Abstract
The pea (Pisum sativum) homolog, PsKO1, of the Arabidopsis GA3 gene was isolated. It codes for a cytochrome P450 from the CYP701A subfamily and has ent-kaurene oxidase (KO) activity, catalyzing the three step oxidation of ent-kaurene to ent-kaurenoic acid in the gibberellin (GA) biosynthetic pathway when expressed in yeast (Saccharomyces cerevisiae). PsKO1 is encoded by the LH gene because in three independent mutant alleles, lh-1, lh-2, and lh-3, PsKO1 has altered sequence, and the lh-1 allele, when expressed in yeast, failed to metabolize ent-kaurene. The lh mutants of pea are GA deficient and have reduced internode elongation and root growth. One mutant (lh-2) also causes a large increase in seed abortion. PsKO1 (LH) is expressed in all tissues examined, including stems, roots, and seeds, and appears to be a single-copy gene. Differences in sensitivity to the GA synthesis inhibitor, paclobutrazol, between the mutants appear to result from the distinct nature of the genetic lesions. These differences may also explain the tissue-specific differences between the mutants.
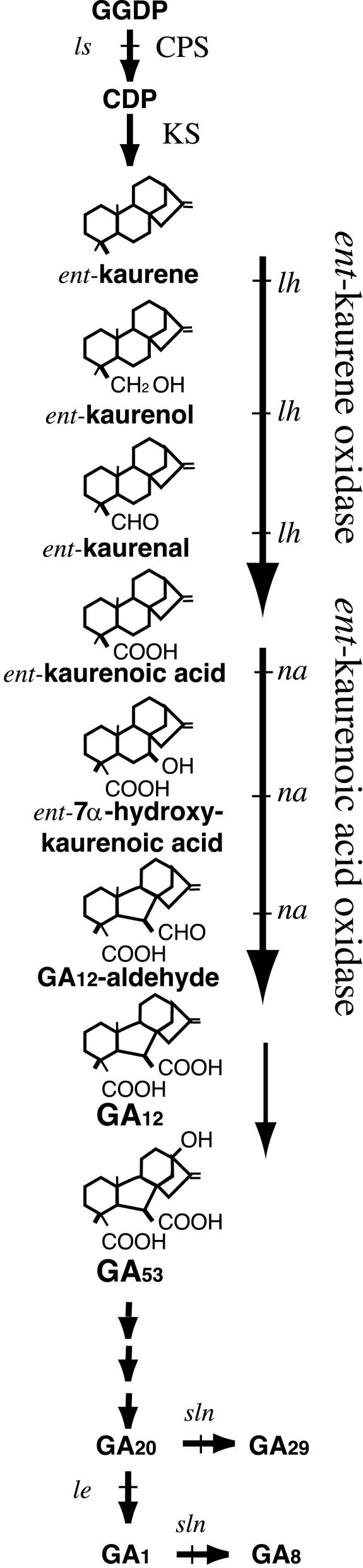
GAs are important plant hormones that regulate many aspects of plant growth including shoot elongation (e.g. Reid and Ross, 1993). GA-deficient mutants have proven important in defining the genes involved in GA biosynthesis (e.g. Ingram et al., 1984; Lester et al., 1997). The lh mutants of pea (Pisum sativum) are GA deficient, and the early internodes are reduced to nearly one-third the length of the wild type (Reid, 1986). They show a pronounced elongation response to the bioactive GA, GA1, which can restore the wild-type tall plant phenotype (Reid and Potts, 1986).
The lh mutation blocks the three-step oxidation of ent-kaurene to ent-kaurenoic acid (Swain et al., 1997). Extracts from developing embryos of the lh-2 mutant were unable to metabolize [14C]-labeled ent-kaurene, ent-kaurenol, and ent-kaurenal (Fig. 1). In contrast, the wild-type extracts readily metabolized ent-kaurene and all the intermediates (Swain et al., 1997). Both the lh-2 mutant and wild type were able to form ent-kaurene from mevalonic acid, geranylgeranyl diphosphate, or copalyl diphosphate and could metabolize ent-kaurenoic acid (Swain et al., 1997). This was consistent with earlier enzyme kinetic studies that suggested the three steps from ent-kaurene to ent-kaurenoic acid were catalyzed by the same enzyme (Coolbaugh et al., 1978). This was confirmed when an Arabidopsis ent-kaurene oxidase gene (AtKO1, GA3) was isolated (Helliwell et al., 1998) and found to be a cytochrome P450 monooxygenase of the subfamily CYP701A. When AtKO1was expressed in yeast (Saccharomyces cerevisiae), it catalyzed the three steps of ent-kaurene to ent-kaurenoic acid (Helliwell et al., 1999). The ent-kaurene oxidase enzyme is thought to play an important role in linking the plastid and endoplasmic reticulum-located steps of GA biosynthesis (Helliwell et al., 2001b). The highly hydrophobic ent-kaurene is produced in the plastid stroma (Sun and Kamiya, 1994; Aach et al., 1995) and is likely to be partitioned into membranes (Hedden, 1997). The Arabidopsis AtKO1 protein is directed to the outer envelope membrane of the chloroplast and, therefore, may channel the less hydrophobic product ent-kaurenoic acid to the ent-kaurenoic acid oxidase enzymes located in the endoplasmic reticulum (Helliwell et al., 2001b).
Figure 1.
The GA biosynthetic pathway in pea shoots. Product structures are represented for the cytochrome P450 monooxygenase-mediated steps showing the steps catalyzed by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). The steps blocked by the pea mutants (ls, lh, na, le, and sln) are indicated. Steps catalyzed by copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) are also indicated.
Besides the lh mutant in pea, there are two other known ent-kaurene oxidase mutants, ga3 in Arabidopsis (Koornneef and van der Veen, 1980) and dx in rice (Oryza sativa; Ogawa et al., 1996). In Arabidopsis, the two alleles ga3-1 and ga3-2 are recessive GA-responsive dwarfs. They accumulate ent-kaurene and show a growth response to applied ent-kaurenoic acid but not ent-kaurene (Helliwell et al., 1998).
The two pea mutant alleles, lh-1 and lh-2, show interesting tissue-specific differences in their phenotypes. This is an unusual phenomenon, and the reasons for such tissue-specific differences between alleles for hormone biosynthesis genes have not been described previously. In the shoot, the lh-1 mutant is slightly more severe with a greater reduction in GA1 and GA20 levels (Swain and Reid, 1992) and slightly shorter seedling stature than the lh-2 mutant (Fig. 2). There is a good correlation between internode length (nodes 4–6) and the log of endogenous GA1 levels for the lh-1, lh-2, LH allelic series (Swain and Reid, 1992). However, in the seed, the lh-1 allele has only a mild effect on seed abortion and transient effect on embryo growth (Swain et al., 1997). In contrast, the lh-2 allele has a large effect on seeds. The seeds are smaller, have delayed development (i.e. take up to 6 d longer to reach contact point; Swain et al., 1993), and have decreased seed survival (less than 50% survived compared with the wild type; Swain et al., 1993, 1997). Again, the phenotypic differences between the alleles reflect the GA content. In young seeds, the GA1 levels of the lh-1 mutant were reduced by only 50%, whereas those of the lh-2 allele were reduced by 90% compared with the wild type (Swain et al., 1995). The lh-2 seeds did not show an early peak in GA levels around 7 d after anthesis, although this was evident in the lh-1 mutant and wild-type seeds. Swain et al. (1995) suggested that because the lh-2 mutant was the only mutation with a strong phenotype affecting seed survival and the only mutation that altered the GA peak early in seed development, then this early peak in GA levels was essential for the development of the seed. At contact point (the 1st d that no liquid endosperm remains in seeds), there is a second peak in GA levels in normal developing seeds (Frydman et al., 1974; Swain et al., 1993). Both lh-1 and lh-2 alleles had decreased GA20 and GA29 levels at contact point compared with the wild type (Swain et al., 1995).
Figure 2.
Photographs of 16-d-old pea seedlings with leaf at fifth node fully expanded.
Another marked difference in the phenotypes of lh-1 and lh-2 plants is their response to the specific ent-kaurene oxidase inhibitor, paclobutrazol (Sugavanam, 1984; Hedden and Graebe, 1985). lh-2 seedlings are 30 times more sensitive to paclobutrazol than lh-1 seedlings (Swain et al., 1997). This inhibition appears to be directly related to GA biosynthesis because GA3 rescues lh-1 and lh-2 seedlings from the dwarfing effect of paclobutrazol (Swain and Reid, 1992).
In this paper, the pea homolog of the Arabidopsis GA3 gene is isolated and shown to encode LH. The previously perplexing differences between the lh-1 and lh-2 mutant phenotypes, including the increased sensitivity of the lh-2 mutant to triazole inhibitors, are then examined and a third allele, lh-3, is characterized.
RESULTS
Characterization of the lh-3 Allele
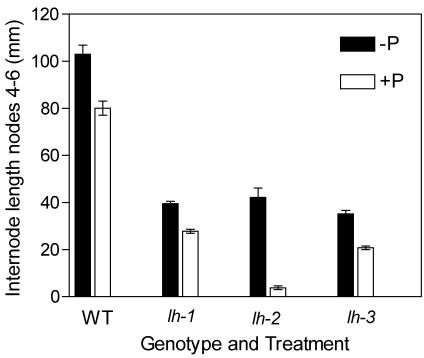
A further mutant with a GA-deficient phenotype was isolated from our mutagenesis program, and allelism testing showed the mutant lh-3 is allelic to lh-1 (J.L. Weller, personal communication). lh-3 seedlings are of similar dwarf stature to the lh-1 and lh-2 seedlings and have internode lengths reduced to approximately 40% of the wild-type cv Torsdag (Fig. 2). The seed survival rates of lh-3 seeds (<5% seed abortions) were similar to that of the wild type and the lh-1 mutant (χ21 = 3.76; P > 0.05) but different from the lh-2 mutant seeds of which 37% aborted (χ21 = 47; P < 0.001; Table I). Similarly, the seed weight of lh-3 was similar to the wild type and lh-1 but greater than seeds of lh-2 (P < 0.001; Table I). The dwarfing response to paclobutrazol (1 μg seed-1) of the lh-3 plants was similar to that of the wild-type cv Torsdag and mutant lh-1 within the range of a 20% to 40% reduction in internode length but markedly less than that for the lh-2 mutant where the same dose of paclobutrazol caused a 90% reduction in internode length (Fig. 3; Swain et al., 1997).
Table I.
Seed wt of healthy seeds and the no. of seed abortions for seeds of wild type (LH), lh-1, lh-2, and lh-3
| Genotype | Healthy Seed Wt | No. of Healthy Seeds | No. of Aborted Seeds |
|---|---|---|---|
| g seed-1 ± se | |||
| Wild type | 0.242 ± 0.005 | 110 | 0 |
| lh-1 | 0.262 ± 0.013 | 210 | 9 |
| lh-2 | 0.198 ± 0.002 | 197 | 116 |
| lh-3 | 0.255 ± 0.005 | 90 | 0 |
Figure 3.
Wild-type (WT) and GA-deficient mutants lh-1, lh-2, and lh-3 were treated (+P) with the GA biosynthesis inhibitor paclobutrazol (1 μg seed-1 in ethanol) or with ethanol alone before planting (-P). The internode length between nodes 4 and 6 of the controls (▪) and paclobutrazol treated (□) plants is shown.
Isolation of Pea CYP701A, the Pea Homolog of Arabidopsis GA3
The pea homolog of the Arabidopsis GA3 gene was isolated by screening a pea seed cDNA library (cv Torsdag) using the full-length GA3 cDNA as probe. The nearly full-length (1,795 bp) clone obtained was extended by 5′-RACE using cDNA prepared from wild type (cv Torsdag) as template (Frohman et al., 1988). The pea gene isolated, PsKO1 (CYP701A10; GenBank accession no. AY245442) showed close homology at the nucleotide level (82-bit score, 84%–90% identities) to the Arabidopsis GA3 gene, AtKO1 (BLASTN; Altschul et al., 1997). At the amino acid level, the full-length putative protein PsKO1 is similar to the pumpkin (Cucurbita maxima) ent-kaurene oxidase, CmKO1 (641-bit score, 65% identities, 80% positives) and the Arabidopsis ent-kaurene oxidase, AtKO1 (592-bit score, 61% identities, 77% positives; National Center for Biotechnology Information BLAST 2 sequences; Fig. 4). No other CYP701A clones were isolated from the pea seed library after a further 125,000 plaque-forming units (pfu) were screened using a 1.3-kb PCR fragment of PsKO1 as probe.
Figure 4.
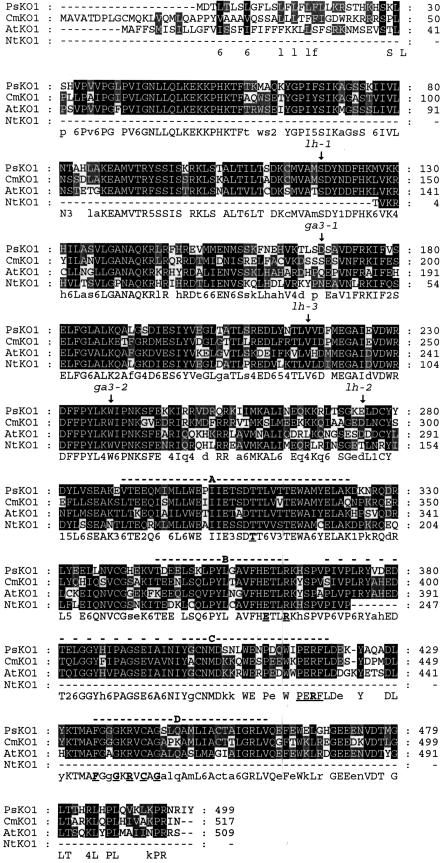
Alignment of the PsKO1 putative protein with the ent-kaurene oxidases of pumpkin (CmKO1; Helliwell et al., 2000), Arabidopsis (AtKO1; Helliwell et al., 1998), and a fragment from tobacco (Nicotiana tabacum; NtKO1; Fukazawa et al., 2000) using GeneDoc conserved base presentation after ClustalW multisequence alignment. The sites of genetic lesions of the pea lh-1, lh-2, and lh-3 mutants and the Arabidopsis ga3-1 and ga3-2 in-frame stop codons are indicated. The catalytic domains A to D (Kalb and Loper, 1988) are labeled, and cytochrome P450 highly conserved amino acids of known significance are underlined. The catalytic A domain is associated with substrate binding (Poulos et al., 1985) and oxygen pocket (Atkins and Sligar, 1988; Imai et al., 1989) and traverses the distal surface of the haem (Poulos et al., 1985). The B domain contains the EXXR section of the highly conserved E,R,R triad involved in positioning the haem-binding pocket (Hasemann et al., 1995). The C domain has the PERF clan designator region (Paquette et al., 2000) that includes the third R of the E,R,R triad (Werck-Reichhart et al., 2002). The D domain has the haem-binding region associated with all cytochrome P450s with the Cys that provides the axial thiolate ligand to the haem iron (Poulos et al., 1986; Porter and Coon, 1991).
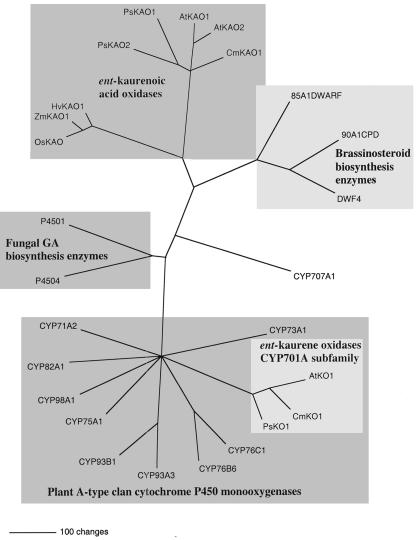
PsKO1 is grouped with the other members of the cytochrome P450 monooxygenase subfamily CYP701A in the plant A-type cytochrome P450s and away from the other GA biosynthesis ent-kaurenoic acid oxidases, the brassinosteroid biosynthesis P450s and the fungal GA biosynthesis cytochrome P450s (Fig. 5). This was confirmed in a phylogram when representatives from the fungal B cytochrome P450 clan were used as the outgroup (data not shown).
Figure 5.
Inferred phylogenetic relationship of ent-kaurene oxidases (CYP701A) and representatives of related cytochrome P450 enzymes. The unrooted phylogram was generated by PAUP 4.0b10 analysis (Swofford, 1999) using putative amino acids of full-length genes (excluding gaps). The ent-kaurene oxidase proteins used in addition to the pea PsKO1 were AtKO1(GA3; AAC39507; Helliwell et al., 1998) and CmKO1(AAG41776; Helliwell et al., 2000). A representative of the plant A-type cytochrome P450 clan subfamilies CYP75A, 93A, 93B, 76B, 76C, 82A, 73A, 98A, and 71A were included. GA biosynthesis kaurenoic acid oxidase proteins used in addition to the pea PsKAO1 and PsKAO2 were from Arabidopsis (AtKAO1 and AtKAO2), pumpkin (CmKAO1), rice (OsKAO1), maize (Zea mays; D3: ZmKAO1), and barley (Hordeum vulgare; Grd5: HvKAO1). The related brassinosteroid biosynthetic enzymes used include Arabidopsis CPD (CYP90A1), brassinosteroid-6-oxidase and DWF4 (CYP90B1), and tomato (Lycopersicon esculentum) DWARF (CYP85A1). The fungal P450–1 and P450–4 GA biosynthesis enzymes with ent-kaurene oxidase and ent-kaurenoic acid oxidase activity were also included.
PsKO1 Sequence Was Altered in All Three Mutant Alleles, lh-1, lh-2, and lh-3
The PsKO1 sequence of the lh-1 mutant plants had a single-base change of guanine to adenine, which translates to a change from Ser to Asn in the putative protein (Fig. 4). These are both amino acids with uncharged polar side chains. However, the Ser hydroxyl group can be available for hydrogen bonding and also can be associated with enzyme active sites (Imai et al., 1989). This Ser is in a highly conserved region in the ent-kaurene oxidase enzymes but not conserved across the cytochrome P450 plant clan A family except in some members of the CYP71A and CYP71D subfamilies. The Ser is not conserved in other cytochrome P450 enzymes including the GA biosynthesis ent-kaurenoic acid oxidases and brassinosteroid biosynthesis enzymes.
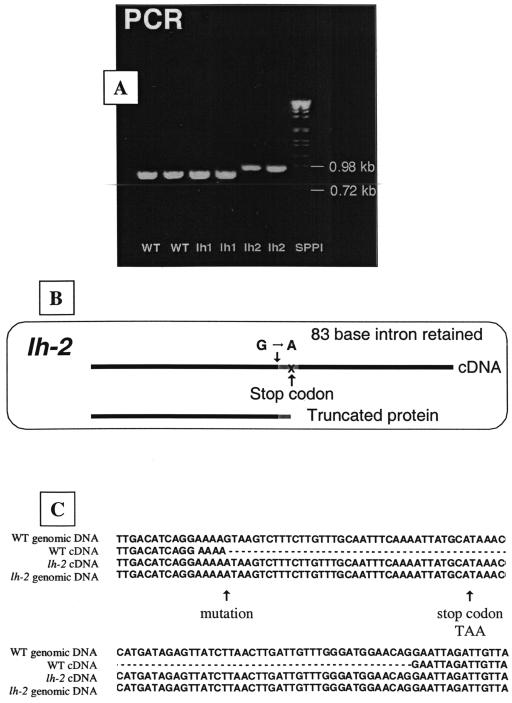
The cDNA PCR fragment from the lh-2 mutant was larger than the equivalent from wild-type and lh-1 plants (Fig. 6A), whereas the genomic PCR fragments using the same primers were the same size. The PsKO1 genomic sequence from lh-2 plants was found to have a single-base substitution of adenine for guanine at the beginning of an intron. This removed the highly conserved G at the beginning of the intron important for forming a lariat and correct pre-mRNA splicing (Fig. 6C; Brown, 1996; Brown et al., 1996). Sequencing of the lh-2 cDNA showed that the whole 83-base intron was retained, which included a stop codon in the reading frame (Fig. 6, B and C). The full sequence of the cDNA showed that no other introns were retained, confirming that this product was not formed from genomic DNA or unspliced RNA. This cDNA (with the 83-base intron) would result in a 275-amino acid truncated putative protein that would not contain the active catalytic domains A to D nor the active haem-binding site common to cytochrome P450s (Fig. 4; Kalb and Loper, 1988) and, therefore, would be expected to be a null mutation.
Figure 6.
The nature of the lh-2 mutation. A, Agarose (2% [w/v])/Tris-acetate EDTA (TAE) electophoresis gel of PCR products with the same primers from wild-type, lh-1, and lh-2 cDNA. B, Schematic diagram of lh-2 mutation. C, Comparison of genomic and cDNA sequence from wild type and lh-2 to illustrate the lh-2 genetic lesion.
The PsKO1 sequence from the lh-3 mutant plants had a single-base change of guanine to adenine, which translates to a change from Val to Met in the putative protein (Fig. 4). Both Val and Met have nonpolar side chains. This replaced Val is conserved in all four ent-kaurene oxidases with sequences available and is conserved in many members of the cytochrome P450 plant clan A family (http:/drnelson.utmem.edu/CytochromeP450.html; http://www.biobase.dk/P450/p450.shtml). This Val is not conserved in the GA biosynthesis ent-kaurenoic acid oxidases or brassinosteroid biosynthesis enzymes.
Yeast Expression
Yeast strains WAT11 and WAT21 (engineered to express one of two Arabidopsis cytochrome P450 reductases; Urban et al., 1997) were transformed with PsKO1 expression constructs, and yeast strains expressing the cDNA were identified by RNA gel blots. Both WAT11 and WAT21 strains expressing PsKO1 converted ent-kaurene to ent-kaurenoic acid (Table II). The identity of the ent-kaurenoic acid produced was confirmed by comparison with an authentic ent-kaurenoic acid standard using GC-MS. Untransformed wild-type yeast did not metabolize ent-kaurene to ent-kaurenoic acid, nor did yeast strains expressing a PsKO1 cDNA isolated from the lh-1 mutant (Table II).
Table II.
Metabolism of ent-kaurene by yeast expressing PsKO cDNA clones
| Yeast Strain | ent-Kaurene | ent-Kaurenol | ent-Kaurenal | ent-Kaurenoic Acida |
|---|---|---|---|---|
| WAT11 PsKO | √ | × | × | √ |
| WAT11 lh-1 | √ | × | × | × |
| WAT11 | √ | × | × | × |
| WAT21 PsKO | √ | × | × | √ |
| WAT21 lh-1 | √ | × | × | × |
| WAT21 | √ | × | × | × |
ent-kaurenoic acid methyl ester product was identified by gas chromatography (GC)-mass spectrometry (MS) and compared with an authentic standard. Kovats retention index for both was 2,342. Comparison of spectra: ion peak heights (percentage relative abundance for WAT21 PsKO product/ent-kaurenoic acid standard): 316 [M+] (29/30), 301 (22/23), 273 (44/43), 257 (87/98), 256 (46/44), 241 (100/100), 213 (45/61), 199 (23/26), 187 (32/27), and 159 (36/41)
Northern-Blot Analysis
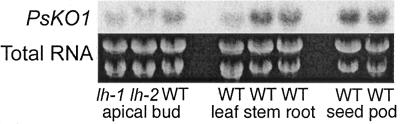
The PsKO1 gene was expressed in all pea organs tested including apical bud, stem, leaf, root, seed, and pod (Fig. 7), although there was less mRNA observed in the leaf than other tissues. The lh-2 PsKO1 mRNA runs slower, consistent with the expected larger mRNA because of the retention of the 83-base intron, and gives confidence in the specificity of the probe.
Figure 7.
PsKO1 transcript level in various parts of wild-type pea and the apical bud from lh-1 and lh-2 mutants. Five micrograms of total RNA from the apical bud (all material above the uppermost fully expanded leaf), leaf (the uppermost fully expanded leaf), stem (internode immediately below the uppermost fully expanded leaf; the internode was 80%–100% fully expanded), and root (50 mm off the end of the tap and lateral roots) of 19-d-old seedlings; also, 5 μg of total RNA from seeds (3 d after contact point) and pods (that originally contained these seeds) from mature plants were loaded on the gel.
Alternate Splicing of the Mutant lh-2 mRNA
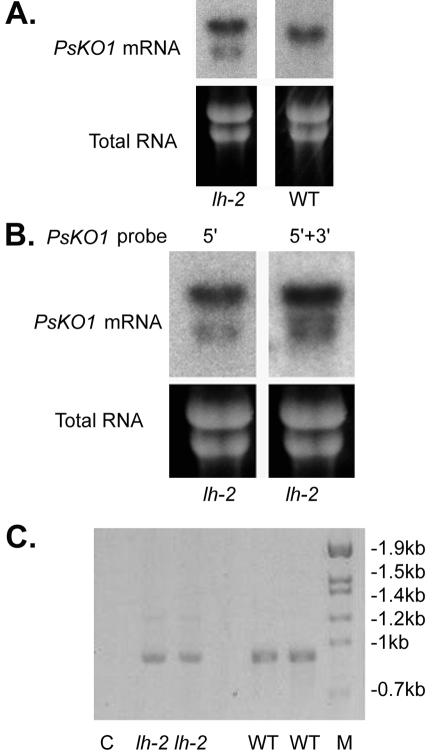
The PsKO1 probe (prepared from a 750-bp template 5′ of the lh-2 lesion) revealed two bands associated with the altered mRNA of the lh-2 mutant seed when expression levels were high (Fig. 8A). One band was larger than wild type, and this was also seen in lh-2 apical buds (Fig. 7) and is expected to be because of the retention of the intron containing the lesion associated with the lh-2 mutation. The other PsKO1 band was smaller than wild type and was only clearly noted in lh-2 seeds when expression levels were high (Fig. 8A). The smaller band is approximately 1 kb in size and was not observed in the wild-type seed with similar expression levels, age, and weight of seeds. When a probe prepared from a 925-bp template spanning the lh-2 lesion (425 bp 5′ and 500 bp 3′ of the lh-2 lesion) was used, a third band of intermediate size (near 1.3 kb) was noted in mRNA from the seeds (Fig. 8B). This band is associated with the region 3′ of the lh-2 lesion and was not observed in the wild-type or lh-1 samples.
Figure 8.
A, Northern-blot analysis of lh-2 and wild-type (LH) seed probed with 32P-labeled 5′ PsKO1 probe. Total RNA (5 μg) from whole seeds 11 d after anthesis was loaded on the gel. B, Northernblot analysis of lh-2 seed probed with 32P-labeled 5′ PsKO1 probe (prepared from 750-bp template 5′ of the lh-2 lesion) or 5′ + 3′ PsKO1 probe prepared from a 925-bp template spanning the lh-2 lesion (425 bp 5′ and 500 bp 3′ of the lh-2 lesion). C, Ethidium bromide-stained 2% (w/v) agarose gel of PCR products showing bands in both WT and lh-2 corresponding to the size of WT-spliced cDNA. The 25-base forward primer was positioned so that the 14 bases at the 5′ end correspond to the final 14 bases at the 3′ end of the exon immediately preceding the lh-2 lesion, and the final 11 bases of the primer correspond to the first 11 bases at the 5′ end of the exon immediately after the lh-2 lesion. This primer was used in conjunction with a reverse primer located 915 bp downstream in the cDNA so that only cDNA with wild-type splicing would amplify. This primer combination encompassed several introns; the size of the PCR product is consistent with the absence of introns. The amounts loaded on the gel were as follows: 12 μL each of PCR product from lh-2 cDNA templates (each from different harvests and different cDNA preparations) and “no reverse transcriptase (RT)” controls (C), whereas only 2 μL of PCR product from wild type (WT) was loaded. M, SPP-1/EcoR1 DNA size marker.
Because alternate splicing is occurring in the lh-2 mutant mRNA, PCR was used to test if any correct splicing occurred in lh-2 plants producing a wild-type mRNA that could encode an enzyme with normal activity. PCR amplification of the cDNA using a primer spanning the intron/exon boundary (of the intron containing the lh-2 lesion) revealed a band of the same size as the wild type in the lh-2 samples (Fig. 8C). For similar intensity on the gel, a larger volume of PCR product from lh-2 and controls was loaded on the gel than wild-type samples, suggesting that markedly less correctly spliced product was present in the lh-2 cDNA than the wild-type cDNA. Two completely independent (with internal replication) real-time quantitative RT-PCR experiments indicated that in the apical buds of wild-type and lh-2 plants, LH expression was similar, but lh-2 plants contained approximately 0.4% to 0.5% of the wild-type level of correctly spliced m RNA. The level of wild-type splicing in seeds harvested at precontact point (seed weight of 0.029 g), contact point (seed weight of 0.20 g), and post-contact (seed weight of 0.29 g) was consistently lower than that found in apical buds and varied from 0.11% to 0.24% of the wild type. Sequencing of a cDNA PCR product using a primer spanning the lh-2 lesion indicated that a wild-type mRNA was produced as predicted from the above data. The overall expression of the lh-2 and LH alleles was similar when checked by quantitative RT-PCR using a region of the alleles unaffected by the lh-2 mutation.
Genomic Southern-Blot Analysis
At high stringency, the PsKO1 gene appears to be a single-copy gene (data not shown). Seven restriction enzymes at high stringency revealed one or two bands indicating hybridization with the PsKO1 probe. The two bands observed when cut by HindIII, SacI, BamHI, and BglII were expected because those restriction sites are present in the cDNA sequence. At low stringency, additional bands were noted.
DISCUSSION
Isolation of the Pea Homolog of Arabidopsis GA3
PsKO1, the pea homolog of the Arabidopsis GA3 gene, was isolated. This gene had high similarity to AtKO1, CmKO1, and NtKO1 of Arabidopsis, pumpkin, and tobacco, respectively (Fig. 4), and is grouped with the ent-kaurene oxidases (AtKO1 and CmKO1) in the cytochrome P450 CYP701A subfamily (Fig. 5). The CYP701A subfamily is found within the plant A-type clan and away from the other cytochrome P450 GA and brassinosteroid biosynthetic enzymes. This coincides with the cytochrome P450 alignment of David R. Nelson (http://drnelson.utmem.edu/CytochromeP450.html) and the Arabidopsis alignments (Paquette et al., 2000; http://www.biobase.dk/P450/p450.shtml).
The Pea Gene LH Encodes PsKO1
The evidence indicates that the pea LH gene encodes PsKO1 because the PsKO1 sequence is altered compared with the wild-type progenitor cv Torsdag in tissue from all three GA-responsive dwarf mutants, lh-1, lh-2, and lh-3 (Fig. 4), and yeast expressing a PsKO1 cDNA metabolizes ent-kaurene to ent-kaurenoic acid (Table II). The mutations lh-1, lh-2, and lh-3 are allelic (Swain and Reid, 1992; J.L. Weller, personal communication) and produced by independent mutational events. Also, the predominant mRNA of the lh-2 mutant seen on northern blots is larger than wild type, consistent with the predicted larger size because of retention of an 83-base intron (Figs. 7 and 8).
Consistent with this interpretation, PsKO1 is expressed in the organs where an lh phenotype is observed. PsKO1 was expressed in the shoot including the apical bud, stem, and leaf (Fig. 7), and all three lh alleles cause decreased stature compared with the wild type (Fig. 2). The GA1 levels in the shoot are also reduced in the lh-1 and lh-2 mutants (Swain and Reid, 1992). PsKO1 is expressed in the roots (Fig. 7), and the roots of lh-2 mutant seedlings have decreased tap root length (Batge et al., 1999; Yaxley et al., 2001) and lateral root length (Yaxley et al., 2001), coinciding with reduced GA1 and GA19 levels in the root (Yaxley et al., 2001). PsKO1 is also expressed in the pod (Fig. 7), and lh-2 plants have reduced pod GA1, GA20, GA29, and GA19 levels (Swain et al., 1993; MacKenzie-Hose et al., 1998). PsKO1 is expressed in the seed (Fig. 7), and both lh-1 and lh-2 seeds have reduced GA1 levels in young seeds and reduced GA20 and GA29 levels at contact point (Swain et al., 1995). The lh-1 mutant has a transient effect on seed growth and development (Swain et al., 1997), and the lh-2 mutant has decreased seed size and survival (Swain et al., 1993). Finally, extracts from lh-2 plants have been shown previously to be unable to metabolize ent-kaurene, ent-kaurenol, or ent-kaurenal (Swain et al., 1997). Therefore, the lh mutation blocked the same three-step oxidation of ent-kaurene to ent-kaurenoic acid catalyzed by other CYP701A (ent-kaurene oxidase) enzymes such as Arabidopsis AtKO1 (GA3; Helliwell et al., 1999) and pumpkin CmKO1 (Fig. 2; Helliwell et al., 2001a).
Now that the LH gene has been cloned and the nature of the genetic lesions of the three lh alleles are known, we can explain some of the previously perplexing phenotypic differences between plants carrying the lh-1 and lh-2 alleles.
Sensitivity to Paclobutrazol
One dramatic difference between the lh-1 and lh-2 seedlings is in their response to triazole inhibitors (Swain et al., 1997). The lh-2 mutant is 30 times more sensitive to the ent-kaurene oxidase-specific inhibitor, paclobutrazol, than the lh-1 or lh-3 mutants or the LH wild type (Fig. 3; Swain et al., 1997). Swain et al. (1997) proposed “the changed structure (of the lh-2 gene product) both decreases enzyme activity and increases the affinity for inhibitors, possibly by altering the active site.” However, evidence presented here shows that the putative lh-2 aberrant protein is truncated (Fig. 6) before the cytochrome P450 catalytic domains and would not be expected to have an active site. Therefore, there must be another explanation. Paclobutrazol is a competitive inhibitor specific for ent-kaurene oxidase (Sugavanam, 1984; Hedden and Graebe, 1985), and the lh-1 and lh-3 aberrant proteins have a substrate-binding site (Fig. 4). Therefore, the lh-1 and lh-3 aberrant proteins may bind paclobutrazol and, therefore, dilute the effect of the inhibitor on residual or alternate KO activity and, thus, have a similar sensitivity to paclobutrazol as the wild type. The primary lh-2 aberrant protein, however, is truncated before the cytochrome P450 substrate-binding sites (Fig. 6), leaving all the applied inhibitor free to affect a small quantity of alternative splice products or the activity of another ent-kaurene oxidase.
Since ent-kaurene and ent-kaurenoic acid are part of the GA biosynthesis pathway common to all plant species (Hedden and Phillips, 2000), some form of KO activity is expected in the lh-1, lh-2, and lh-3 mutants because they germinate, grow to one-third the height of the wild type, and set seed. A mutation causing a plant to be devoid of GAs may be expected to be lethal (Koornneef and van der Veen, 1980; Hooley, 1994; Swain et al., 1997; Yamaguchi and Kamiya, 2002) or at least very much shorter than lh-1, lh-2, and lh-3 plants (Reid, 1986). Hence, the ent-kaurene oxidation step of GA biosynthesis in lh mutants appears to be “leaky.” This is confirmed because although the putative protein from the lh-1 and lh-3 mutants are altered from wild-type PsKO1, the GA biosynthetic inhibitor, paclobutrazol, further reduced the internode lengths of the lh-1 and lh-3 mutants (Fig. 3). Other GA biosynthetic inhibitors including a copalyl diphosphate synthase inhibitor (AMO-1618) and a 2-oxoglutarate-dependent dioxygenase inhibitor (BX-112) also reduced the stature of the lh-1 mutant (Swain et al., 1997). Further, the lh-1 double recessive with other GA biosynthetic mutants (ls lh-1 and na lh-1) have such short internodes that the plants appear rosette-like (Reid, 1986; Reid and Ross, 1993). Thirdly, Ingram and Reid (1987) found that the lh-1 mutant still responded to added ent-kaurene when endogenous production of ent-kaurene was prevented by the inhibitor AMO-1618, although the elongation response was less than that achieved in the ls mutant or wild-type (cv Torsdag) plants.
It cannot be ruled out that the “leaky” nature of the ent-kaurene oxidation step may be because of limited activity by other KO enzymes in pea shoots (i.e. not PsKO1). However, PsKO1 appears to be a single-copy gene from the results of the Southern blot at high stringency, and additional library screening with PsKO1 did not reveal another CYP701A gene. The extra bands present in the Southern-blot analysis at low stringency may indicate some similar genes but probably indicate cross-hybridization with another closely related cytochrome P450 subfamily. PsKO1 is expressed in all the tissue tested so far (Fig. 7), so it is possible for it to be the sole ent-kaurene oxidase activity in pea as appears to be the case in Arabidopsis (Helliwell et al., 1998).
The lh-1 and lh-3 putative proteins are full length and contain the substrate-binding and catalytic domains common to cytochrome P450 monooxygenases (Kalb and Loper, 1988; Fig. 4) and, therefore, may have some residual ent-kaurene oxidase activity even though this was not observed during expression of lh-1 in yeast (Table II). However, the lh-2 mutant, which would be expected to produce a severely truncated protein (Fig. 6), still appears to contain some residual KO activity. Alternative splicing may explain this apparent contradiction. The premature stop codon associated with the lh-2 allele resulted from incorrect splicing with retention of an intron (Fig. 6). Ait-Ali et al. (1997) found that the ls-1 mutant with a base substitution at the 3′ acceptor splice site produced a series of alternately spliced products. Some of these products may have activity because a more severe dwarf allele ls-3 has been found, suggesting that the ls-1 mutation was not null. Further, alternative splicing has been shown for the ga3 (AtKO1) gene in Arabidopsis (Helliwell et al., 1998).
Evidence of intron retention in mRNA from the lh-2 mutant is presented in PsKO1 northern-blot analysis because the predominant band in lh-2 mutant tissue is larger than wild type (Figs. 7 and 8). If some small percentage of the mutant RNA splicing is not prevented and occurs normally, then protein prepared from this correctly spliced mRNA would be full length with catalytic domains and have enzymatic activity. In the seed, where LH expression levels are high, there is another smaller band than the wild-type mRNA present (approximately 1-kb compared with 1.8-kb wild type; Fig. 8). This may be the product of a different splicing event, perhaps skipping the mutated 5′ splice site and removing several exons to attain the observed size. Alternatively, it may be because of impaired splicing. Normal removal of introns is a two-step cleavage ligation reaction. The first step involves cleavage of the 5′ splice site with the formation of intron lariat with the G (changed to A in the lh-2 mutant; Fig. 6) binding to a branch point in the intron. In the second step, the 3′ splice site is cleaved, the exons ligate, and the intron lariat is released to be debranched and degraded (Brown et al., 1996). The GT to AT mutation at the 5′ intron splicing site in genes in Arabidopsis (Orozco et al., 1993; Bradley et al., 1995) and maize (Lal et al., 1999) formed a lariat but did not proceed to the second step, and the 5′ exons and an intermediate (consisting of the intron lariat and 3′ exons) were detected. The smaller band of lh-2 mRNA (Fig. 8) could correspond to the cleaved-off 5′ exons because it is approximately the correct size and hybridized with a PsKO1 probe prepared from the template 5′ of the lh-2 genetic lesion. Another PsKO1 probe template spanning the lh-2 lesion revealed a minor additional band of intermediate size to the bands detected by the probe 5′ of the lh-2 lesion (Fig. 8). This suggests that the first step of intron removal has occurred on some occasions. Liu and Filipowicz (1996) found that a small percentage (10%) of the lariat-exon intermediates of a synthetic gene with the GT to AT mutation could go on to complete the second step. If any lh-2 product was correctly spliced, then it would have ent-kaurene oxidase activity.
Northern-blot analysis of lh-2 may not be sensitive enough to show a small percentage of wild-type mRNA, and PCR may be a more sensitive tool. PCR amplification of the cDNA with a primer spanning the intron/exon boundary (of the intron containing the lh-2 lesion) detected a small percentage of wild-type (correctly spliced) product in the lh-2 samples (Fig. 8). Real-time PCR results indicate that lh-2 apical bud samples contained approximately 0.5% correctly spliced mRNA compared with wild-type apical buds. The increased sensitivity of the lh-2 mutant to paclobutrazol, therefore, probably occurs because the small quantity of residual activity provided by correct splicing in the lh-2 mutant was exposed to all the applied paclobutrazol.
Phenotypes of lh-1, lh-2, and lh-3 Differ in a Tissue-Specific Manner
In the seed, the lh-2 allele has a more severe phenotype than lh-1 and lh-3, whereas the shoot phenotypes of all three alleles are similar (Fig. 2). The lh-2 mutant causes a severe decrease in seed survival compared with WT, and seeds have delayed development, taking up to 6 d longer to achieve contact point. Mature lh-2 seeds are smaller and lighter than wild-type and lh-1 seeds (Swain et al., 1993). In contrast, the lh-1 allele has only a transient effect on embryo and seed growth and a very mild increase in seed abortion (Swain et al., 1997). The lh-3 allele seed survival was similar to lh-1 (Table I).
The differences between the lh-2 and the lh-1 and lh-3 seed phenotypes, therefore, follow the differences in severity of the genetic lesion. The young seeds of the lh-2 mutant, which is expected to have a severely truncated PsKO1 putative protein, had GA1 levels reduced to 10% of the wild-type value (Swain et al., 1993; 1995). However, the GA1 levels were only reduced to approximately 50% in young lh-1 seeds (Swain et al., 1993) with a full-length putative protein containing only a single-base substitution. A log-linear relationship exists between endogenous GA1 levels and internode elongation in pea shoots (Ingram et al., 1986; Ross et al., 1989), and if a similar relationship is present in seeds, then a 90% reduction in GA1 levels may cause a large and observable phenotype, whereas 50% reduction produces only a transient effect.
In contrast to the large difference in the phenotypes between lh-2 and lh-1 and lh-3 seeds, all three alleles are dwarfs of similar stature and have an approximate 40% reduction in stature from the wild type (Fig. 2). The difference could result from the different efficiencies of the wild-type splicing in the seeds (0.11%–0.24%) compared with shoot apical buds (0.4%–0.5%) as shown by quantitative RT-PCR. Alternatively, the environment presented to the ent-kaurene oxidase enzyme within the cells of the seeds and vegetative tissue may differ, accounting for the phenotypic differences. This could include different plastid types in these tissues because kaurene oxidase activity is associated with the plastids, or the higher flux of GA precursors through the pathway in developing seeds compared with shoots.
In conclusion, the pea LH gene encodes an ent-kaurene oxidase. PsKO1 is expressed in all tissues tested including apical bud, stem, leaf, root, seed, and pod. This contrasts with the tissue-specific expression of most other GA biosynthetic genes examined (e.g. Davidson et al., 2003). The phenotypes of the lh-1, lh-2, and lh-3 mutants differ in their sensitivity to paclobutrazol and in a tissue-specific manner. This is probably explained by differences in the putative aberrant proteins.
MATERIALS AND METHODS
Plant Material and Growing Conditions
Three independent mutational events in pea (Pisum sativum L. cv Torsdag) resulted in the alleles lh-1, lh-2, and lh-3. The lh-1 mutant (line K511) was ethylmethane sulfonate induced, and the lh-2 mutant (line NGB5843) was ethyleneimine induced by Dr. Klavdiya K. Sidorova (Institute of Cytology and Genetics, Novosibirsk, Russia; Reid, 1986; Swain and Reid, 1992). The lh-3 mutant line ABO4 was ethylmethane sulfonate induced from cv Torsdag by Dr. Jim L. Weller (University of Tasmania, Hobart, Australia; Weller et al., 1997). Hobart L107, a selection from cv Torsdag (Hobart line 107), was used as wild type. Plants were grown two per pot in a heated greenhouse under an 18-h photoperiod (Beveridge and Murfet, 1996). Seeds treated with paclobutrazol received 5 μL of a solution of 0.2 μg paclobutrazol. μL ethanol-1 applied to a nick in the testa before planting.
Library Screening
A total of 280,000 pfu were screened from a pea seed cDNA library to isolate the pea homolog of the Arabidopsis GA3 gene. A further 125,000 pfu were screened from that library in an attempt to find other genes similar to the pea CYP701A gene initially isolated. The seed cDNA library used was constructed in Lambda ZAPII (Stratagene, La Jolla, CA) with cDNA prepared from pea cv Torsdag seeds at contact point (Ait-Ali et al., 1997). The library screening and the isolation of clones was according to methods recommended by the manufacturer (Stratagene).
The probe template for the initial middle stringency screening (65°C hybridization and 65°C washes with 2× SSC + 0.1% [w/v] SDS) was the full-length (1.5-kb) Arabidopsis GA3 cDNA supplied by Dr. Elizabeth S. Dennis (CSIRO Plant Industry, Canberra, Australia; Helliwell et al., 1998). It was digested out of a pYE22mcs vector with SalI and BamHI then gel purified from 1% (w/v) agarose/TAE gel (QIAquick gel extraction kit, Qiagen, Hilden, Germany) and then 32P labeled (Gigaprime DNA labeling kit, Geneworks, Adelaide, SA, Australia). The probe template for the subsequent low-stringency screening (50°C hybridization and 60°C washes with 2× SSC + 0.1% [w/v] SDS) was a 1.3-kb PCR product amplified from cDNA prepared from cv Torsdag apical buds using specific primers of the PsKO1 sequence cDNA obtained from the initial library screening. The PCR product was gel purified on 1% (w/v) agarose/TAE (QIAquick gel extraction kit, Qiagen USA), then gel filtered (CENTRISPIN 10, Princeton Separations, Adelphia, NJ), and then 32P labeled as above.
Genomic Southern Blots
Genomic DNA was isolated (Ellis, 1994), digested, and run on a 0.7% (w/v) agarose/TAE gel for 24.5 h at 20 V. This was blotted to Genesceen Plus (DuPont/NEN, Boston, MA) in 2 × SSC and hybridized in a hybridization solution (0.5 m sodium phosphate [pH 7.2], 1 mm EDTA, and 7% [w/v] SDS; Ausubel et al., 1994). For high stringency, the membrane was hybridized at 65°C with 65°C (0.2× SSC and 0.1% [w/v] SDS) washes. The same blots were then hybridized at low stringency (50°C) with initial washes at 50°C, then final washes at 60°C with 2× SSC and 0.1% (w/v) SDS. The PsKO1 cDNA (1.8 kb) obtained from the library screening (above) was cut out of the Lambda ZapII vector with XhoI and EcoRI and then gel purified and labeled with 32P (Gigaprime DNA labeling kit, Geneworks) to use as the probe.
Northern-Blot Analysis
Total RNA was extracted using the phenol/SDS method (Ausubel et al., 1994), the RNeasy Plant Kit (Qiagen), or the Tri Reagent RNA extraction (Sigma-Aldrich, St. Louis) consistent within the blot. The RNA (5 μg lane-1) was fractionated in 1.5% (w/v) agarose gel containing formaldehyde and transferred to Genescreen Plus hybridization transfer membrane (DuPont/NEN) using 10× SSC. The membrane was hybridized at 42°C in 5× SSC, 5× Denhardts, 50% (w/v) formamide, 1% (w/v) SDS, and 200 μg mL-1 salmon sperm with a cDNA 32P probe (see above). The membrane was washed in 2× SSC and 0.1% (w/v) SDS and then 0.2× SSC and 0.1% (w/v) SDS at 65°C and exposed to Kodak biomax x-ray film (Eastman-Kodak, Rochester, NY) at -70°C.
The PsKO1probe template was a PCR fragment amplified from the clone isolated by the library screening (above) using vector primers and then nested with PsKO1-specific primers to give a 750-bp fragment covering the 5′ region of the gene. The fragment 5′ of the lh-2 genetic lesion was chosen as template to allow direct comparison of the lh-1, lh-2, and wild-type expression patterns. In addition, another PsKO1 probe (5′ + 3′ PsKO1) was prepared from a 925-bp template spanning the lh-2 lesion (425 bp 5′ and 500 bp 3′ of the lh-2 lesion), initially PCR amplified from the clone isolated by the library screening (above) using vector primers and then nested twice with PsKO1 specific primers.
PCR of Correctly Spliced Wild-Type mRNA in lh-2 Mutant Tissue
RNA was extracted using the phenol/SDS method (Ausubel et al., 1994), and cDNA was prepared with Superscript III (Life Technologies/Gibco-BRL, Cleveland). PCR amplification of the cDNA was performed with a 25-base forward primer that spans the splice junction in the cDNA resulting from the removal of the intron with the lh-2 lesion (so that only correctly spliced wild-type cDNA would be amplified). The primers used produced a 915-bp amplicon. The forward primer was 5′-TGACATCAGGAAAAGAATTAGATTG-3′, and the reverse primer was 5′-AGACAAACAAAACTAACTAGCCTCTCTT-3′ (Fig. 8C).
Real-time quantitative PCR using the Quantitect SYBR Green PCR kit (Qiagen USA) was performed on a Rotorgene 2000 (Corbett Research, Melbourne, Australia) to quantify wild-type splicing in lh-2 plants, i.e. removal of the intron with the lh-2 lesion. A 104-bp amplicon was produced using the forward primer described above, with reverse primer 5′-AATAATTGGCTCCCAGAGCA-3′.
To check the overall expression of the lh-2 and LH alleles in lh-2 and wild-type plants, we amplified a 102-bp region downstream of the lh-2 lesion using real-time quantitative PCR as described above. This region is the same in wild type and lh-2. The following primers were used: forward primer, 5′-CTGCTCTGGGAGCCAATTAT-3′; and reverse primer, 5′-CAGACGGTCCTGACGATTTT-3′.
Sequence Analysis
The putative amino acids of full-length genes were aligned using ClustalW (http://www.searchlauncher.bcm.tmc.edu/multi-align; Thompson et al., 1994) and then presented in GeneDoc (http://www.psc.edu/biomed/genedoc/; Nicholas et al., 1997) or analyzed by PAUP 4.0b10 analysis (Swofford, 1999). The PAUP analysis used the bootstrap method (1,000 replicates) with heuristic search on the aligned sequences with gaps excluded. CYP701A sequences used (in addition to the pea PsKO1) included AtKO1 (GA3, GenBank accession no. AAC39507; Helliwell et al., 1998) from Arabidopsis and CmKO1 (AAG41776; Helliwell et al., 2000) from pumpkin (Cucurbita maxima).
Yeast (Saccharomyces cerevisiae) Expression
The constructs were prepared in the pYEDP 60 plasmid vector (Pompon et al., 1996) as specified previously (Davidson et al., 2003). The PsKO1 cDNA was prepared from m RNA extracted from cv Torsdag. Preparation of yeast and analysis by GC-MS was as described previously (Davidson et al., 2003) with product confirmation by comparison of spectra and Kovats' retention index to an authentic ent-kaurenoic acid standard.
Acknowledgments
We thank Ian Cummings (University of Tasmania), Tracey Jackson (University of Tasmania), and Sue Allen (CSIRO, Canberra, Australia) for technical assistance; Dr. Denis Pompon (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for the WAT11 and WAT21 yeast strains; Dr. Jim Weller (University of Tasmania) for seeds; and Dr. Bruce Twitchin (Australian National University, Canberra) and Professor Lewis Mander (Australian National University, Canberra) for the provision of authentic GA standards. We would also like to thank Dr. Bob Elliott (University of Tasmania), Dr. L. Huub Kerckhofs (University of Tasmania, Hobart, Australia), and Dr. Steve Swain (CSIRO) for helpful discussions.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.032706.
This work was supported by the Australian Research Council (grants to J.B.R.) and by an Australian Postgraduate Award (to S.E.D.).
References
- Aach H, Bose G, Graebe JE (1995) ent-Kaurene biosynthesis in a cell-free system from wheat (Triticum aestivum L.) seedlings and the localization of ent-kaurene synthetase in plastids of 3 species. Planta 197: 333-342 [Google Scholar]
- Ait-Ali T, Swain SM, Reid JB, Sun TP, Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11: 443-454 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins WM, Sligar SG (1988) The roles of active site hydrogen bonding in cytochrome P-450cam as revealed by site-directed mutagenesis. J Biol Chem 263: 18842-18849 [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JG, Struhl K (1994) Current Protocols in Molecular Biology 1. Wiley Inter-science, New York
- Batge SL, Ross JJ, Reid JB (1999) Abscisic acid levels in seeds of the gibberellin-deficient mutant lh-2 of pea (Pisum sativum). Physiol Plant 105: 485-490 [Google Scholar]
- Beveridge CA, Murfet IC (1996) The gigas mutant in pea is deficient in the floral stimulus. Physiol Plant 96: 637-645 [Google Scholar]
- Bradley JM, Whitelam GC, Harberd NP (1995) Impaired splicing of phytochrome-B pre-messenger RNA in a novel Phyb mutant of Arabidopsis. Plant Mol Biol 27: 1133-1142 [DOI] [PubMed] [Google Scholar]
- Brown JWS (1996) Arabidopsis intron mutations and pre-mRNA splicing. Plant J 10: 771-780 [DOI] [PubMed] [Google Scholar]
- Brown JWS, Smith P, Simpson CG (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 32: 531-535 [DOI] [PubMed] [Google Scholar]
- Coolbaugh RC, Hirano SS, West CA (1978) Studies on the specificity and site of action of α-cyclopropyl-α-[p-methoxyphenyl]-5-pyrimidine methyl alcohol (ancymidol), a plant growth regulator. Plant Physiol 62: 571-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis THN (1994) Approaches to the genetic mapping of pea. In HF Linskens, JF Jackson, eds, Modern Methods of Plant Analysis, Vol 16, Vegetables and Vegetable Products. Springer Verlag, Berlin, pp 117-160
- Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998-9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman VM, Gaskin P, MacMillan J (1974) Qualitative and quantitative analyses of gibberellins throughout seed maturation in Pisum sativum cv. progress no. 9. Planta 118: 123-132 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Sakai F, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901-915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J (1995) Structure and function of cytochrome-P450: a comparative analysis of 3 crystal structures. Structure 3: 41-62 [DOI] [PubMed] [Google Scholar]
- Hedden P (1997) The oxidases of gibberellin biosynthesis: their function and mechanism. Physiol Plant 101: 709-719 [Google Scholar]
- Hedden P, Graebe JE (1985) Inhibition of gibberellin biosynthesis by paclobutrazol in cell-free homogenates of Cucurbita maxima endosperm and Malus pumila embryos. Plant Growth Regul 4: 111-122 [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523-530 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001a) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065-2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Olive MR, Gebbie L, Forster R, Peacock WJ, Dennis ES (2000) Isolation of an ent-kaurene oxidase cDNA from Cucurbita maxima. Aust J Plant Physiol 27: 1141-1149 [Google Scholar]
- Helliwell CA, Poole A, Peacock WJ, Dennis ES (1999) Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol 119: 507-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JAD, Peacock WJ, Dennis ES (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95: 9019-9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sullivan JA, Mould RM, Gray JC, Peacock WJ, Dennis ES (2001b) A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J 28: 201-208 [DOI] [PubMed] [Google Scholar]
- Hooley R (1994) Gibberellins: perception, transduction and responses. Plant Mol Biol 26: 1529-1555 [DOI] [PubMed] [Google Scholar]
- Imai M, Shimada H, Watanabe Y, Matsushima-Hibiya Y, Makino R, Koga H, Horiuchi T, Ishimura Y (1989) Uncoupling of the cytochrome P-450cam monooxygenase reaction by a single mutation, threonine-252 to alanine or valine: possible role of the hydroxy amino acid in oxygen activation. Proc Natl Acad Sci USA 86: 7823-7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB (1987) Internode length in Pisum: I. Gene na may block gibberellin synthesis between ent-7-alpha-hydroxykaurenoic acid and gibberellin A12-aldehyde. Plant Physiol 83: 1048-1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB, MacMillian J (1986) The quantitative relationship between gibberellin A1 and internode growth in Pisum sativum L. Planta 168: 414-420 [DOI] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB, Murfet IC, Gaskin P, Willis CL, MacMillian J (1984) Internode Length in Pisum. Planta 160: 455-463 [DOI] [PubMed] [Google Scholar]
- Kalb VF, Loper JC (1988) Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity. Proc Natl Acad Sci USA 85: 7221-7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257-263 [DOI] [PubMed] [Google Scholar]
- Lal SL, Choi JH, Shaw JR, Hannah LC (1999) A splice site mutant of maize activates cryptic splice sites, elicits intron inclusion and exon exclusion, and permits branch point elucidation. Plant Physiol 121: 411-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendels stem length gene (Le) encodes a gibberellin 3-beta-hydroxylase. Plant Cell 9: 1435-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Filipowicz W (1996) Mapping of branchpoint nucleotides in mutant pre-mRNAs expressed in plant cells. Plant J 9: 381-389 [DOI] [PubMed] [Google Scholar]
- MacKenzie-Hose AK, Ross JJ, Davies NW, Swain SM (1998) Expression of gibberellin mutations in fruits of Pisum sativum L. Planta 204: 397-403 [Google Scholar]
- Nicholas KB Jr, Nicholas HB, Deerfield DWII (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW. NEWS 4: 14 [Google Scholar]
- Ogawa S, Toyomasu T, Yamane H, Murofushi N, Ikeda R, Morimoto Y, Nishimura Y, Omori T (1996) A step in the biosynthesis of gibberellins that is controlled by the mutation in the semi-dwarf rice cultivar Tanginbozu. Plant Cell Physiol 37: 363-368 [Google Scholar]
- Orozco BM, McClung CR, Werneke JM, Ogren WL (1993) Molecular basis of the ribulose-1,5-bisphosphate carboxylase oxygenase activase mutation in Arabidopsis thaliana is a guanine-to-adenine transition at the 5′-splice junction of intron 3. Plant Physiol 102: 227-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19: 307-317 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51-64 [DOI] [PubMed] [Google Scholar]
- Porter TD, Coon MJ (1991) Cytochrome-P-450: multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem 266: 13469-13472 [PubMed] [Google Scholar]
- Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J (1985) The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem 260: 16122-16130 [PubMed] [Google Scholar]
- Poulos TL, Finzel BC, Howard AJ (1986) Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry 25: 5314-5322 [DOI] [PubMed] [Google Scholar]
- Reid JB (1986) Internode length in Pisum. Three further loci, lh, ls and lk. Ann Bot 57: 577-592 [Google Scholar]
- Reid JB, Potts BM (1986) Internode length in Pisum. Two further mutants, lh and ls, with reduced gibberellin synthesis, and a gibberellin insensitive mutant, lk. Plant Physiol 66: 417-426 [Google Scholar]
- Reid JB, Ross JJ (1993) A mutant based approach, using Pisum sativum, to understand plant growth. Int J Plant Sci 154: 22-34 [Google Scholar]
- Ross JJ, Reid JB, Gaskin P, MacMillian J (1989) Internode length in Pisum: estimation of GA1 levels in genotypes Le, le and led. Physiol Plant 76: 173-176 [Google Scholar]
- Sugavanam B (1984) Diastereoisomers and enantiomers of paclobutrazol: their preparation and biological activity. Pestic Sci 15: 296-302 [Google Scholar]
- Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509-1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Reid JB (1992) Internode length in Pisum: a new allele at the Lh locus. Physiol Plant 86: 124-130 [Google Scholar]
- Swain SM, Reid JB, Kamiya Y (1997) Gibberellins are required for embryo growth and seed development in pea. Plant J 12: 1329-1338 [Google Scholar]
- Swain SM, Reid JB, Ross JJ (1993) Seed development in Pisum: the lhi allele reduces gibberellin levels in developing seeds, and increases seed abortion. Planta 191: 482-488 [Google Scholar]
- Swain SM, Ross JJ, Reid JB, Kamiya Y (1995) Gibberellins and pea seed development: expression of the lhi, ls and le5839 mutations. Planta 195: 426-433 [Google Scholar]
- Swofford DL (1999) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176-19186 [DOI] [PubMed] [Google Scholar]
- Weller JL, Murfet IC, Reid JB (1997) Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol 114: 1225-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochromes P450. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0028,http://www.aspb.org/publications/arabidopsis
- Yamaguchi S, Kamiya Y (2002) Gibberellins and light-stimulated seed germination. J Plant Growth Regul 20: 369-376 [DOI] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB (2001) Gibberellin biosynthesis mutations and root development in pea. Plant Physiol 125: 627-633 [DOI] [PMC free article] [PubMed] [Google Scholar]