Abstract
Background
Most diseases are thought to arise from interactions between environmental factors and the host genotype. To detect gene–environment interactions in the development of lifestyle-related diseases, and especially cancer, the Japan Multi-institutional Collaborative Cohort (J-MICC) Study was launched in 2005.
Methods
We initiated a cross-sectional study to examine associations of genotypes with lifestyle and clinical factors, as assessed by questionnaires and medical examinations. The 4519 subjects were selected from among participants in the J-MICC Study in 10 areas throughout Japan. In total, 108 polymorphisms were chosen and genotyped using the Invader assay.
Results
The study group comprised 2124 men and 2395 women with a mean age of 55.8 ± 8.9 years (range, 35–69 years) at baseline. Among the 108 polymorphisms examined, 4 were not polymorphic in our study population. Among the remaining 104 polymorphisms, most variations were common (minor allele frequency ≥0.05 for 96 polymorphisms). The allele frequencies in this population were comparable with those in the HapMap-JPT data set for 45 Japanese from Tokyo. Only 5 of 88 polymorphisms showed allele-frequency differences greater than 0.1. Of the 108 polymorphisms, 32 showed a highly significant difference in minor allele frequency among the study areas (P < 0.001).
Conclusions
This comprehensive data collection on lifestyle and clinical factors will be useful for elucidating gene–environment interactions. In addition, it is likely to be an informative reference tool, as free access to genotype data for a large Japanese population is not readily available.
Key words: allele frequency, cross-sectional studies, gene–environment interactions, Japan Multi-institutional Collaborative Cohort Study, polymorphism
INTRODUCTION
Although the etiology of many diseases is not completely understood, most are likely to be caused by interactions between hazardous environmental factors and the host genome. Recent advances in genotyping techniques have allowed many epidemiologic studies to investigate gene–environment interactions in chronic diseases.1–4 Cohort and case–control studies focusing on such interactions are ongoing worldwide, and these investigations use DNA from established and new cohorts.5–8 Understanding gene–environment interactions requires long-term cohort studies to clarify the temporality of associations and to avoid information and selection biases that are inevitable in cross-sectional and case–control studies.9 For most multifactorial diseases, such cohort studies must be conducted on a large scale to ensure significant results.
The Japan Multi-institutional Collaborative Cohort (J-MICC) Study is a new cohort study that was launched in 2005 to examine gene–environment interactions in lifestyle-related diseases, especially cancers. It is supported by a research grant for Scientific Research on Special Priority Areas of Cancer from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT).10,11 The J-MICC Study group is composed of 10 cohorts surveyed by 10 independent research teams.12,13
Although the long-term aim of this study is to elucidate gene–environment interactions in the whole cohort, some of its research objectives will be achieved by cross-sectional studies. In 2009, we started a cross-sectional study to examine correlations between lifestyle and medical factors, as assessed using questionnaires and medical examinations, and the distribution of possible related genotypes. Here we describe the recruitment and profile of the participants, including their genotype analysis, and selected demographic, lifestyle, and medical characteristics.
METHODS
Study participants, data collection, and blood sampling
The participants in this study completed questionnaires on lifestyle factors and diseases and donated blood samples at the time of the baseline survey for the J-MICC Study. The details of the J-MICC Study have been described elsewhere.10 The participants were enrolled in 9 study areas throughout Japan between 2005 and 2008, and in 1 area in 2004, under the supervision of an associate member of the J-MICC Study. The study participants were enrolled from the community by mailing invitation letters or distributing leaflets (3 areas), or by recruiting patients at their first visit to a cancer hospital (1 area) or at health checkups (6 areas). The response rates for the baseline survey were 7.0%, 36.5%, 25.9%, 58.4%, 60.1%, 37.6%, 14.0%, 24.0%, 19.7%, and 65.5% for the Chiba, Shizuoka, Okazaki, Aichi Cancer Center, Takashima, Kyoto, Tokushima, Fukuoka, Saga, and Amami areas, respectively. For cases in which the baseline survey is still ongoing in a cohort, the latest response rate (as of 30 September 2010 or later) was used. Anthropometry, blood pressure, and blood chemistry data obtained from health checkups were available in 8 of the study areas. The subjects for the cross-sectional study comprised 500 to 600 participants enrolled consecutively in each area of the J-MICC Study, except in 2 areas, where fewer participants had been recruited. The recruitment period for the present study, however, was arbitrarily defined by the researchers in each area after the enrollment.
Of the 5108 men and women initially selected, we excluded participants for whom we did not have sufficient DNA (n = 442), appropriate informed consent (n = 8), questionnaire data (n = 9), or local government registration of residence in the study area (n = 7), as well as anyone who had declined follow up (n = 2) or withdrew from the study (n = 1), and the 120 participants who were younger than 35 years or older than 69 years. Thus, our final study group comprised 4519 participants aged 35 to 69 years.
All the participants included in this analysis had provided written informed consent. The ethics committees of Nagoya University School of Medicine (the affiliation of the former principal investigator, Nobuyuki Hamajima) and the other participating institutions approved the protocol for the J-MICC Study.
Genotyping
We chose 107 single nucleotide polymorphisms (SNPs) and 1 insertion/deletion polymorphism for genotyping, based on their potential relevance to the lifestyle and medical factors described in the next section (“Lifestyle and clinical data”). Researchers from all participating cohorts proposed potentially relevant polymorphisms, and those selected for inclusion in the present study were determined through discussion among the members of the J-MICC Study Group.
In all study areas except Fukuoka, buffy coat fractions were prepared from blood samples and stored at −80°C at the central J-MICC Study office. DNA was extracted from all buffy coat fractions using a BioRobot M48 Workstation (Qiagen Group, Tokyo, Japan) at the central study office. For the samples from the Fukuoka area, DNA was extracted locally from samples of whole blood, using an automatic nucleic acid isolation system (NA-3000, Kurabo, Co., Ltd, Osaka, Japan). The buffy coat fractions or DNA samples were anonymized in a linkable manner14 and then sent to the central office.
The selected polymorphisms were genotyped using the multiplex polymerase chain reaction (PCR)-based Invader assay15 (Third Wave Technologies, Madison, WI, USA) at the Laboratory for Genotyping Development, Center for Genomic Medicine, RIKEN.
Lifestyle and clinical characteristics
The lifestyle factors considered were smoking and drinking habits, coffee consumption, sleep, and mental stress, while the clinical characteristics were height, weight, blood pressure, blood glucose, glycated hemoglobin (HbA1c), serum triglyceride, total and high-density lipoprotein (HDL) cholesterol, uric acid, aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyltransferase (γ-GT), C-reactive protein (CRP), creatinine, and bone mineral density. Ages at menarche and menopause were also ascertained.
We used a standard questionnaire in all study areas except the Fukuoka area, where some questions are slightly different from those of other areas. Furthermore, a validated food-frequency questionnaire was used for the dietary assessment.16–19 We were unable to directly control the quality of information from health examinations because most data were obtained at routine health checkups offered by other institutions. However, the J-MICC Study Group is now collecting information on participation in the Japan Medical Association’s quality control program for clinical laboratories and the instruction manuals used for measurement of blood pressure, height, and weight. For the current report, participants whose blood was drawn less than 3 hours after their last meal were excluded from the analysis of serum lipids and blood glucose.
Statistical analysis
We tabulated selected baseline characteristics by sex and 10-year age group or by sex and study area. In this analysis, body mass index (BMI; kg/m2) was calculated on the basis of self-reported height and body weight, as independent measurements were not available in some study areas. In the case of educational attainment, participants from the Fukuoka area were excluded from the analysis because the questionnaire used there had not included this item. Participants who consumed alcohol at least once a week were classified as drinkers. To compare characteristics among participating cohorts, we attempted to adjust for age by using the direct method (for proportions) or the general linear model (for means). The variations among study areas, however, were not significantly altered after adjusting for age. Thus, in this report, we present only crude figures by sex and study area. The difference in the minor allele frequency (MAF) among the cohorts was tested by the chi-square test for contingency tables. The MAF of the ABCC11 Arg180Gly (T/C) polymorphism by study area is not presented here because the inter-area variation in the distribution of this genotype will be reported in a separate article.
Genotypes with distributions that departed from the Hardy–Weinberg equilibrium were assessed using the exact test20 with the genhwi command of Stata version 8.0 (Stata Corp, College Station, TX, USA). Other statistical analyses were performed using Statistical Analysis System version 9.1 (SAS Institute Inc, Cary, NC, USA).21 To compare the allele frequencies of genotypes in our study with those in another Japanese population, we used data from HapMap, which is an open access database that includes allele frequencies for 45 Japanese in Tokyo (HapMap-JPT, http://www.ncbi.nlm.nih.gov/snp). Of the 108 polymorphisms of interest, we made comparisons for 88. The 20 polymorphisms excluded from our analysis showed no minor alleles in our study group (n = 4), were not represented (n = 15), or had invalid data (n = 1, 100% heterozygotes) in the HapMap-JPT data set.
RESULTS
Our analysis included 2124 men (47.0%) and 2395 women (53.0%) with a mean age ± standard deviation at baseline of 55.8 ± 8.9 years (range, 35–69 years). There were considerable differences in the age and sex distributions of different study areas (Table 1). In Fukuoka and Saga, the participants originally enrolled in the J-MICC Study were limited to adults aged 50 years or older and 40 years or older, respectively.
Table 1. Sex and age distribution of study participants by study area.
| Study area | Men | Women | ||||||||||||||||||
| Age (years) | Total | Age (years) | Total | |||||||||||||||||
| 35–39 | 40–49 | 50–59 | 60–69 | 35–39 | 40–49 | 50–59 | 60–69 | |||||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| Chiba | 4 | 2.7 | 22 | 14.8 | 56 | 37.6 | 67 | 45.0 | 149 | 100.0 | 30 | 8.4 | 138 | 38.7 | 118 | 33.1 | 71 | 19.9 | 357 | 100.0 |
| Shizuoka | 21 | 5.0 | 122 | 29.3 | 175 | 42.1 | 98 | 23.6 | 416 | 100.0 | 16 | 10.1 | 35 | 22.0 | 70 | 44.0 | 38 | 23.9 | 159 | 100.0 |
| Okazaki | 13 | 4.8 | 29 | 10.6 | 66 | 24.2 | 165 | 60.4 | 273 | 100.0 | 12 | 4.7 | 45 | 17.6 | 85 | 33.3 | 113 | 44.3 | 255 | 100.0 |
| Aichi Cancer Center | 12 | 4.4 | 32 | 11.6 | 115 | 41.8 | 116 | 42.2 | 275 | 100.0 | 33 | 10.9 | 88 | 29.0 | 102 | 33.7 | 80 | 26.4 | 303 | 100.0 |
| Takashima | 7 | 4.2 | 18 | 10.7 | 45 | 26.8 | 98 | 58.3 | 168 | 100.0 | 27 | 7.2 | 59 | 15.8 | 102 | 27.3 | 186 | 49.7 | 374 | 100.0 |
| Kyoto | 37 | 30.3 | 31 | 25.4 | 48 | 39.3 | 6 | 4.9 | 122 | 100.0 | 9 | 23.7 | 19 | 50.0 | 9 | 23.7 | 1 | 2.6 | 38 | 100.0 |
| Tokushima | 8 | 11.0 | 21 | 28.8 | 24 | 32.9 | 20 | 27.4 | 73 | 100.0 | 1 | 4.5 | 9 | 40.9 | 10 | 45.5 | 2 | 9.1 | 22 | 100.0 |
| Fukuoka | 0 | 0.0 | 0 | 0.0 | 60 | 31.9 | 128 | 68.1 | 188 | 100.0 | 0 | 0.0 | 0 | 0.0 | 96 | 37.5 | 160 | 62.5 | 256 | 100.0 |
| Saga | 0 | 0.0 | 31 | 12.7 | 82 | 33.5 | 132 | 53.9 | 245 | 100.0 | 0 | 0.0 | 64 | 19.3 | 127 | 38.4 | 140 | 42.3 | 331 | 100.0 |
| Amami | 1 | 0.5 | 53 | 24.7 | 82 | 38.1 | 79 | 36.7 | 215 | 100.0 | 1 | 0.3 | 77 | 25.7 | 135 | 45.0 | 87 | 29.0 | 300 | 100.0 |
| Total | 103 | 4.8 | 359 | 16.9 | 753 | 35.5 | 909 | 42.8 | 2124 | 100.0 | 129 | 5.4 | 534 | 22.3 | 854 | 35.7 | 878 | 36.7 | 2395 | 100.0 |
Table 2 summarizes selected demographic, lifestyle, and medical characteristics of the participants by sex and age. Within our sample, 29.1% of men and 7.1% of women were current smokers. More than two thirds (71.4%) of men drank alcoholic beverages at least once a week, as did 27.7% of the women. Table 3 presents data on selected lifestyle and medical variables of the participants by sex and study area. Considerable variations were found among the participating cohorts.
Table 2. Selected demographic, lifestyle, and medical characteristics of participants by sex and age.
| Men | Women | |||||||||
| Age (years) | Total | Age (years) | Total | |||||||
| 35–39 | 40–49 | 50–59 | 60–69 | 35–39 | 40–49 | 50–59 | 60–69 | |||
| n | 103 | 359 | 753 | 909 | 2124 | 129 | 534 | 854 | 878 | 2395 |
| Educational attainment (%)a | ||||||||||
| Elementary/junior high school |
1.0 | 3.1 | 10.0 | 20.9 | 12.6 | 2.3 | 2.1 | 9.8 | 31.6 | 14.7 |
| High school | 42.7 | 37.8 | 41.9 | 43.7 | 41.9 | 41.9 | 38.4 | 50.7 | 46.4 | 45.6 |
| Vocational school | 14.6 | 9.5 | 6.8 | 3.2 | 6.3 | 15.5 | 13.9 | 12.7 | 10.6 | 12.5 |
| Junior college | 3.9 | 4.8 | 3.9 | 2.2 | 3.4 | 24.8 | 26.5 | 16.1 | 7.4 | 16.3 |
| University | 33.0 | 39.5 | 34.8 | 27.6 | 32.7 | 15.5 | 17.7 | 10.2 | 3.8 | 10.2 |
| Postgraduate school | 4.9 | 5.0 | 2.2 | 1.7 | 2.6 | 0.0 | 1.5 | 0.5 | 0.0 | 0.6 |
| Others | 0.0 | 0.3 | 0.4 | 0.7 | 0.5 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 |
| Current smokers (%) | 39.8 | 32.3 | 34.8 | 21.9 | 29.1 | 11.6 | 10.3 | 7.4 | 4.1 | 7.1 |
| Ex-smokers (%) | 25.2 | 35.7 | 41.8 | 48.6 | 42.9 | 5.4 | 7.9 | 4.8 | 2.8 | 4.8 |
| Current drinkers (%)b | 60.2 | 70.8 | 75.1 | 69.9 | 71.4 | 32.6 | 34.6 | 27.5 | 22.8 | 27.7 |
| Exercise ≥1/month (%) | 75.7 | 81.6 | 79.7 | 86.8 | 82.9 | 62.0 | 70.4 | 74.9 | 82.5 | 76.0 |
| Body mass index ≥25.0 (%) |
37.9 | 32.2 | 32.6 | 24.9 | 29.5 | 9.5 | 16.5 | 20.0 | 21.8 | 19.3 |
| History of disease (%) | ||||||||||
| Diabetes | 1.9 | 4.5 | 8.4 | 13.9 | 9.7 | 0.0 | 0.6 | 3.4 | 7.1 | 3.8 |
| Hypertension | 3.9 | 8.9 | 23.2 | 36.2 | 25.2 | 0.0 | 4.3 | 17.1 | 30.5 | 17.9 |
| Coronary heart disease |
0.0 | 1.1 | 2.1 | 7.3 | 4.0 | 0.8 | 0.6 | 2.8 | 5.3 | 3.0 |
| Stroke | 0.0 | 1.1 | 2.7 | 3.9 | 2.8 | 2.3 | 0.9 | 1.6 | 2.9 | 2.0 |
| Cancer | 6.1 | 2.0 | 7.7 | 12.6 | 8.6 | 2.8 | 5.0 | 9.3 | 6.4 | 6.9 |
| Blood pressure and blood chemistryc | ||||||||||
| Systolic blood pressure (mm Hg) |
120.4 ± 14.3 | 121.1 ± 15.4 | 129.7 ± 17.9 | 135.4 ± 19.3 | 130.1 ± 18.8 | 109.6 ± 11.4 | 116.8 ± 18.3 | 125.0 ± 18.9 | 133.4 ± 19.0 | 126.5 ± 19.9 |
| Diastolic blood pressure (mm Hg) |
73.6 ± 10.3 | 77.6 ± 11.7 | 81.9 ± 12.6 | 82.1 ± 11.1 | 80.8 ± 11.9 | 65.4 ± 7.4 | 72.0 ± 11.9 | 76.7 ± 11.6 | 79.0 ± 10.8 | 76.4 ± 11.7 |
| Total cholesterol (mg/dl) |
191.3 ± 28.9 | 205.3 ± 30.2 | 206.6 ± 32.5 | 205.2 ± 33.1 | 205.2 ± 32.3 | 185.6 ± 25.2 | 202.4 ± 31.0 | 223.3 ± 34.2 | 223.1 ± 33.5 | 217.9 ± 34.6 |
| HDL-cholesterol (mg/dl) |
56.2 ± 16.0 | 57.6 ± 15.4 | 59.1 ± 15.8 | 59.7 ± 16.2 | 58.9 ± 15.9 | 73.1 ± 15.7 | 68.7 ± 15.3 | 68.9 ± 15.0 | 66.2 ± 16.0 | 68.0 ± 15.5 |
| Triglyceride (mg/dl) | 128.6 ± 74.9 | 146.5 ± 96.5 | 135.1 ± 93.0 | 132.0 ± 92.9 | 135.9 ± 93.0 | 64.8 ± 28.4 | 88.0 ± 61.2 | 104.9 ± 64.2 | 113.7 ± 69.2 | 103.4 ± 65.7 |
| Blood glucose (mg/dl) | 98.0 ± 27.2 | 98.3 ± 15.7 | 103.2 ± 17.6 | 103.9 ± 23.3 | 102.1 ± 20.3 | 86.8 ± 7.1 | 92.4 ± 19.7 | 95.9 ± 16.8 | 97.1 ± 15.8 | 95.1 ± 16.9 |
| HbA1c (%) | 4.96 ± 0.32 | 5.12 ± 0.48 | 5.32 ± 0.86 | 5.30 ± 0.75 | 5.27 ± 0.74 | 4.86 ± 0.27 | 4.96 ± 0.36 | 5.17 ± 0.64 | 5.26 ± 0.57 | 5.17 ± 0.57 |
Plus-minus values are means ± SDs.
aParticipants in Fukuoka area were excluded from the analysis because they were not asked about educational attainment in the questionnaire.
bIndividuals who drank alcoholic beverages ≥1 day/week.
cNot available in some study areas, as shown in Table 3.
Table 3. Selected lifestyle and medical characteristics of participants by sex and study area.
| n | Study area | |||||||||||
| Total | Chiba | Shizuoka | Okazaki | ACC | Takashima | Kyoto | Tokushima | Fukuoka | Saga | Amami | ||
| Men | ||||||||||||
| Current smokers (%) | 2123 | 29.1 | 23.5 | 23.6 | 25.6 | 31.0 | 35.7 | 41.8 | 31.5 | 33.0 | 34.3 | 23.3 |
| Ex-smokers (%) | 2123 | 42.9 | 42.3 | 48.3 | 50.2 | 44.2 | 33.3 | 33.6 | 38.4 | 39.4 | 42.9 | 39.1 |
| Current drinkers (%)a | 2121 | 71.4 | 78.5 | 73.4 | 62.6 | 66.9 | 73.8 | 68.0 | 61.6 | 70.2 | 71.4 | 84.1 |
| Exercise ≥1/month (%) | 2109 | 82.9 | 87.1 | 88.7 | 81.0 | 83.5 | 60.1 | 81.0 | 94.5 | 89.9 | 85.3 | 75.9 |
| Body mass index ≥25.0 (%) | 2114 | 29.5 | 29.1 | 25.2 | 26.4 | 22.6 | 27.0 | 23.8 | 41.1 | 31.4 | 27.7 | 52.8 |
| History of hypertension (%) | 2052 | 25.2 | 28.2 | 14.7 | 29.5 | 24.0 | 28.7 | 9.9 | 21.9 | 44.4 | 28.6 | 32.1 |
| Systolic blood pressure (mm Hg) | 1698 | 130.1 ± 18.8 | NA | 121.9 ± 14.8 | 128.7 ± 15.2 | NA | 133.0 ± 19.5 | 122.8 ± 16.0 | 119.8 ± 17.6 | 144.0 ± 19.6 | 138.3 ± 19.3 | 132.0 ± 17.7 |
| Diastolic blood pressure (mm Hg) |
1698 | 80.8 ± 11.9 | NA | 76.5 ± 10.5 | 81.4 ± 9.1 | NA | 81.2 ± 11.2 | 74.3 ± 11.9 | 73.7 ± 12.8 | 88.0 ± 11.1 | 86.0 ± 12.8 | 81.9 ± 11.1 |
| Total cholesterol (mg/dl) | 1296 | 205.2 ± 32.3 | NA | 201.3 ± 29.3 | 205.9 ± 32.0 | NA | 208.3 ± 35.6 | —b | 209.0 ± 31.4 | 209.9 ± 31.5 | 203.6 ± 33.6 | 208.0 ± 35.7 |
| HDL-cholesterol (mg/dl) | 1378 | 58.9 ± 15.9 | NA | 58.3 ± 15.4 | 64.1 ± 17.5 | NA | 59.4 ± 17.0 | 61.4 ± 17.6 | 52.5 ± 11.6 | 54.5 ± 16.1 | 55.3 ± 14.2 | 57.7 ± 13.4 |
| Triglyceride (mg/dl) | 1377 | 135.9 ± 93.0 | NA | 122.8 ± 68.6 | 120.8 ± 86.5 | NA | 116.9 ± 60.0 | 127.9 ± 85.7 | 126.7 ± 64.2 | 181.5 ± 131.9 | 164.9 ± 93.6 | 165.7 ± 130.8 |
| Blood glucose (mg/dl) | 1175 | 102.1 ± 20.3 | NA | 101.1 ± 14.3 | 100.0 ± 17.0 | NA | 97.9 ± 16.3 | 98.7 ± 25.0 | 104.2 ± 26.4 | NA | NA | 110.3 ± 28.8 |
| HbA1c (%) | 1354 | 5.27 ± 0.74 | NA | 5.27 ± 0.56 | 5.36 ± 0.64 | NA | 5.24 ± 0.79 | NA | 5.17 ± 0.52 | 5.17 ± 0.82 | 5.28 ± 1.04 | 5.31 ± 0.42 |
| Women | ||||||||||||
| Current smokers (%) | 2390 | 7.1 | 7.6 | 5.0 | 6.3 | 12.4 | 5.6 | 15.8 | 0.0 | 9.0 | 5.4 | 4.4 |
| Ex-smokers (%) | 2390 | 4.8 | 6.7 | 4.4 | 5.9 | 8.4 | 1.3 | 7.9 | 4.6 | 4.7 | 5.7 | 1.0 |
| Current drinkers (%)a | 2383 | 27.7 | 36.1 | 27.2 | 22.4 | 31.7 | 24.6 | 34.2 | 22.7 | 25.0 | 28.2 | 23.3 |
| Exercise ≥1/month (%) | 2381 | 76.0 | 82.8 | 81.8 | 83.9 | 70.9 | 55.6 | 44.7 | 81.8 | 88.3 | 83.4 | 73.1 |
| Body mass index ≥25.0 (%) | 2359 | 19.3 | 12.8 | 15.5 | 17.3 | 14.0 | 21.9 | 15.8 | 27.3 | 19.5 | 16.5 | 36.2 |
| History of hypertension (%) | 2289 | 17.9 | 10.4 | 15.8 | 19.3 | 12.9 | 18.5 | 7.9 | 18.2 | 37.9 | 17.6 | 22.0 |
| Systolic blood pressure (mm Hg) | 1732 | 126.5 ± 19.9 | NA | 115.7 ± 16.8 | 122.7 ± 15.6 | NA | 124.6 ± 19.5 | 120.4 ± 19.5 | 111.5 ± 11.7 | 137.5 ± 21.9 | 129.5 ± 19.5 | 126.8 ± 19.0 |
| Diastolic blood pressure (mm Hg) |
1732 | 76.4 ± 11.7 | NA | 70.3 ± 10.5 | 76.7 ± 9.5 | NA | 73.6 ± 11.4 | 71.8 ± 11.1 | 68.2 ± 10.3 | 84.0 ± 11.3 | 77.9 ± 12.2 | 76.0 ± 10.4 |
| Total cholesterol (mg/dl) | 1318 | 217.9 ± 34.6 | NA | 206.4 ± 31.9 | 213.5 ± 34.1 | NA | 222.6 ± 36.5 | NA | 210.0 ± 35.7 | 230.1 ± 31.4 | 220.3 ± 33.5 | 218.0 ± 33.7 |
| HDL-cholesterol (mg/dl) | 1347 | 68.0 ± 15.5 | NA | 72.3 ± 16.1 | 75.0 ± 16.9 | NA | 67.0 ± 15.0 | 69.3 ± 13.7 | 65.5 ± 12.0 | 66.1 ± 16.6 | 64.3 ± 14.1 | 63.4 ± 12.6 |
| Triglyceride (mg/dl) | 1347 | 103.4 ± 65.7 | NA | 80.0 ± 40.9 | 97.3 ± 60.9 | NA | 96.8 ± 54.9 | 68.6 ± 27.8 | 84.3 ± 41.2 | 128.0 ± 66.0 | 129.2 ± 83.4 | 109.4 ± 74.2 |
| Blood glucose (mg/dl) | 1072 | 95.1 ± 16.9 | NA | 91.9 ± 8.3 | 94.7 ± 13.7 | NA | 91.0 ± 10.2 | 89.6 ± 7.1 | 101.9 ± 31.9 | NA | NA | 101.8 ± 24.3 |
| HbA1c (%) | 1396 | 5.17 ± 0.57 | NA | 5.15 ± 0.35 | 5.32 ± 0.57 | NA | 5.11 ± 0.54 | NA | —b | 5.16 ± 0.65 | 5.10 ± 0.59 | —b |
Abbreviations: ACC, Aichi Cancer Center; HDL, high-density lipoprotein; NA, not available.
Plus-minus values are means ± SDs.
aIndividuals who drank alcoholic beverages ≥1 day/week.
bData available for fewer than 20 participants.
The genotype distributions and allele frequencies of the analyzed genetic polymorphisms are summarized in Table 4. The call rate ranged from 99.40% to 99.98%. Of the 108 polymorphisms, the 4 SNPs for which we found no different alleles were APOA1 Arg184Pro (G/C), ESR1 IVS1-351A/G (Xba I), LCAT/SLC12A4 Ser232Thr (T/A), and SCARB1 Val135Ile (G/A). For the remaining 104 polymorphisms, the MAF varied from 0.016 (PTGS2(COX2) C-163G) to 0.492 (CETP Ile405Val (A/G)), and most of the variations were common (MAF ≥0.05 for 96 polymorphisms).
Table 4. Genotype distributions and allele frequencies of 108 selected genetic polymorphisms (107 SNPs and 1 insertion/deletion polymorphism).
| Gene | Polymorphism | rs number | Genotypea | na | Frequency (proportion) |
P for HWE |
MAF | ||||||||||
| Observed | Expectedb | ||||||||||||||||
| AA | Aa | aa | AA | Aa | aa | XX | AA | Aa | aa | AA | Aa | aa | |||||
| ABCA1 | C-565T | rs2422493 | CC | CT | TT | 1634 | 2137 | 746 | 2 | 0.362 | 0.473 | 0.165 | 0.358 | 0.481 | 0.161 | 0.29 | 0.402 |
| ABCA1 | G-191C | rs1800976 | GG | GC | CC | 1628 | 2144 | 746 | 1 | 0.360 | 0.475 | 0.165 | 0.357 | 0.481 | 0.162 | 0.37 | 0.402 |
| ABCA1 | G-17C | rs2740483 | GG | GC | CC | 2211 | 1898 | 409 | 1 | 0.489 | 0.420 | 0.091 | 0.489 | 0.420 | 0.090 | 0.94 | 0.301 |
| ABCA1 | Val825Ile (G/A) | rs2066715 | GG | GA | AA | 1877 | 2064 | 577 | 1 | 0.415 | 0.457 | 0.128 | 0.415 | 0.459 | 0.127 | 0.80 | 0.356 |
| ABCA1 | Val771Met (G/A) | rs2066718 | GG | GA | AA | 3945 | 550 | 23 | 1 | 0.873 | 0.122 | 0.005 | 0.872 | 0.123 | 0.004 | 0.40 | 0.066 |
| ABCA1 | Arg1587Lys (G/A) | rs2230808 | GG | GA | AA | 1673 | 2154 | 689 | 3 | 0.370 | 0.477 | 0.153 | 0.371 | 0.476 | 0.153 | 0.95 | 0.391 |
| ABCC11 | Arg180Gly (T/C) | rs17822931 | TT | TC | CC | 3368 | 1051 | 95 | 5 | 0.746 | 0.233 | 0.021 | 0.744 | 0.237 | 0.019 | 0.23 | 0.137 |
| ACE | Insertion/Deletion (I/D) | rs1799752 | I/I | I/D | D/D | 1854 | 2021 | 634 | 10 | 0.411 | 0.448 | 0.141 | 0.404 | 0.463 | 0.133 | 0.029 | 0.365 |
| ADD1 | Trp460Gly (T/G) | rs4961 | TT | TG | GG | 1399 | 2163 | 956 | 1 | 0.310 | 0.479 | 0.212 | 0.301 | 0.495 | 0.203 | 0.026 | 0.451 |
| ADH1B | His47Arg (A/G) | rs1229984 | AA | AG | GG | 2607 | 1659 | 250 | 3 | 0.577 | 0.367 | 0.055 | 0.579 | 0.364 | 0.057 | 0.54 | 0.239 |
| ADH1C | Arg272Gln (C/T) | rs1693482 | CC | CT | TT | 3882 | 591 | 43 | 3 | 0.860 | 0.131 | 0.010 | 0.856 | 0.139 | 0.006 | 0.0005 | 0.075 |
| ADIPOQ | C-11377G | rs266729 | CC | CG | GG | 2553 | 1663 | 301 | 2 | 0.565 | 0.368 | 0.067 | 0.561 | 0.376 | 0.063 | 0.18 | 0.251 |
| ADIPOQ | Gly15Gly (T/G) | rs2241766 | TT | TG | GG | 2314 | 1789 | 415 | 1 | 0.512 | 0.396 | 0.092 | 0.504 | 0.412 | 0.084 | 0.011 | 0.290 |
| ADIPOQ | G276T | rs1501299 | GG | GT | TT | 2498 | 1674 | 346 | 1 | 0.553 | 0.371 | 0.077 | 0.545 | 0.387 | 0.069 | 0.006 | 0.262 |
| ADIPOQ | IVS-3971A/G | rs822396 | AA | AG | GG | 3984 | 517 | 16 | 2 | 0.882 | 0.114 | 0.004 | 0.882 | 0.114 | 0.004 | 1.00 | 0.061 |
| ADIPOR1 | G-8503A | rs6666089 | GG | GA | AA | 4234 | 272 | 7 | 6 | 0.938 | 0.060 | 0.002 | 0.938 | 0.061 | 0.001 | 0.22 | 0.032 |
| ADIPOR1 | C5843T | rs1342387 | CC | CT | TT | 1230 | 2206 | 1073 | 10 | 0.273 | 0.489 | 0.238 | 0.268 | 0.499 | 0.233 | 0.17 | 0.483 |
| ADIPOR1 | C10224G | rs7539542 | CC | CG | GG | 2788 | 1505 | 225 | 1 | 0.617 | 0.333 | 0.050 | 0.614 | 0.339 | 0.047 | 0.24 | 0.216 |
| ADRB2 | Gln27Glu (C/G) | rs1042714 | CC | CG | GG | 3943 | 551 | 20 | 5 | 0.874 | 0.122 | 0.004 | 0.873 | 0.122 | 0.004 | 0.81 | 0.065 |
| ADRB3 | Trp64Arg (T/C) | rs4994 | TT | TC | CC | 2932 | 1397 | 182 | 8 | 0.650 | 0.310 | 0.040 | 0.648 | 0.314 | 0.038 | 0.34 | 0.195 |
| AGT | Thr174Met (C/T) | rs4762 | CC | CT | TT | 3615 | 848 | 52 | 4 | 0.801 | 0.188 | 0.012 | 0.800 | 0.189 | 0.011 | 0.75 | 0.105 |
| AGT | Thr235Met (C/T) | rs699 | CC | CT | TT | 2989 | 1372 | 157 | 1 | 0.662 | 0.304 | 0.035 | 0.662 | 0.304 | 0.035 | 1.00 | 0.187 |
| AGTR1(ATR1) | A1166C (at 3′UTR) | rs5186 | AA | AC | CC | 3777 | 695 | 45 | 2 | 0.836 | 0.154 | 0.010 | 0.834 | 0.159 | 0.008 | 0.048 | 0.087 |
| ALDH2 | Glu487Lys (G/A) | rs671 | GG | GA | AA | 2544 | 1649 | 321 | 5 | 0.564 | 0.365 | 0.071 | 0.557 | 0.379 | 0.064 | 0.018 | 0.254 |
| APOA1 | Ala61Thr (C/T) | rs12718465 | CC | CT | TT | 4036 | 463 | 18 | 2 | 0.894 | 0.103 | 0.004 | 0.893 | 0.104 | 0.003 | 0.25 | 0.055 |
| APOA1 | Arg184Pro (G/C) | rs5078 | GG | GC | CC | 4512 | 0 | 0 | 7 | 1.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 1.00 | 0.000 |
| APOA5 | T-1131C | rs662799 | TT | TC | CC | 1955 | 2001 | 556 | 7 | 0.433 | 0.443 | 0.123 | 0.429 | 0.452 | 0.119 | 0.21 | 0.345 |
| APOA5 | Gly185Cys (G553T) | rs2075291 | GG | GT | TT | 3920 | 559 | 32 | 8 | 0.869 | 0.124 | 0.007 | 0.867 | 0.129 | 0.005 | 0.020 | 0.069 |
| APOE | T-203G | rs405509 | TT | TG | GG | 2213 | 1847 | 451 | 8 | 0.491 | 0.409 | 0.100 | 0.483 | 0.424 | 0.093 | 0.025 | 0.305 |
| APOE | Arg158Cys (C/T) (at exon2) | rs7412 | CC | CT | TT | 4140 | 369 | 9 | 1 | 0.916 | 0.082 | 0.002 | 0.916 | 0.082 | 0.002 | 0.72 | 0.043 |
| APOE | Cys112Arg (T/C) (at exon4) | rs429358 | TT | TC | CC | 3669 | 782 | 67 | 1 | 0.812 | 0.173 | 0.015 | 0.808 | 0.182 | 0.010 | 0.001 | 0.101 |
| ARNTL(BMAL1) | A/G | rs7950226 | AA | AG | GG | 1638 | 2102 | 773 | 6 | 0.363 | 0.466 | 0.171 | 0.355 | 0.482 | 0.163 | 0.028 | 0.404 |
| ART4(DOK1) | Leu208Leu (G/A) | rs3088189 | GG | GA | AA | 3595 | 875 | 47 | 2 | 0.796 | 0.194 | 0.010 | 0.797 | 0.192 | 0.012 | 0.48 | 0.107 |
| ART4(DOK1) | Asp265Asn (G/A) | rs11276 | GG | GA | AA | 3597 | 872 | 48 | 2 | 0.796 | 0.193 | 0.011 | 0.797 | 0.191 | 0.011 | 0.59 | 0.107 |
| BHMT | Arg239Gln (G742A) | rs3733890 | GG | GA | AA | 2765 | 1511 | 241 | 2 | 0.612 | 0.335 | 0.053 | 0.607 | 0.344 | 0.049 | 0.069 | 0.221 |
| CBS | Tyr233Tyr (C699T) | rs234706 | CC | CT | TT | 4302 | 212 | 2 | 3 | 0.953 | 0.047 | 0.000 | 0.953 | 0.047 | 0.001 | 1.00 | 0.024 |
| CD14 | T-260C/T-159C | rs2569190 | TT | TC | CC | 1304 | 2256 | 958 | 1 | 0.289 | 0.499 | 0.212 | 0.290 | 0.497 | 0.213 | 0.79 | 0.462 |
| CDKAL1 | G/C | rs7754840 | GG | GC | CC | 1581 | 2195 | 740 | 3 | 0.350 | 0.486 | 0.164 | 0.352 | 0.483 | 0.166 | 0.64 | 0.407 |
| CDKN2A/B | T/C | rs10811661 | TT | TC | CC | 1379 | 2205 | 934 | 1 | 0.305 | 0.488 | 0.207 | 0.302 | 0.495 | 0.203 | 0.34 | 0.451 |
| CETP | A-629C | rs1800775 | AA | AC | CC | 1384 | 2242 | 891 | 2 | 0.306 | 0.496 | 0.197 | 0.308 | 0.494 | 0.198 | 0.76 | 0.445 |
| CETP | Ile405Val (A/G) | rs5882 | AA | AG | GG | 1236 | 2116 | 1166 | 1 | 0.274 | 0.468 | 0.258 | 0.258 | 0.500 | 0.242 | <0.0001 | 0.492 |
| CETP | TaqIB (C/T) | rs708272 | CC | CT | TT | 1603 | 2174 | 741 | 1 | 0.355 | 0.481 | 0.164 | 0.354 | 0.482 | 0.164 | 0.93 | 0.405 |
| CETP | G/T | rs3764261 | GG | GT | TT | 2707 | 1512 | 298 | 2 | 0.599 | 0.335 | 0.066 | 0.588 | 0.358 | 0.054 | <0.0001 | 0.233 |
| CHRNB2 | G-42A | rs2072658 | GG | GA | AA | 2837 | 1518 | 161 | 3 | 0.628 | 0.336 | 0.036 | 0.634 | 0.324 | 0.042 | 0.015 | 0.204 |
| CHRNB2 | C/T (at 3′UTR) | rs2072660 | CC | CT | TT | 2601 | 1641 | 275 | 2 | 0.576 | 0.363 | 0.061 | 0.574 | 0.367 | 0.059 | 0.44 | 0.243 |
| COMT | Val158Met (G/A) | rs4680 | GG | GA | AA | 2006 | 1983 | 528 | 2 | 0.444 | 0.439 | 0.117 | 0.440 | 0.446 | 0.113 | 0.27 | 0.336 |
| CYP1A1 | Ile462Val (A/G) | rs1048943 | AA | AG | GG | 2630 | 1620 | 268 | 1 | 0.582 | 0.359 | 0.059 | 0.580 | 0.363 | 0.057 | 0.39 | 0.239 |
| CYP1A2 | A734C | rs762551 | AA | AC | CC | 1878 | 2043 | 592 | 6 | 0.416 | 0.453 | 0.131 | 0.413 | 0.459 | 0.128 | 0.33 | 0.358 |
| CYP11B2 | T-344C | rs1799998 | TT | TC | CC | 2031 | 1976 | 507 | 5 | 0.450 | 0.438 | 0.112 | 0.447 | 0.443 | 0.110 | 0.42 | 0.331 |
| CYP17A1 | T-34C | rs743572 | TT | TC | CC | 1281 | 2257 | 977 | 4 | 0.284 | 0.500 | 0.216 | 0.285 | 0.498 | 0.217 | 0.79 | 0.466 |
| ESR1 | IVS1-351A/G (Xba I) | rs11155816 | AA | AG | GG | 4515 | 0 | 0 | 4 | 1.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 1.00 | 0.000 |
| ESR1 | IVS1-397T/C (Pvu II) | rs2234693 | TT | TC | CC | 1508 | 2172 | 834 | 5 | 0.334 | 0.481 | 0.185 | 0.330 | 0.489 | 0.181 | 0.30 | 0.425 |
| ESR2 | Val328Val (G1082A) (Rsa I) | rs1256049 | GG | GA | AA | 2320 | 1823 | 373 | 3 | 0.514 | 0.404 | 0.083 | 0.512 | 0.407 | 0.081 | 0.58 | 0.284 |
| FTO | T/A | rs9939609 | TT | TA | AA | 2881 | 1454 | 183 | 1 | 0.638 | 0.322 | 0.041 | 0.638 | 0.322 | 0.041 | 1.00 | 0.201 |
| GCK | G-30A | rs1799884 | GG | GA | AA | 2989 | 1348 | 181 | 1 | 0.662 | 0.298 | 0.040 | 0.657 | 0.307 | 0.036 | 0.065 | 0.189 |
| GCKR | A/G | rs780094 | AA | AG | GG | 1379 | 2221 | 917 | 2 | 0.305 | 0.492 | 0.203 | 0.304 | 0.495 | 0.201 | 0.67 | 0.449 |
| GCKR | Leu446Pro (T/C) | rs1260326 | TT | TC | CC | 1402 | 2214 | 902 | 1 | 0.310 | 0.490 | 0.200 | 0.308 | 0.494 | 0.198 | 0.61 | 0.445 |
| GNAS1 | T393C | rs7121 | TT | TC | CC | 1466 | 2233 | 817 | 3 | 0.325 | 0.494 | 0.181 | 0.327 | 0.490 | 0.183 | 0.52 | 0.428 |
| IGF2BP2 | G/T (at intron) | rs4402960 | GG | GT | TT | 2215 | 1866 | 436 | 2 | 0.490 | 0.413 | 0.097 | 0.486 | 0.422 | 0.092 | 0.14 | 0.303 |
| IL-1B | T-31C | rs1143627 | TT | TC | CC | 1331 | 2196 | 990 | 2 | 0.295 | 0.486 | 0.219 | 0.289 | 0.497 | 0.214 | 0.14 | 0.462 |
| IL-2 | T-330G | rs2069762 | TT | TG | GG | 2033 | 1986 | 494 | 6 | 0.450 | 0.440 | 0.109 | 0.450 | 0.442 | 0.109 | 0.79 | 0.329 |
| IL-4 | T-33C | rs2070874 | TT | TC | CC | 1997 | 1989 | 531 | 2 | 0.442 | 0.440 | 0.118 | 0.439 | 0.447 | 0.114 | 0.30 | 0.338 |
| IL-6 | C-634G | rs1800796 | CC | CG | GG | 2611 | 1607 | 298 | 3 | 0.578 | 0.356 | 0.066 | 0.572 | 0.369 | 0.059 | 0.019 | 0.244 |
| IL-8 | T-251A | rs4073 | TT | TA | AA | 2077 | 1952 | 463 | 27 | 0.462 | 0.435 | 0.103 | 0.462 | 0.435 | 0.103 | 0.89 | 0.320 |
| IL-10 | T-819C | rs1800871 | TT | TC | CC | 1942 | 2009 | 557 | 11 | 0.431 | 0.446 | 0.124 | 0.427 | 0.453 | 0.120 | 0.29 | 0.346 |
| IL-13 | C-1111T | rs1800925 | CC | CT | TT | 3009 | 1359 | 147 | 4 | 0.666 | 0.301 | 0.033 | 0.667 | 0.299 | 0.034 | 0.69 | 0.183 |
| KCNJ11 | Glu23Lys (C/T) | rs5219 | CC | CT | TT | 1822 | 2081 | 615 | 1 | 0.403 | 0.461 | 0.136 | 0.401 | 0.464 | 0.134 | 0.59 | 0.366 |
| LCAT/SLC12A4 | Ser232Thr (T/A) | rs4986970 | TT | TA | AA | 4516 | 0 | 0 | 3 | 1.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 1.00 | 0.000 |
| LIPC | T-514C | rs1800588 | TT | TC | CC | 1178 | 2255 | 1085 | 1 | 0.261 | 0.499 | 0.240 | 0.260 | 0.500 | 0.240 | 0.93 | 0.490 |
| LIPC | Val95Met (G/A) | rs6078 | GG | GA | AA | 2659 | 1606 | 252 | 2 | 0.589 | 0.356 | 0.056 | 0.587 | 0.358 | 0.055 | 0.65 | 0.234 |
| MPO | G-463A | rs2333227 | GG | GA | AA | 3612 | 852 | 52 | 3 | 0.800 | 0.189 | 0.012 | 0.800 | 0.189 | 0.011 | 0.81 | 0.106 |
| MTHFD1 | Arg134Lys (C401T) | rs1950902 | CC | CT | TT | 2773 | 1517 | 228 | 1 | 0.614 | 0.336 | 0.050 | 0.611 | 0.341 | 0.048 | 0.28 | 0.218 |
| MTHFD1 | Arg653Gln (G1958A) | rs2236225 | GG | GA | AA | 2340 | 1790 | 384 | 5 | 0.518 | 0.397 | 0.085 | 0.514 | 0.406 | 0.080 | 0.12 | 0.283 |
| MTHFR | Ala222Val (C677T) | rs1801133 | CC | CT | TT | 1763 | 2060 | 694 | 2 | 0.390 | 0.456 | 0.154 | 0.382 | 0.472 | 0.146 | 0.023 | 0.382 |
| MTHFR | Glu429Ala (A1298C) | rs1801131 | AA | AC | CC | 3023 | 1323 | 169 | 4 | 0.670 | 0.293 | 0.037 | 0.666 | 0.300 | 0.034 | 0.11 | 0.184 |
| MTR | Asp919Gly (A/G) | rs1805087 | AA | AG | GG | 2883 | 1446 | 185 | 5 | 0.639 | 0.320 | 0.041 | 0.638 | 0.321 | 0.040 | 0.82 | 0.201 |
| MTRR | Ile22Met (A66G) | rs1801394 | AA | AG | GG | 2155 | 1925 | 436 | 3 | 0.477 | 0.426 | 0.097 | 0.477 | 0.428 | 0.096 | 0.83 | 0.310 |
| NOS3 | T-786C | rs2070744 | TT | TC | CC | 3602 | 863 | 48 | 6 | 0.798 | 0.191 | 0.011 | 0.799 | 0.190 | 0.011 | 0.70 | 0.106 |
| PPARD | T-842C (at exon3) | rs2267668 | TT | TC | CC | 2922 | 1421 | 172 | 4 | 0.647 | 0.315 | 0.038 | 0.647 | 0.315 | 0.038 | 1.00 | 0.195 |
| PPARD | T-48444C (at exon3) | rs6902123 | TT | TC | CC | 4343 | 173 | 2 | 1 | 0.961 | 0.038 | 0.000 | 0.961 | 0.038 | 0.000 | 0.69 | 0.020 |
| PPARD | Asn163Asn (A65G) (at exon7) | rs2076167 | AA | AG | GG | 2780 | 1530 | 204 | 5 | 0.616 | 0.339 | 0.045 | 0.617 | 0.337 | 0.046 | 0.76 | 0.215 |
| PPARG | Pro12Ala (C/G) | rs1801282 | CC | CG | GG | 4236 | 274 | 5 | 4 | 0.938 | 0.061 | 0.001 | 0.938 | 0.061 | 0.001 | 0.80 | 0.031 |
| PPARG | His477His (C161T) | rs3856806 | CC | CT | TT | 3231 | 1198 | 86 | 4 | 0.716 | 0.265 | 0.019 | 0.720 | 0.257 | 0.023 | 0.043 | 0.152 |
| PPARGC1A | Thr394Thr (C/T) | rs2970847 | CC | CT | TT | 2755 | 1539 | 221 | 4 | 0.610 | 0.341 | 0.049 | 0.609 | 0.343 | 0.048 | 0.76 | 0.219 |
| PPARGC1A | Gly482Ser (G/A) | rs8192678 | GG | GA | AA | 1317 | 2247 | 952 | 3 | 0.292 | 0.498 | 0.211 | 0.292 | 0.497 | 0.211 | 0.93 | 0.460 |
| PRKAA2 | IVS4+961T/C | rs1418442 | TT | TC | CC | 2677 | 1581 | 251 | 10 | 0.594 | 0.351 | 0.056 | 0.591 | 0.355 | 0.053 | 0.38 | 0.231 |
| PRKAA2 | IVS7+81T/C | rs932447 | TT | TC | CC | 2679 | 1585 | 252 | 3 | 0.593 | 0.351 | 0.056 | 0.591 | 0.356 | 0.053 | 0.38 | 0.231 |
| PRKAA2 | A/T (at 3′UTR) | rs1342382 | AA | AT | TT | 2739 | 1556 | 218 | 6 | 0.607 | 0.345 | 0.048 | 0.607 | 0.344 | 0.049 | 0.90 | 0.221 |
| PTGS2(COX2) | G-765C | rs20417 | GG | GC | CC | 4260 | 247 | 6 | 6 | 0.944 | 0.055 | 0.001 | 0.943 | 0.056 | 0.001 | 0.27 | 0.029 |
| PTGS2(COX2) | C-163G | rs5270 | CC | CG | GG | 4379 | 136 | 3 | 1 | 0.969 | 0.030 | 0.001 | 0.969 | 0.031 | 0.000 | 0.10 | 0.016 |
| PTPN11 | G33861A | rs2301756 | GG | GA | AA | 2926 | 1418 | 174 | 1 | 0.648 | 0.314 | 0.039 | 0.647 | 0.314 | 0.038 | 0.89 | 0.195 |
| RETN | C-420G | rs1862513 | CC | CG | GG | 1956 | 1977 | 585 | 1 | 0.433 | 0.438 | 0.129 | 0.425 | 0.454 | 0.121 | 0.015 | 0.348 |
| SCARB1 | Val135Ile (G/A) | rs5891 | GG | GA | AA | 4518 | 0 | 0 | 1 | 1.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 1.00 | 0.000 |
| SCARB1 | Ala350Ala (C1119T) | rs5888 | CC | CT | TT | 2678 | 1619 | 221 | 1 | 0.593 | 0.358 | 0.049 | 0.596 | 0.352 | 0.052 | 0.25 | 0.228 |
| SCARB1 | G/A (at intron) | rs3782287 | GG | GA | AA | 2676 | 1574 | 268 | 1 | 0.592 | 0.348 | 0.059 | 0.588 | 0.358 | 0.055 | 0.074 | 0.234 |
| SERPINC1 | C/G | rs677 | CC | CG | GG | 2762 | 1527 | 227 | 3 | 0.612 | 0.338 | 0.050 | 0.609 | 0.342 | 0.048 | 0.41 | 0.219 |
| SHMT1 | Leu435Phe (C1420T) | rs1979277 | CC | CT | TT | 3755 | 722 | 41 | 1 | 0.831 | 0.160 | 0.009 | 0.830 | 0.162 | 0.008 | 0.36 | 0.089 |
| SLC19A(RFC1) | His27Arg (A80G) | rs1051266 | AA | AG | GG | 1536 | 2187 | 793 | 3 | 0.340 | 0.484 | 0.176 | 0.339 | 0.486 | 0.175 | 0.76 | 0.418 |
| SLC30A8 | Arg325Trp (C/T) | rs13266634 | CC | CT | TT | 1543 | 1992 | 976 | 8 | 0.342 | 0.442 | 0.216 | 0.317 | 0.492 | 0.191 | <0.0001 | 0.437 |
| SRD5A2 | Leu89Val (C/G) | rs523349 | CC | CG | GG | 1334 | 2286 | 896 | 3 | 0.295 | 0.506 | 0.198 | 0.301 | 0.495 | 0.204 | 0.15 | 0.452 |
| TAS2R38 | Pro49Ala (C/G) | rs713598 | CC | CG | GG | 1399 | 2219 | 898 | 3 | 0.310 | 0.491 | 0.199 | 0.309 | 0.494 | 0.198 | 0.74 | 0.445 |
| TAS2R38 | Ala262Val (C/T) | rs1726866 | CC | CT | TT | 1401 | 2218 | 899 | 1 | 0.310 | 0.491 | 0.199 | 0.309 | 0.494 | 0.198 | 0.70 | 0.444 |
| TAS2R38 | Val296Ile (C/T) | rs10246939 | CC | CT | TT | 1401 | 2218 | 898 | 2 | 0.310 | 0.491 | 0.199 | 0.309 | 0.494 | 0.197 | 0.72 | 0.444 |
| TCF7L2 | C/T (at intron) | rs7903146 | CC | CT | TT | 4174 | 333 | 11 | 1 | 0.924 | 0.074 | 0.002 | 0.923 | 0.075 | 0.002 | 0.11 | 0.039 |
| TNF-A | T-1031C | rs1799964 | TT | TC | CC | 3147 | 1244 | 127 | 1 | 0.697 | 0.275 | 0.028 | 0.696 | 0.277 | 0.027 | 0.75 | 0.166 |
| TNF-A | C-857T | rs1799724 | CC | CT | TT | 2990 | 1365 | 162 | 2 | 0.662 | 0.302 | 0.036 | 0.661 | 0.304 | 0.035 | 0.70 | 0.187 |
| USF1 | C7131T | rs3737787 | CC | CT | TT | 2793 | 1506 | 218 | 2 | 0.618 | 0.333 | 0.048 | 0.616 | 0.338 | 0.046 | 0.43 | 0.215 |
| VDR | Met1Thr (Fok I) (C/T) | rs2228570 | CC | CT | TT | 1768 | 2105 | 643 | 3 | 0.391 | 0.466 | 0.142 | 0.390 | 0.469 | 0.141 | 0.68 | 0.375 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
aAA, Aa, aa, and XX indicate homozygotes of major alleles, heterozygotes, homozygotes of minor alleles, and samples for which the genotype could not be determined, respectively.
bBased on the Hardy–Weinberg equilibrium.
The P value for departures from the Hardy–Weinberg equilibrium was less than 0.05 for 19 polymorphisms. However, the only genotypes for which the difference between the observed and expected frequencies exceeded 3% were the CETP Ile405Val (A/G) heterozygote and the SLC30A8 Arg325Trp (C/T) heterozygote. As shown in Table 5, some polymorphisms demonstrated a considerable difference in MAF among the participating cohorts; for 32 of the 108 polymorphisms, including ABCC11 Arg180Gly (T/C), there was a highly significant difference in MAF among study areas (P < 0.001).
Table 5. Minor allele frequencies of 107 selected genetic polymorphisms (106 SNPs and 1 insertion/deletion polymorphism) by study area.
| Gene | Polymorphism | rs number | Minor allele frequency by study area | Pa | ||||||||||
| Total | Chiba | Shizuoka | Okazaki | ACC | Takashima | Kyoto | Tokushima | Fukuoka | Saga | Amami | ||||
| ABCA1 | C-565T | rs2422493 | 0.402 | 0.371 | 0.429 | 0.406 | 0.426 | 0.401 | 0.394 | 0.468 | 0.404 | 0.405 | 0.355 | 0.005 |
| ABCA1 | G-191C | rs1800976 | 0.402 | 0.370 | 0.429 | 0.406 | 0.425 | 0.403 | 0.403 | 0.468 | 0.408 | 0.405 | 0.356 | 0.006 |
| ABCA1 | G-17C | rs2740483 | 0.301 | 0.363 | 0.296 | 0.300 | 0.271 | 0.294 | 0.300 | 0.247 | 0.313 | 0.313 | 0.271 | 0.0002 |
| ABCA1 | Val825Ile (G/A) | rs2066715 | 0.356 | 0.354 | 0.358 | 0.331 | 0.354 | 0.350 | 0.347 | 0.353 | 0.360 | 0.358 | 0.389 | 0.51 |
| ABCA1 | Val771Met (G/A) | rs2066718 | 0.066 | 0.067 | 0.060 | 0.067 | 0.059 | 0.061 | 0.034 | 0.047 | 0.073 | 0.068 | 0.088 | 0.041 |
| ABCA1 | Arg1587Lys (G/A) | rs2230808 | 0.391 | 0.393 | 0.391 | 0.384 | 0.392 | 0.394 | 0.347 | 0.326 | 0.369 | 0.388 | 0.441 | 0.027 |
| ACE | Insertion/Deletion (I/D) | rs1799752 | 0.365 | 0.373 | 0.369 | 0.346 | 0.363 | 0.365 | 0.313 | 0.353 | 0.357 | 0.341 | 0.425 | 0.003 |
| ADD1 | Trp460Gly (T/G) | rs4961 | 0.451 | 0.454 | 0.436 | 0.424 | 0.455 | 0.430 | 0.463 | 0.463 | 0.429 | 0.464 | 0.509 | 0.006 |
| ADH1B | His47Arg (A/G) | rs1229984 | 0.239 | 0.220 | 0.232 | 0.200 | 0.247 | 0.220 | 0.250 | 0.184 | 0.266 | 0.223 | 0.318 | <0.0001 |
| ADH1C | Arg272Gln (C/T) | rs1693482 | 0.075 | 0.053 | 0.068 | 0.054 | 0.178 | 0.056 | 0.053 | 0.058 | 0.065 | 0.053 | 0.072 | <0.0001 |
| ADIPOQ | C-11377G | rs266729 | 0.251 | 0.245 | 0.239 | 0.254 | 0.238 | 0.252 | 0.300 | 0.279 | 0.257 | 0.250 | 0.255 | 0.60 |
| ADIPOQ | Gly15Gly (T/G) | rs2241766 | 0.290 | 0.285 | 0.304 | 0.285 | 0.309 | 0.301 | 0.316 | 0.295 | 0.287 | 0.284 | 0.251 | 0.18 |
| ADIPOQ | G276T | rs1501299 | 0.262 | 0.290 | 0.284 | 0.282 | 0.147 | 0.266 | 0.263 | 0.289 | 0.286 | 0.308 | 0.236 | <0.0001 |
| ADIPOQ | IVS-3971A/G | rs822396 | 0.061 | 0.053 | 0.049 | 0.069 | 0.057 | 0.079 | 0.050 | 0.047 | 0.073 | 0.057 | 0.056 | 0.063 |
| ADIPOR1 | G-8503A | rs6666089 | 0.032 | 0.029 | 0.040 | 0.034 | 0.028 | 0.027 | 0.044 | 0.026 | 0.029 | 0.024 | 0.040 | 0.29 |
| ADIPOR1 | C5843T | rs1342387 | 0.483 | 0.484 | 0.473 | 0.476 | 0.481 | 0.480 | 0.478 | 0.426 | 0.495 | 0.483 | 0.504 | 0.78 |
| ADIPOR1 | C10224G | rs7539542 | 0.216 | 0.226 | 0.230 | 0.201 | 0.202 | 0.223 | 0.219 | 0.263 | 0.233 | 0.215 | 0.194 | 0.21 |
| ADRB2 | Gln27Glu (C/G) | rs1042714 | 0.065 | 0.073 | 0.051 | 0.061 | 0.066 | 0.093 | 0.081 | 0.079 | 0.072 | 0.063 | 0.039 | 0.0001 |
| ADRB3 | Trp64Arg (T/C) | rs4994 | 0.195 | 0.184 | 0.173 | 0.195 | 0.184 | 0.195 | 0.147 | 0.168 | 0.193 | 0.218 | 0.240 | 0.001 |
| AGT | Thr174Met (C/T) | rs4762 | 0.105 | 0.104 | 0.106 | 0.101 | 0.106 | 0.092 | 0.094 | 0.095 | 0.123 | 0.105 | 0.115 | 0.68 |
| AGT | Thr235Met (C/T) | rs699 | 0.187 | 0.181 | 0.183 | 0.208 | 0.189 | 0.195 | 0.219 | 0.211 | 0.173 | 0.178 | 0.170 | 0.31 |
| AGTR1(ATR1) | A1166C (at 3′UTR) | rs5186 | 0.087 | 0.089 | 0.093 | 0.083 | 0.088 | 0.076 | 0.081 | 0.089 | 0.080 | 0.082 | 0.104 | 0.61 |
| ALDH2 | Glu487Lys (G/A) | rs671 | 0.254 | 0.232 | 0.274 | 0.316 | 0.298 | 0.254 | 0.226 | 0.300 | 0.269 | 0.267 | 0.112 | <0.0001 |
| APOA1 | Ala61Thr (C/T) | rs12718465 | 0.055 | 0.051 | 0.049 | 0.056 | 0.061 | 0.055 | 0.088 | 0.016 | 0.046 | 0.043 | 0.079 | 0.0005 |
| APOA1 | Arg184Pro (G/C) | rs5078 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| APOA5 | T-1131C | rs662799 | 0.345 | 0.355 | 0.336 | 0.322 | 0.337 | 0.326 | 0.338 | 0.342 | 0.342 | 0.337 | 0.412 | 0.002 |
| APOA5 | Gly185Cys (G553T) | rs2075291 | 0.069 | 0.084 | 0.062 | 0.057 | 0.073 | 0.068 | 0.063 | 0.074 | 0.064 | 0.079 | 0.065 | 0.36 |
| APOE | T-203G | rs405509 | 0.305 | 0.289 | 0.297 | 0.294 | 0.302 | 0.256 | 0.306 | 0.295 | 0.318 | 0.308 | 0.383 | <0.0001 |
| APOE | Arg158Cys (C/T) (at exon2) | rs7412 | 0.043 | 0.045 | 0.048 | 0.048 | 0.035 | 0.042 | 0.050 | 0.042 | 0.044 | 0.048 | 0.030 | 0.49 |
| APOE | Cys112Arg (T/C) (at exon4) | rs429358 | 0.101 | 0.105 | 0.105 | 0.101 | 0.123 | 0.101 | 0.078 | 0.068 | 0.109 | 0.102 | 0.075 | 0.026 |
| ARNTL(BMAL1) | A/G | rs7950226 | 0.404 | 0.424 | 0.405 | 0.420 | 0.395 | 0.402 | 0.391 | 0.347 | 0.427 | 0.431 | 0.344 | 0.002 |
| ART4(DOK1) | Leu208Leu (G/A) | rs3088189 | 0.107 | 0.114 | 0.114 | 0.130 | 0.084 | 0.113 | 0.078 | 0.068 | 0.105 | 0.108 | 0.109 | 0.022 |
| ART4(DOK1) | Asp265Asn (G/A) | rs11276 | 0.107 | 0.114 | 0.113 | 0.130 | 0.084 | 0.113 | 0.078 | 0.068 | 0.105 | 0.108 | 0.109 | 0.023 |
| BHMT | Arg239Gln (G742A) | rs3733890 | 0.221 | 0.235 | 0.236 | 0.241 | 0.229 | 0.203 | 0.209 | 0.226 | 0.243 | 0.217 | 0.164 | 0.0005 |
| CBS | Tyr233Tyr (C699T) | rs234706 | 0.024 | 0.027 | 0.033 | 0.026 | 0.020 | 0.021 | 0.028 | 0.011 | 0.034 | 0.017 | 0.017 | 0.073 |
| CD14 | T-260C/T-159C | rs2569190 | 0.462 | 0.460 | 0.467 | 0.467 | 0.439 | 0.499 | 0.428 | 0.395 | 0.468 | 0.448 | 0.471 | 0.088 |
| CDKAL1 | G/C | rs7754840 | 0.407 | 0.428 | 0.390 | 0.430 | 0.392 | 0.411 | 0.363 | 0.374 | 0.424 | 0.435 | 0.367 | 0.009 |
| CDKN2A/B | T/C | rs10811661 | 0.451 | 0.465 | 0.450 | 0.466 | 0.446 | 0.470 | 0.453 | 0.411 | 0.441 | 0.452 | 0.419 | 0.42 |
| CETP | A-629C | rs1800775 | 0.445 | 0.445 | 0.463 | 0.442 | 0.436 | 0.441 | 0.469 | 0.405 | 0.462 | 0.422 | 0.458 | 0.52 |
| CETP | Ile405Val (A/G) | rs5882 | 0.492 | 0.537 | 0.376 | 0.452 | 0.523 | 0.498 | 0.509 | 0.458 | 0.495 | 0.556 | 0.506 | <0.0001 |
| CETP | TaqIB (C/T) | rs708272 | 0.405 | 0.404 | 0.401 | 0.419 | 0.419 | 0.384 | 0.375 | 0.363 | 0.378 | 0.443 | 0.398 | 0.067 |
| CETP | G/T | rs3764261 | 0.233 | 0.183 | 0.340 | 0.295 | 0.203 | 0.188 | 0.209 | 0.158 | 0.204 | 0.223 | 0.240 | <0.0001 |
| CHRNB2 | G-42A | rs2072658 | 0.204 | 0.236 | 0.214 | 0.185 | 0.215 | 0.222 | 0.203 | 0.253 | 0.202 | 0.203 | 0.140 | <0.0001 |
| CHRNB2 | C/T (at 3′UTR) | rs2072660 | 0.243 | 0.258 | 0.225 | 0.264 | 0.224 | 0.241 | 0.216 | 0.237 | 0.214 | 0.233 | 0.292 | 0.001 |
| COMT | Val158Met (G/A) | rs4680 | 0.336 | 0.310 | 0.328 | 0.336 | 0.323 | 0.359 | 0.303 | 0.295 | 0.336 | 0.315 | 0.406 | <0.0001 |
| CYP1A1 | Ile462Val (A/G) | rs1048943 | 0.239 | 0.222 | 0.235 | 0.256 | 0.222 | 0.244 | 0.250 | 0.268 | 0.222 | 0.225 | 0.274 | 0.069 |
| CYP1A2 | A734C | rs762551 | 0.358 | 0.336 | 0.349 | 0.356 | 0.344 | 0.367 | 0.352 | 0.384 | 0.351 | 0.363 | 0.391 | 0.38 |
| CYP11B2 | T-344C | rs1799998 | 0.331 | 0.334 | 0.318 | 0.339 | 0.289 | 0.294 | 0.346 | 0.284 | 0.329 | 0.325 | 0.434 | <0.0001 |
| CYP17A1 | T-34C | rs743572 | 0.466 | 0.453 | 0.460 | 0.449 | 0.485 | 0.492 | 0.500 | 0.353 | 0.467 | 0.452 | 0.482 | 0.018 |
| ESR1 | IVS1-351A/G (Xba I) | rs11155816 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| ESR1 | IVS1-397T/C (Pvu II) | rs2234693 | 0.425 | 0.436 | 0.422 | 0.422 | 0.416 | 0.452 | 0.334 | 0.421 | 0.430 | 0.438 | 0.415 | 0.062 |
| ESR2 | Val328Val (G1082A) (Rsa I) | rs1256049 | 0.284 | 0.268 | 0.281 | 0.297 | 0.272 | 0.308 | 0.272 | 0.284 | 0.314 | 0.293 | 0.250 | 0.054 |
| FTO | T/A | rs9939609 | 0.201 | 0.172 | 0.195 | 0.220 | 0.190 | 0.188 | 0.169 | 0.158 | 0.206 | 0.186 | 0.277 | <0.0001 |
| GCK | G-30A | rs1799884 | 0.189 | 0.193 | 0.179 | 0.183 | 0.186 | 0.182 | 0.138 | 0.147 | 0.304 | 0.173 | 0.159 | <0.0001 |
| GCKR | A/G | rs780094 | 0.449 | 0.428 | 0.436 | 0.440 | 0.439 | 0.425 | 0.350 | 0.453 | 0.450 | 0.445 | 0.562 | <0.0001 |
| GCKR | Leu446Pro (T/C) | rs1260326 | 0.445 | 0.430 | 0.434 | 0.439 | 0.438 | 0.416 | 0.353 | 0.421 | 0.447 | 0.432 | 0.559 | <0.0001 |
| GNAS1 | T393C | rs7121 | 0.428 | 0.448 | 0.427 | 0.408 | 0.463 | 0.422 | 0.469 | 0.342 | 0.454 | 0.418 | 0.391 | 0.002 |
| IGF2BP2 | G/T (at intron) | rs4402960 | 0.303 | 0.328 | 0.306 | 0.292 | 0.298 | 0.296 | 0.309 | 0.268 | 0.300 | 0.304 | 0.306 | 0.81 |
| IL-1B | T-31C | rs1143627 | 0.462 | 0.444 | 0.463 | 0.477 | 0.439 | 0.454 | 0.491 | 0.484 | 0.477 | 0.462 | 0.475 | 0.52 |
| IL-2 | T-330G | rs2069762 | 0.329 | 0.330 | 0.336 | 0.316 | 0.311 | 0.323 | 0.309 | 0.305 | 0.350 | 0.321 | 0.365 | 0.21 |
| IL-4 | T-33C | rs2070874 | 0.338 | 0.317 | 0.337 | 0.315 | 0.333 | 0.291 | 0.313 | 0.284 | 0.336 | 0.337 | 0.457 | <0.0001 |
| IL-6 | C-634G | rs1800796 | 0.244 | 0.247 | 0.206 | 0.250 | 0.234 | 0.226 | 0.225 | 0.232 | 0.223 | 0.240 | 0.338 | <0.0001 |
| IL-8 | T-251A | rs4073 | 0.320 | 0.334 | 0.321 | 0.318 | 0.320 | 0.304 | 0.331 | 0.266 | 0.339 | 0.329 | 0.306 | 0.56 |
| IL-10 | T-819C | rs1800871 | 0.346 | 0.329 | 0.346 | 0.320 | 0.340 | 0.313 | 0.356 | 0.268 | 0.354 | 0.359 | 0.425 | <0.0001 |
| IL-13 | C-1111T | rs1800925 | 0.183 | 0.182 | 0.178 | 0.197 | 0.185 | 0.178 | 0.213 | 0.158 | 0.172 | 0.190 | 0.176 | 0.73 |
| KCNJ11 | Glu23Lys (C/T) | rs5219 | 0.366 | 0.386 | 0.364 | 0.382 | 0.343 | 0.374 | 0.363 | 0.316 | 0.361 | 0.366 | 0.368 | 0.52 |
| LCAT/SLC12A4 | Ser232Thr (T/A) | rs4986970 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| LIPC | T-514C | rs1800588 | 0.490 | 0.522 | 0.478 | 0.496 | 0.484 | 0.497 | 0.522 | 0.484 | 0.502 | 0.503 | 0.429 | 0.006 |
| LIPC | Val95Met (G/A) | rs6078 | 0.234 | 0.232 | 0.229 | 0.221 | 0.269 | 0.259 | 0.247 | 0.237 | 0.208 | 0.253 | 0.182 | <0.0001 |
| MPO | G-463A | rs2333227 | 0.106 | 0.107 | 0.105 | 0.107 | 0.102 | 0.113 | 0.100 | 0.100 | 0.106 | 0.093 | 0.119 | 0.84 |
| MTHFD1 | Arg134Lys (C401T) | rs1950902 | 0.218 | 0.214 | 0.216 | 0.218 | 0.212 | 0.229 | 0.184 | 0.195 | 0.217 | 0.239 | 0.215 | 0.66 |
| MTHFD1 | Arg653Gln (G1958A) | rs2236225 | 0.283 | 0.271 | 0.273 | 0.269 | 0.265 | 0.280 | 0.284 | 0.279 | 0.278 | 0.301 | 0.331 | 0.036 |
| MTHFR | Ala222Val (C677T) | rs1801133 | 0.382 | 0.387 | 0.402 | 0.388 | 0.411 | 0.393 | 0.375 | 0.442 | 0.410 | 0.364 | 0.288 | <0.0001 |
| MTHFR | Glu429Ala (A1298C) | rs1801131 | 0.184 | 0.195 | 0.189 | 0.202 | 0.193 | 0.193 | 0.225 | 0.191 | 0.147 | 0.222 | 0.105 | <0.0001 |
| MTR | Asp919Gly (A/G) | rs1805087 | 0.201 | 0.188 | 0.210 | 0.186 | 0.183 | 0.179 | 0.184 | 0.232 | 0.193 | 0.176 | 0.299 | <0.0001 |
| MTRR | Ile22Met (A66G) | rs1801394 | 0.310 | 0.292 | 0.314 | 0.298 | 0.315 | 0.296 | 0.306 | 0.337 | 0.318 | 0.297 | 0.346 | 0.24 |
| NOS3 | T-786C | rs2070744 | 0.106 | 0.111 | 0.125 | 0.117 | 0.112 | 0.098 | 0.119 | 0.122 | 0.109 | 0.102 | 0.068 | 0.004 |
| PPARD | T-842C (at exon3) | rs2267668 | 0.195 | 0.175 | 0.199 | 0.201 | 0.183 | 0.205 | 0.213 | 0.221 | 0.192 | 0.190 | 0.210 | 0.55 |
| PPARD | T-48444C (at exon3) | rs6902123 | 0.020 | 0.015 | 0.020 | 0.030 | 0.025 | 0.017 | 0.019 | 0.016 | 0.019 | 0.019 | 0.012 | 0.15 |
| PPARD | Asn163Asn (A65G) (at exon7) | rs2076167 | 0.215 | 0.190 | 0.212 | 0.231 | 0.206 | 0.221 | 0.225 | 0.234 | 0.209 | 0.205 | 0.236 | 0.30 |
| PPARG | Pro12Ala (C/G) | rs1801282 | 0.031 | 0.031 | 0.027 | 0.028 | 0.037 | 0.047 | 0.022 | 0.011 | 0.029 | 0.032 | 0.025 | 0.064 |
| PPARG | His477His (C161T) | rs3856806 | 0.152 | 0.153 | 0.159 | 0.143 | 0.149 | 0.167 | 0.159 | 0.147 | 0.177 | 0.166 | 0.099 | 0.0002 |
| PPARGC1A | Thr394Thr (C/T) | rs2970847 | 0.219 | 0.229 | 0.223 | 0.237 | 0.219 | 0.222 | 0.209 | 0.237 | 0.208 | 0.243 | 0.168 | 0.005 |
| PPARGC1A | Gly482Ser (G/A) | rs8192678 | 0.460 | 0.472 | 0.447 | 0.444 | 0.431 | 0.448 | 0.463 | 0.442 | 0.471 | 0.438 | 0.538 | <0.0001 |
| PRKAA2 | IVS4+961T/C | rs1418442 | 0.231 | 0.216 | 0.228 | 0.232 | 0.206 | 0.217 | 0.184 | 0.237 | 0.205 | 0.237 | 0.318 | <0.0001 |
| PRKAA2 | IVS7+81T/C | rs932447 | 0.231 | 0.216 | 0.228 | 0.232 | 0.207 | 0.218 | 0.184 | 0.237 | 0.205 | 0.239 | 0.318 | <0.0001 |
| PRKAA2 | A/T (at 3′UTR) | rs1342382 | 0.221 | 0.238 | 0.220 | 0.200 | 0.250 | 0.225 | 0.204 | 0.211 | 0.233 | 0.197 | 0.212 | 0.072 |
| PTGS2(COX2) | G-765C | rs20417 | 0.029 | 0.028 | 0.033 | 0.031 | 0.033 | 0.027 | 0.025 | 0.053 | 0.026 | 0.030 | 0.017 | 0.22 |
| PTGS2(COX2) | C-163G | rs5270 | 0.016 | 0.017 | 0.023 | 0.017 | 0.020 | 0.015 | 0.009 | 0.011 | 0.009 | 0.011 | 0.016 | 0.32 |
| PTPN11 | G33861A | rs2301756 | 0.195 | 0.194 | 0.199 | 0.172 | 0.167 | 0.185 | 0.184 | 0.205 | 0.173 | 0.159 | 0.320 | <0.0001 |
| RETN | C-420G | rs1862513 | 0.348 | 0.351 | 0.328 | 0.330 | 0.343 | 0.330 | 0.334 | 0.316 | 0.340 | 0.366 | 0.411 | 0.002 |
| SCARB1 | Val135Ile (G/A) | rs5891 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| SCARB1 | Ala350Ala (C1119T) | rs5888 | 0.228 | 0.240 | 0.212 | 0.226 | 0.228 | 0.225 | 0.253 | 0.205 | 0.240 | 0.240 | 0.213 | 0.59 |
| SCARB1 | G/A (at intron) | rs3782287 | 0.234 | 0.225 | 0.223 | 0.243 | 0.227 | 0.249 | 0.297 | 0.189 | 0.207 | 0.219 | 0.263 | 0.008 |
| SERPINC1 | C/G | rs677 | 0.219 | 0.220 | 0.197 | 0.209 | 0.238 | 0.223 | 0.238 | 0.211 | 0.220 | 0.227 | 0.217 | 0.58 |
| SHMT1 | Leu435Phe (C1420T) | rs1979277 | 0.089 | 0.072 | 0.089 | 0.071 | 0.093 | 0.092 | 0.084 | 0.095 | 0.084 | 0.091 | 0.117 | 0.026 |
| SLC19A(RFC1) | His27Arg (A80G) | rs1051266 | 0.418 | 0.415 | 0.435 | 0.412 | 0.426 | 0.423 | 0.397 | 0.395 | 0.430 | 0.454 | 0.352 | 0.001 |
| SLC30A8 | Arg325Trp (C/T) | rs13266634 | 0.437 | 0.439 | 0.449 | 0.443 | 0.438 | 0.418 | 0.472 | 0.453 | 0.434 | 0.451 | 0.411 | 0.53 |
| SRD5A2 | Leu89Val (C/G) | rs523349 | 0.452 | 0.437 | 0.468 | 0.430 | 0.458 | 0.458 | 0.391 | 0.399 | 0.449 | 0.471 | 0.464 | 0.13 |
| TAS2R38 | Pro49Ala (C/G) | rs713598 | 0.445 | 0.448 | 0.439 | 0.419 | 0.427 | 0.454 | 0.434 | 0.479 | 0.445 | 0.435 | 0.491 | 0.080 |
| TAS2R38 | Ala262Val (C/T) | rs1726866 | 0.444 | 0.449 | 0.438 | 0.419 | 0.427 | 0.454 | 0.434 | 0.479 | 0.444 | 0.435 | 0.491 | 0.078 |
| TAS2R38 | Val296Ile (C/T) | rs10246939 | 0.444 | 0.448 | 0.438 | 0.419 | 0.427 | 0.454 | 0.434 | 0.479 | 0.444 | 0.435 | 0.491 | 0.079 |
| TCF7L2 | C/T (at intron) | rs7903146 | 0.039 | 0.051 | 0.039 | 0.042 | 0.043 | 0.037 | 0.034 | 0.042 | 0.025 | 0.034 | 0.043 | 0.27 |
| TNF-A | T-1031C | rs1799964 | 0.166 | 0.175 | 0.148 | 0.165 | 0.157 | 0.153 | 0.153 | 0.237 | 0.151 | 0.180 | 0.188 | 0.024 |
| TNF-A | C-857T | rs1799724 | 0.187 | 0.172 | 0.171 | 0.173 | 0.166 | 0.161 | 0.194 | 0.184 | 0.203 | 0.199 | 0.255 | <0.0001 |
| USF1 | C7131T | rs3737787 | 0.215 | 0.229 | 0.223 | 0.220 | 0.234 | 0.217 | 0.263 | 0.216 | 0.196 | 0.204 | 0.178 | 0.020 |
| VDR | Met1Thr (Fok I) (C/T) | rs2228570 | 0.375 | 0.344 | 0.393 | 0.368 | 0.380 | 0.381 | 0.391 | 0.437 | 0.402 | 0.377 | 0.343 | 0.048 |
Abbreviations: ACC, Aichi Cancer Center; SNP, single nucleotide polymorphism.
aP for difference among study areas.
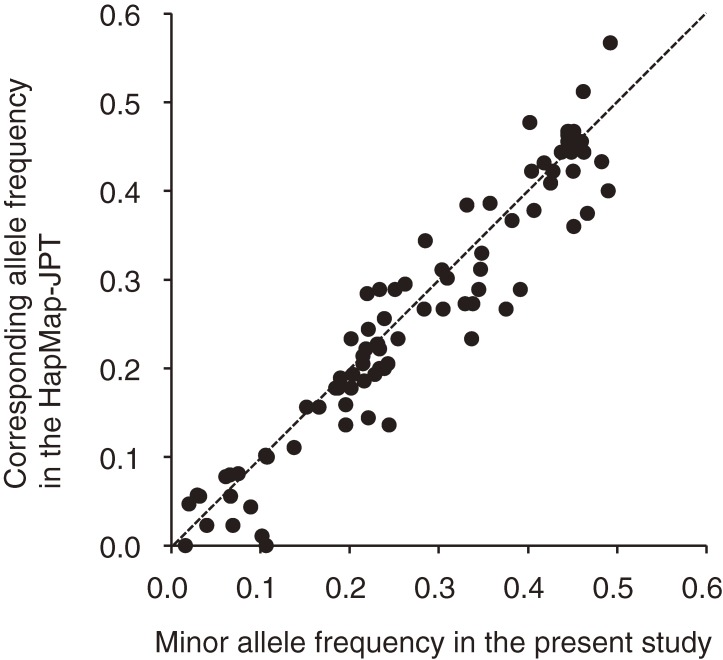
The Figure shows a comparison of the allele frequencies in our study population and the HapMap-JPT data set. Among 88 polymorphisms, only 5 (ABCA1 rs2230808, COMT rs4680, IL-6 rs1800796, NOS3 rs2070744, and VDR rs2228570) showed a difference in allele frequencies of more than 0.1 between the 2 populations.
Figure. Scatter plot of allele frequencies for 88 polymorphisms in the data set from the Japan Multi-institutional Collaborative Cohort (J-MICC) Study and the HapMap-JPT data set registered at the US National Library of Medicine (http://www.ncbi.nlm.nih.gov/snp). Points on the identity (dotted) line represent allele frequencies that are identical in the 2 populations.
DISCUSSION
The present report describes the profiles of participants in a cross-sectional study within the J-MICC Study data set and the allele frequencies of 108 polymorphisms, with potential relevance to lifestyle and clinical factors, in their genomes. The allele frequencies for most polymorphisms in our study population were comparable to those in the HapMap-JPT data set.
It has been suggested that polymorphisms for APOA1 184Pro (C), ESR1 IVS1-351G, LCAT/SLC12A4 232Thr (A), and SCARB1 135Ile (A) do not exist in the Japanese population (http://www.ncbi.nlm.nih.gov/snp and personal communication); however, we included them in the present study to test this notion in a large sample (>4000 people). Our results confirmed that these minor alleles were indeed absent among Japanese.
Of the remaining 104 polymorphisms, 19 showed departures from the Hardy–Weinberg equilibrium with P values <0.05. In most cases, however, the absolute differences between the actual and expected frequencies were minimal. Thus, these apparently small P values could be accounted for by the large sample size and the multiple tests used in our study, and any errors in genotyping seemed unlikely to have resulted in substantial misclassification.
Although genotype data for only 45 people, at most, are available in the HapMap-JPT data set, the allele frequencies in the HapMap-JPT population and our study population were remarkably similar for most of the polymorphisms examined (Figure). For 45 individuals, the 95% confidence intervals were 0.047 to 0.181, 0.208 to 0.406, and 0.393 to 0.607 for MAF values of 0.1, 0.3, and 0.5, respectively, based on a binomial distribution.
A major strength of the current study was that it provided a comprehensive collection of data on lifestyle and clinical factors. Because it is not easy to gain access to data on genotype distributions in a large Japanese population, our data might also be useful as a reference tool. However, because the participants in this study were recruited from various sources throughout Japan, associations of genotypes with lifestyle and clinical factors might vary between populations. There might also have been differences between institutions in terms of the measurement methods used in the clinical examinations, because we could not directly control the quality of the health examinations. These differences must be taken into consideration when analyzing and interpreting the data. In addition, some polymorphisms showed a substantial difference in MAF among the participating cohorts (Table 5). Yamaguchi-Kabata et al suggested that individuals from the Ryukyu Islands, including the Amami Islands, had genetic characteristics that differed considerably from those of individuals from the main islands of Japan,22 which was consistent with our present results. Genetic variations among study areas should be taken into account in the data analysis. Furthermore, the generalizability of the study findings should be considered because the response rates were low in some areas. In most cases, however, the underlying biological mechanisms are unlikely to differ between the respondents and members of the general population. The low response rate might have been due to the recruitment methods (mailing invitation letters or distributing leaflets to the general populations of 3 areas) or the strict procedures used to obtain informed consent.
In conclusion, this comprehensive data collection on lifestyle and clinical factors will be useful in elucidating gene–environment interactions and could provide an informative reference tool, particularly because free access to genotype data for a large Japanese population is not readily available.
ACKNOWLEDGMENTS
The authors thank Kyota Ashikawa, Tomomi Aoi, and the other members of the Laboratory for Genotyping Development, Center for Genomic Medicine, RIKEN, for support with genotyping, Yoko Mitsuda, Keiko Shibata, and Etsuko Kimura at the Department of Preventive Medicine, Nagoya University Graduate School of Medicine, Yasushi Yatabe at the Department of Pathology and Molecular Diagnostics, Aichi Cancer Center Hospital, Fusako Katsurada at the Department of Health Science, Shiga University of Medical Science, and Mitsuhiko Matsushita and Yasunobu Sagara at the Tokushima Prefecture Health Examination Center for their cooperation, technical assistance, and valuable comments. This study was supported, in part, by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (Nos. 17015018 and 221S0001).
Conflicts of interest: None declared.
REFERENCES
- 1.Yang CX , Matsuo K , Ito H , Hirose K , Wakai K , Saito T , et al. Esophageal cancer risk by ALDH2 and ADH2 polymorphisms and alcohol consumption: exploration of gene-environment and gene-gene interactions . Asian Pac J Cancer Prev. 2005;6:256–62 [PubMed] [Google Scholar]
- 2.Taioli E Gene-environment interaction in tobacco-related cancers . Carcinogenesis. 2008;29:1467–74 10.1093/carcin/bgn062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassi MG Metabolic syndrome, diabetes and atherosclerosis: influence of gene-environment interaction . Mutat Res. 2009;667:35–43 10.1016/j.mrfmmm.2008.10.018 [DOI] [PubMed] [Google Scholar]
- 4.Vercelli D Gene-environment interactions in asthma and allergy: the end of the beginning? Curr Opin Allergy Clin Immunol. 2010;10:145–8 10.1097/ACI.0b013e32833653d7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins FS The case for a US prospective cohort study of genes and environment . Nature. 2004;429:475–7 10.1038/nature02628 [DOI] [PubMed] [Google Scholar]
- 6.Potter JD Toward the last cohort . Cancer Epidemiol Biomarkers Prev. 2004;13:895–7 [PubMed] [Google Scholar]
- 7.Ollier W , Sprosen T , Peakman T. UK Biobank: from concept to reality . Pharmacogenomics. 2005;6:639–46 10.2217/14622416.6.6.639 [DOI] [PubMed] [Google Scholar]
- 8.Manolio TA , Bailey-Wilson JE , Collins FS. Genes, environment and the value of prospective cohort studies . Nat Rev Genet. 2006;7:812–20 10.1038/nrg1919 [DOI] [PubMed] [Google Scholar]
- 9.Rothman KJ. Modern Epidemiology, 1st ed. Boston: Little, Brown and Company; 1986. [Google Scholar]
- 10.The J-MICC Study Group The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer . Asian Pac J Cancer Prev. 2007;8:317–23 [PubMed] [Google Scholar]
- 11.Naito M , Eguchi H , Okada R , Ishida Y , Nishio K , Hishida A , et al. Controls for monitoring the deterioration of stored blood samples in the Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) . Nagoya J Med Sci. 2008;70:107–15 [PubMed] [Google Scholar]
- 12.Asai Y , Naito M , Suzuki M , Tomoda A , Kuwabara M , Fukada Y , et al. Baseline data of Shizuoka area in the Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) . Nagoya J Med Sci. 2009;71:137–44 [PMC free article] [PubMed] [Google Scholar]
- 13.Hara M , Higaki Y , Imaizumi T , Taguchi N , Nakamura K , Nanri H , et al. Factors influencing participation rate in a baseline survey of a genetic cohort in Japan . J Epidemiol. 2010;20:40–5 10.2188/jea.JE20090062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamajima N , Atsuta Y , Niwa Y , Nishio K , Tanaka D , Yamamoto K , et al. Precise definition of anonymization in genetic polymorphism studies . Asian Pac J Cancer Prev. 2004;5:83–8 [PubMed] [Google Scholar]
- 15.Ohnishi Y , Tanaka T , Ozaki K , Yamada R , Suzuki H , Nakamura Y. A high-throughput SNP typing system for genome-wide association studies . J Hum Genet. 2001;46:471–7 10.1007/s100380170047 [DOI] [PubMed] [Google Scholar]
- 16.Tokudome S , Goto C , Imaeda N , Tokudome Y , Ikeda M , Maki S. Development of a data-based short food frequency questionnaire for assessing nutrient intake by middle-aged Japanese . Asian Pac J Cancer Prev. 2004;5:40–3 [PubMed] [Google Scholar]
- 17.Tokudome Y , Goto C , Imaeda N , Hasegawa T , Kato R , Hirose K , et al. Relative validity of a short food frequency questionnaire for assessing nutrient intake versus three-day weighed diet records in middle-aged Japanese . J Epidemiol. 2005;15:135–45 10.2188/jea.15.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto C , Tokudome Y , Imaeda N , Takekuma K , Kuriki K , Igarashi F , et al. Validation study of fatty acid consumption assessed with a short food frequency questionnaire against plasma concentration in middle-aged Japanese people . Scand J Nutr. 2006;50:77–82 10.1080/11026480600568720 [DOI] [Google Scholar]
- 19.Imaeda N , Goto C , Tokudome Y , Hirose K , Tajima K , Tokudome S. Reproducibility of a short food frequency questionnaire for Japanese general population . J Epidemiol. 2007;17:100–7 10.2188/jea.17.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wigginton JE , Cutler DJ , Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium . Am J Hum Genet. 2005;76:887–93 10.1086/429864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SAS Institute Inc. SAS/STAT 9.1 User’s Guide. Cary, NC: SAS Institute Inc; 2004. [Google Scholar]
- 22.Yamaguchi-Kabata Y , Nakazono K , Takahashi A , Saito S , Hosono N , Kubo M , et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies . Am J Hum Genet. 2008;83:445–56 10.1016/j.ajhg.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]