, Taguchi R. Efficient Identification and Quantification of Proteins Using Isotope-Coded 1-(6-methylnicotinoyloxy)succinimides by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Rapid Commun. Mass Spectrom. 2007;21:3815–3824. doi: 10.1002/rcm.3279. [DOI] [PubMed] [Google Scholar]
- [10].Madsen JA, Brodbelt JS. Simplifying Fragmentation Patterns of Multiply Charged Peptides by N-Terminal Derivatization and Electron Transfer Collision Activated Dissociation. Anal. Chem. 2009;81:3645–3653. doi: 10.1021/ac9000942. [DOI] [PubMed] [Google Scholar]
- [11].Stutzman JR, Hassel KM, McLuckey SA. Dissociation Behavior of Tryptic and Intramolecular Disulfide-Linked Peptide Ions Modified in the Gas Phase via Ion/Ion Reactions. Int. J. Mass Spectrom. 2012;312:195–200. doi: 10.1016/j.ijms.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vasicek L, Brodbelt JS. Enhancement of Ultraviolet Photodissociation Eficiencies through Attachment of Aromatic Chromophores. Anal. Chem. 2010;82:9441–9446. doi: 10.1021/ac102126s. [DOI] [PubMed] [Google Scholar]
- [13].Wang P, Hadjar O, Gassman P, Laskin J. Reactive Landing of Peptide Ions on Self-Assembled Monolayer Surfaces: An Alternative Approach for Covalent Immobilization of Peptides on Surfaces. Phys. Chem. Chem. Phys. 2008;10:1512–1522. doi: 10.1039/b717617a. [DOI] [PubMed] [Google Scholar]
- [14].Han H, McLuckey SA. Selective covalent Bond Formation in Polypeptide Ions via Gas-Phase Ion/Ion Reaction Chemistry. J. Am. Chem. Soc. 2009;131:12884–12885. doi: 10.1021/ja904812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hassell KM, Stutzman JR, McLuckey SA. Gas-Phase Bioconjugation of Peptides via Ion/Ion Charge Inversion: Schiff Base Formation on the Conversion of Cations to Anions. Anal. Chem. 2010;82:1594–1597. doi: 10.1021/ac902732v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mentinova M, McLuckey SA. Covalent Modification of Gaseous Peptide Ions with N-Hydroxysuccinimide Ester Reagent Ions. J. Am. Chem. Soc. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mentinova M, Barefoot NZ, McLuckey SA. Solution versus Gas-Phase Modification of Peptide Cations with NHS-Ester Reagents. J. Am. Soc. Mass Spectrom. 2011;23:282–289. doi: 10.1007/s13361-011-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stutzman JR, Luongo CA, McLuckey SA. Covalent and Non-Covalent Binding in the Ion/Ion Charge Inversion of Peptide Cations with Benzene-Disulfonic Acid Anions. J. Mass Spectrom. 2012;47:669–675. doi: 10.1002/jms.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stutzman JR, McLuckey SA. Ion/Ion Reactions of MALDI-Derived Peptide Ions: Increased Sequence Coverage via Covalent and Electrostatic Modifications upon Charge Inversion. Anal. Chem. 2012;84:10679–10685. doi: 10.1021/ac302374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McGee WM, Mentinova M, McLuckey SA. Gas-Phase Conjugation to Arginine Residues in Polypeptide Ions via N-Hydroxysuccinimide Ester-Based Reagent Ions. J. Am. Chem. Soc. 2012;134:11412–11414. doi: 10.1021/ja304778j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hermanson GT. Bioconjugate Techniques. 2nd ed. Academic Press; Amsterdam: 1998. [Google Scholar]
- [22].Miller BT, Collins TJ, Rogers ME, Kurosky A. Peptide Biotinylation with Amine-Reactive Esters: Differential Side Chain Reactivity. Peptides. 1997;18:1585–1595. doi: 10.1016/s0196-9781(97)00225-8. [DOI] [PubMed] [Google Scholar]
- [23].Liang X, Xia Y, McLuckey SA. Alternately Pulsed Nano-electrospray Ionization/Atmospheric Pressure Chemical Ionization for Ion/Ion Reactions in an Electrodynamic Ion Trap. Anal. Chem. 2006;78:3788–3793. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xia Y, Wu J, Londry FA, Hager JW, McLuckey s. A. Mutual Storage Mode Ion/Ion Reactions in Hybrid Linear Ion Trap. J. Am. Soc. Mass Spectrom. 2005;16:71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- [25].Londry FA, Hager JW. Mass Selective Axial Ion Ejection from a Linear Quadrupole Ion Trap. J. Am. Soc. Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- [26].Schug KA, Lindner W. Noncovalent Binding between Guanidinium and Anionic Groups: Focus on biological- and Synthetic-Based Arginine/Guanidinium Interactions with Phosph[on]ate and Sulf[on]ate Residues. Chem. Rev. 2005;105:67–113. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- [27].McGee WM, McLuckey SA. Selective Cleavages Induced by Ornithine Residues in Peptides. J. Mass Spectrom. Submitted. [Google Scholar]
- [28].Bythell BJ, Csonka IP, Suhai S, Barofsky DF, Paizs B. Gas-Phase Structure and Fragmentation Pathways of Singly Protonated Peptides with N-Terminal Arginine. J. Phys. Chem. B. 2010;114:15092–15105. doi: 10.1021/jp108452y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.