Abstract
Evidence for the in planta defensive function of trypsin protease inhibitors (TPIs) comes from observations of enhanced herbivore resistance after heterologous TPI expression or the manipulation of signal cascades that activate numerous defense responses, including TPI production; no studies have altered the expression of an endogenous pi gene to examine defensive function. We isolated two genes with seven- and six-repeat TPI domains from Nicotiana attenuata from the potato (Solanum tuberosum) PI-II family. To determine whether endogenous TPIs in N. attenuata function defensively against the native herbivores, hornworm (Manduca sexta) and mirids (Tupiocoris notatus), we expressed 175 bp of the seven-domain pi from N. attenuata in an antisense orientation in a TPI-producing genotype to reduce TPI expression and expressed the full-length seven-domain pi in a sense orientation under control of a constitutive promoter to restore TPI activity in a natural genotype from Arizona unable to produce TPIs. Constitutive and inducible TPI production in two antisense lines were diminished by 80% to 90% and 33% to 52%, respectively, and sense expression restored 67% of the activity found in the TPI-producing genotype after caterpillar attack in the TPI-deficient A genotype. Hornworm larvae fed on genotypes with low or no TPI activity grew faster, had higher survivorship, and produced heavier pupae than those that fed on genotypes with high TPI activity. T. notatus showed higher preference for genotypes with low or no TPI activity than for genotypes with high TPI levels. We conclude that endogenous TPIs are an effective defense against these native herbivores.
Plant proteinase inhibitors (PIs) are polypeptides or proteins that occur naturally in a wide range of plants and are considered to be an essential part of the plant's natural defense system against herbivores (Ryan, 1990; Jongsma and Bolter, 1997; Schuler et al., 1998). Evidence for the in planta defense function of PIs comes from two types of experiments: heterologous expression of pi genes (Hilder et al., 1987; Johnson et al., 1989; McManus et al., 1994; Duan et al., 1996) and manipulation of signal cascades affecting PI elicitors (e.g. Orozco-Cardenas et al., 1993). PIs have been found to reduce herbivore growth in plants that were transformed with a heterologous pi gene in at least 45 studies (e.g. Hilder et al., 1987; Johnson et al., 1989; McManus et al., 1994; Duan et al., 1996). Surprisingly, we know of no studies that have altered the expression of an endogenous pi gene to examine its defensive function.
Although heterologous expression studies have demonstrated reduced herbivore performance, two complications temper the extrapolation of these results to the conclusion that PIs are natural defenses against herbivores that normally feed on plants (Johnson et al., 1989; McManus et al., 1994; Broadway, 1996). First, as a result of the coevolutionary interaction between insects and their natural hosts, the gut proteases of specialist insects are frequently resistant to the PIs of their host plants (Broadway, 1995; Jongsma et al., 1995; Winterer, 2002). Hence, when an insect feeds on a transgenic plant expressing a novel pi gene, the results may not reflect the effect of endogenous PIs (e.g. Gatehouse et al., 1999; De Leo et al., 2001; Winterer and Bergelson, 2001) because the larvae are not adapted to the novel PI. Second, the effects of PIs on insect performance can be strongly influenced by other nutritional and defensive factors resulting in PI-diet and PI-plant interactions (Broadway, 1997; McManus et al., 1999). For example, when tobacco (Nicotiana tabacum) and potato (Solanum tuberosum), transformed with the same heterologous pi gene that resulted in similar trypsin PI (TPI) contents, were challenged with Spodoptera litoralis larvae, the tobacco plants killed 95% of the attacking larvae, whereas the potato plants killed only 36% (Marchetti et al., 2000). PI-diet interactions are clearly seen in artificial diet studies in which manipulations of PI content do not reflect changes in insect performance on plant material (Broadway, 1996). An additional complication for the interpretation of the heterologous expression studies is that high levels of endogenous TPIs can be elicited by herbivore attack (Green and Ryan, 1972; Jongsma et al., 1994), and these endogenous inhibitors may interact with introduced inhibitors.
The second line of evidence for PI defensive function comes from experiments in which transgenic suppression of the wound signal cascades that elicit PI production enhances the performance of herbivores compared with those herbivores that feed on plants with intact signaling (McGurl et al., 1992; Orozco-Cardenas et al., 1993; Howe et al., 1996; Royo et al., 1999). These signaling cascades regulate many other traits in addition to PIs (Bergey et al., 1996), and it is not possible to attribute the changes in performance observed in these studies solely to changes in PI expression.
Ideally, one should determine the benefits of an endogenous pi in plants that differ only in a gene that controls the expression of a resistance trait but are otherwise identical (Bergelson and Purrington, 1996). There are at least two different ways to test the defensive function of an endogenous pi gene: produce a defenseless plant (one deficient in defense signaling) and overexpress the pi gene; or down- or up-regulate the pi gene, keeping the other defense traits and their signaling intact. The first approach is a weaker test of the defensive value of TPIs than the second and has similarities to tests of defensive function by heterologous expression because the function of the pi is examined without the context of the other defenses present in the plant. Interactions with other defenses may profoundly determine the defensive function of TPIs and these interactions would be lost in the first approach. Here, we provide a test of the defensive value of endogenous TPIs with the second approach in a native tobacco species.
Nicotiana attenuata Torr. Ex Wats., a postfire annual inhabiting the Great Basin Desert, has a number of well-described herbivore-induced direct and indirect defenses (Baldwin, 2001) that increase the fitness of plants under attack in natural populations (Baldwin, 1998; Kessler and Baldwin, 2001). In addition to nicotine, the plants also produce TPIs after herbivore attack (van Dam et al., 2001), which reduce the performance of hornworm (Manduca sexta) larvae (Glawe et al., 2003), a specialized lepidopteran herbivore from which three trypsin-like cDNA sequence have been cloned (Peterson et al., 1994). Mirids (Tupiocoris notatus) are the second most commonly found herbivore on N. attenuata (Glawe et al., 2003), and feed by a “lacerate and flush” mechanism in which the stylets and watery saliva are used to lacerate and flush out pockets of cells (Miles, 1972). Recently, a chymotrypsin-like protease was detected in the salivary glands of the mirids (Colebatch et al., 2001), and one chymotrypsin-like cDNA sequence has been sequenced from the midgut (Colebatch et al., 2002).
Here, we report the cloning of two multidomain TPI genes belonging to potato PI-II family from N. attenuata plants collected in Utah, and we provide a critical test of whether endogenous TPIs in N. attenuata function defensively against two specialist herbivores, hornworm and mirids. We conducted bioassay experiments comparing hornworm larval performance and mirid colonization preference for plants with low or no TPI activity. We used two independently transformed N. attenuata lines in which the expression of the pi gene was down-regulated by antisense expression of a 175-bp fragment of the N. attenuata pi gene (AS–and AS-), one line independently transformed with an empty-vector construct (C), and untransformed wild-type plants (WT) of the same genetic background (an inbred line collected from Utah). In addition, we used a natural N. attenuata genotype collected from Arizona that has a mutation in the endogenous seven-domain pi gene and does not produce pi transcripts or TPI activity (A) and that we transformed with the full-length cDNA of the seven-domain pi gene in a sense orientation under control of a constitutive promotor (S++) so that it produced TPIs at 67% of the level found in the WT Utah genotype after caterpillar attack.
RESULTS
Tobacco Produces a Seven-Domain TPI Belonging to the Potato PI-II Family
Earlier results with PI activity assays of N. attenuata plants revealed that TPI activity is strongly increased after methyl jasmonate (MeJA) elicitation (van Dam and Baldwin, 2001). To isolate the TPI gene(s) responsible for the MeJA-elicited TPI activity, N-terminal sequencing of partially purified extracts revealed a high degree of similarity with the Nicotiana alata multidomain PIs (Atkinson et al., 1993). A set of homologous primers were synthesized and used in PCR of chromosomal DNA of N. attenuata. The amplified product (pUCPI2/14) was sequenced and used to screen the cDNA library of caterpillar-attacked N. attenuata leaves (Hermsmeier et al., 2001) to isolate the full-length clones. Four clones were isolated and sequenced, and all four yielded the identical sequence. The largest clone (pPI1/14) was 1,546 bp long with a 1,368-bp open reading frame (Supplemental Fig. 1, available in the online version of this article at http://www.plantphysiol.org: GenBank accession no. AF 542547). The TPI precursor contains a signal peptide at the N-terminal followed by seven repeat domains encoding TPIs and a vacuolar targeting sequence at the C-terminal. The seven domains are linked by a five-amino acid linker peptide EE-KKN (Fig. 1).
Figure 1.
Organization of the PI gene isolated (GenBank accession no. AF 542547). The gene contains a 29-amino acid signal peptide region (SP) followed by six or seven repeated trypsin inhibitor domains (T1–T6 or T7) followed by vacuolar targeting region. The box delineates the amino acids of the TPI reactive site; the eight conserved cysteines (labeled with a dot) and the conserved Pro (labeled with a diamond on the first repeat) are all essential for the conformation of PI-II repeat domains (Antcheva et al., 2001). The missing repeat from the six-repeat gene is marked with an underline and the signal peptide of this gene differs only by a Met (overlined) to Leu substitution. Bold letters signify the amino acids differing among the repeats.
Multiple sequence alignments of a single PI repeat (T2) with the corresponding repeats of potato PI-II family members found in other Solanaceous species reveal a high degree of amino acid identity, including the conserved cysteines involved in the four disulfide bridges and the conserved Pro (Fig. 1) that characterize all 77 known PI-II repeat sequences (Antcheva et al., 2001; Barta et al., 2002). The precursor PI has 93% homology with TPI isolated from N. alata stigmas (Atkinson et al., 1993).
N. attenuata TPI Belongs to a Multigene PI Family
Genomic DNA of N. attenuata was completely digested by EcoRI, HindIII, EcoRV, and SspI enzymes and was hybridized with a radiolabeled plasmid containing the repeat domain of the tobacco PI precursor (Supplemental Fig. 2). The analysis revealed at least two bands and suggested that TPIs exist in a multigene family in N. attenuata.
To isolate the additional gene(s), primer pairs corresponding to the 5′ and 3′ regions were synthesized and used in reverse transcriptase-PCR of mRNA extracted from MeJA-elicited leaves. The resultant clone was sequenced and found to encode a six-domain PI highly homologous to the earlier isolated PI. In addition to the deletion of one repeat domain, the “new” PI also differed by a single amino acid in the signal peptide region (Fig. 1).
TPI Activity and Transcripts
To determine the constitutive and caterpillar-inducible levels of TPI mRNA of the transformed lines, northern-blot analysis was performed on total RNA from transformed lines (AS-, AS–, S+, S++, C, and AC) and untransformed genotypes (WT and A). Analysis of unattacked leaves by larvae from untransformed WT and the line transformed with empty vector construct (C) revealed a 1.4-kb TPI transcript that increased 4-fold 24 h after the larvae started to feed on the leaf (Fig. 2A). Although TPI transcripts were not detectable in AS–, not even after caterpillar damage, intermediate levels were found in the AS-line (Fig. 2A). TPI mRNA in A as well as in the AC line (A independently transformed with an empty-vector construct), which lacks the ability to produce TPIs (Glawe et al., 2003), was not detectable. Constitutive TPI mRNA levels in A plants transformed with the full-length N. attenuata pi gene in the sense orientation and low levels of TPI (S+) were not detectable, but in a line with high TPI activity (S++), transcript levels were similar to the constitutive levels found in WT plants (Fig. 2, A and B). TPI mRNA accumulation correlated well with TPI activity levels.
Figure 2.
Northern-blot analysis of TPI mRNA and TPI activity (mean ± sem) in leaves growing at node +1. RNA gel-blot analysis of pi gene transcripts of unelicited control (CON) and hornworm-elicited (CAT) plants 24 h after the start of feeding (top band, TPI mRNA: 1.4 kb, bottom band, 18S rRNA: 3.4 kb). TPI activity (mean ± sem) in CON- and CAT-elicited plants 3 d after elicitation (n.d., Not detectable in A genotype). A, Untransformed WT N. attenuata plants of the Utah genotype (WT), two homozygous T3 independently transformed lines of the Utah genotype that had been transformed with a construct containing a 175-bp pi gene fragment in an antisense orientation (AS–and AS-), or with an empty vector construct (C). B, Untransformed plants of the A genotype, and plants of the Arizona genotype transformed with a construct containing the full-length pi gene in a sense orientation (S+ and S++), or with an empty vector construct (AC). Hornworm caterpillars fed on leaves growing at node +1 (one position older than the source-sink transition leaf: node 0) at the rosette stage.
Endogenous leaf TPI activity was determined before and 3 d after larvae started to feed on the node +1 leaf of plants from transformed and untransformed genotypes. Compared with the constitutive levels of TPI activity in the WT and C plants (which did not differ significantly; F1,38= 0.04; P = 0.8434), levels in AS–and AS-plants were 90% and 33% lower, respectively (Fig. 2A; F3,76= 90.640; P < 0.0001). Caterpillar damage increased TPI activity 4-fold in WT and C plants, whereas AS–and AS-TPI levels were also increased after caterpillar damage and TPI levels in these two genotypes were 17% and 48% of those found in attacked WT plants (Fig. 2A; F3,76= 49.434; P < 0.0001). Caterpillar attack did not alter TPI activity in S++ and S+ plants (F1,38-S++= 3.744; P = 0.06; F1,38-S+= 0.015; P = 0.9044), which remained at approximately 67% and 4% of the induced WT plants, respectively (Fig. 2, A and B; F2,57= 122.655; P < 0.0001). As expected, the untransformed A and the transformed AC genotypes showed no TPI activity even after caterpillars had fed on the plant for 24 h (Fig. 2B). Protein levels were not significantly different among genotypes.
Because chymotrypsin can be inhibited by TPI (Moura and Ryan, 2001), we measured the qualitative TPI and chymotrypsin proteinase inhibitor (CPI) activities of untransformed (WT and A) and transformed (AS–and S++) genotypes by radial diffusion assay after 3 d of W + oral secretion (OS) treatment (Supplemental Table I). W + OS increased TPI activity in WT plants, but did not affect TPI activity in the S++ genotype, which was high before and after induction (Supplemental Table I). CPI activity in WT and S++ genotypes was low even after elicitation with W + OS (Supplemental Table I). The AS–geno-type showed low TPI and CPI activities in all treatments, whereas the A genotype did not show any activity even after induction with W + OS (Supplemental Table I).
Real-time PCR analysis was used to quantify the increases in the TPI transcripts in response to caterpillar elicitation. WT and C plants showed similar responses to caterpillar attack; TPI mRNA expression increased 7-fold after 24 h of caterpillar damage in the attacked leaf (+1) and 4-fold in the undamaged systemic leaf (-1), compared with undamaged WT plants (Fig. 3A). Constitutive expression of TPI mRNA was lower in AS–(30% of the undamaged WT) than in AS-(50% of the undamaged WT; Fig. 3B). After caterpillars fed on antisense plants for 24 h, the expression of TPI mRNA increased 2- and 1.5-fold in AS–and 5-and 2-fold in AS-lines in the local and systemic leaves, respectively, compared with undamaged WT (Fig. 3B). Caterpillar attack had a small effect on TPI mRNA transcripts in S++ (2-fold) and S+ (0.5-fold) genotypes during the 48-h feeding period, compared with the levels found in undamaged WT leaves growing at the same nodal positions (Fig. 3C). Surprisingly, 24 h after caterpillar damage, the relative expression of TPI mRNA in the systemic leaves decreased transiently in the S++ genotype to only 1.1-fold of that found in the undamaged WT (Fig. 3C), perhaps due to general metabolic stress resulting from caterpillar attack.
Figure 3.
Fold induction (mean ± sem) of the TPI transcripts by real-time PCR in local (+1; ○ and •) and systemic (-1; □ and ▪) leaves after hornworm elicitation relative to that of unelicited (control; ▵ and ▴) WT leaves from three replicate plants harvested and analyzed separately at 0, 6, 12, 24, and 48 h after elicitation. A, WT and C genotypes. B, AS–and AS- genotypes. C, S++ and S+ genotypes.
Endogenous TPIs As a Defense against Herbivores
To determine whether endogenous TPIs in tobacco function defensively against hornworm and mirids, we assessed caterpillar performance and colonization preference of mirids on transformed and untransformed genotypes with low or no TPI activity (A, AC, S+, AS–, and AS-) and high TPI activity (WT, C, and S++). Hornworm larval mass gain per day, survivorship, and pupal mass differed significantly between caterpillars fed on genotypes with high or low TPI activity (repeated-measures analysis of variance [ANOVA] on larval mass gain, F4,190-A-AS–AS-WT-C = 5.069; P = 0.0023; F4,170-A-AC-S+-S++-WT = 3.910; P = 0.0102; survivorship analysis in Table I; ANOVA on pupal mass, F4,51-A-AS–AS-WT-C = 32.370; P < 0.0001; F4,51-A-AC-S+-S++-WT = 8.613; P < 0.0001; Fig. 4, A and B). Within the first group of genotype comparisons (A, AS–, AS-, WT, and C), larvae fed on genotypes with low or no TPI activity (repeated-measures ANOVA, F2, 130-A-AS–AS- = 1.234; P = 0.3075) grew faster, had higher survivorship (Table I), and produced heavier pupae than those fed on genotypes with high TPI activity (repeated-measures ANOVA, F1, 60-C-WT = 0.024; P 0.8787; Fig. 4A). Between the 1st and 2nd d after caterpillar attack, the larval mass gain per day was 3.89 ± 0.41 mg d-1 (AS–), 4.22 ± 0.45 mg d-1 (AS-), 1.42 ± 0.31 mg d-1 (WT), 3.71 ± 0.33 mg d-1 (A), and 1.57 ± 0.23 mg d-1 (C); data obscured by symbols (Fig. 4A). Larvae fed on AS-lines showed intermediate percentage survivorship (Table I); during the first 3 d, survivorship was similar to that on AS–lines, but after d 3 when plants had accumulated TPIs in response to larval feeding, survivorship was similar to that of larvae fed on WT and C genotypes (Fig. 4A).
Table I.
Global and pairwise survivorship analysis of hornworm larvae that fed on untransformed wild-type N. attenuata plants (WT), two homozygous T3 independently transformed lines transformed with a construct containing a 175-bp pi gene fragment in an antisense orientation (AS—— and AS—), or with an empty vector construct (C); untransformed plants of the Arizona (A) genotype; and A plants transformed with a construct containing the full-length pi gene in a sense orientation (S+ and S++), or with an empty vector construct (AC)
| Chi-Square | d.f. | P | |
|---|---|---|---|
| Global | 46.229 | 4 | <0.0001a |
| Pairwise comparisons | |||
| AS—— versus AS— | 9.001 | 1 | 0.0027a |
| AS—— versus WT | 26.354 | 1 | <0.0001a |
| AS—— versus A | 0.252 | 1 | 0.6157 |
| AS—— versus C | 24.372 | 1 | <0.0001a |
| AS— versus WT | 4.824 | 1 | 0.0281a |
| AS— versus A | 6.291 | 1 | 0.0121a |
| AS— versus C | 3.969 | 1 | 0.0464a |
| WT versus A | 21.697 | 1 | <0.0001a |
| WT versus C | 0.042 | 1 | 0.8372 |
| A versus C | 19.886 | 1 | <0.0001a |
| Global | 51.418 | 4 | <0.0001a |
| Pairwise comparisons | |||
| S++ versus S+ | 4.409 | 1 | 0.0358a |
| S++ versus WT | 0.514 | 1 | 0.4734 |
| S++ versus A | 22.059 | 1 | <0.0001a |
| S++ versus AC | 22.059 | 1 | <0.0001a |
| S+ versus WT | 7.900 | 1 | 0.0049a |
| S+ versus A | 7.000 | 1 | 0.0082a |
| S+ versus AC | 7.000 | 1 | 0.0082a |
| WT versus A | 28.998 | 1 | <0.0001a |
| WT versus AC | 28.998 | 1 | <0.0001a |
| A versus AC | 0.146 | 1 | 0.7026 |
Significant differences at P < 0.05
Figure 4.
Hornworm mass gain per day (mean ± sem), percentage survivorship, and pupal mass (mean ± sem). Neonates started to feed on +1 leaves. A, A, AS–, AS-, WT, and C genotypes. B, A, AC, S+,S++, and WT genotypes. Caterpillars were weighed and counted daily for seven consecutive days (n = 24 caterpillars in each treatment). Larval mass gain per day was calculated as final larval mass– initial larval mass. Bars with the same letter are not significantly different at P < 0.05 as determined by one-way ANOVA.
Similar responses were found within the second group of genotype comparisons (A, AC, S+, S++, and WT). Larvae fed on S+ genotypes showed intermediate percentage survivorship, whereas larvae fed on A and AC showed the highest percentage survivorship, and those fed on S++ and WT genotypes the lowest percentage survivorship (Fig. 4B; Table I). Pupal mass and larval mass gain were higher on A, AC, and S+ (repeated-measures ANOVA, F2, 85-A-AC-S+ = 0.249; P = 0.7824) than on S++ and WT genotypes (repeated-measures ANOVA, F1, 85-S++-WT = 0.421; P = 0.5253; Fig. 4B). Between the 1st and 2nd d after caterpillar attack, the larval mass gain per day was 1.88 ± 0.46 mg d-1 (S++), 3.37 ± 0.63 mg d-1 (S+), 1.42 ± 0.23 mg d-1 (WT), 2.80 ± 0.33 mg d-1 (A), and 3.97 ± 0.31 mg d-1 (AC); data obscured by symbols (Fig. 4B).
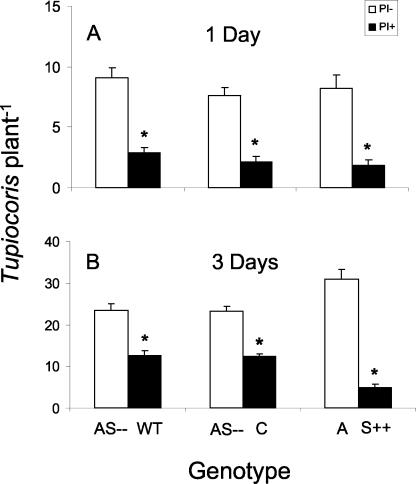
Mirids are usually the first insect species to colonize tobacco in its native habitat, that is, the first growing season after fires in the Great Basin Desert (Glawe et al., 2003). When pairs of N. attenuata genotypes differing in TPI levels (AS–versus WT, AS–versus C, and A versus S++) were exposed to a mirid colony for 24 h, adults and nymphs showed a higher preference for genotypes with low or no TPI activity than for genotypes with high TPI levels (Wilcoxon signed-rank test AS–WT; P = 0.0022; test AS–C; P = 0.0022; test A-S++; P = 0.0022; Fig. 5A). After 3 d of mirid colonization, the number of insects per plant doubled, but the preference for genotypes with low or no TPI activity did not change (Wilcoxon signed-rank test AS–WT; P = 0.0029; test AS–C; P = 0.002; testA-S++; P = 0.002; Fig. 5A).
Figure 5.
Number of adult mirids and nymphs (mean ± sem) per plant on AS–versus WT, AS–versus C, and A versus S++ pairs. Number of mirids 1 d (A) and 3 d (B) after genotypes were exposed to the insects are presented. Asterisks indicate the level of significant differences between genotypes (*P < 0.05); genotypes with no or low TPI activity are depicted by white bars and those with high levels of TPI activity are depicted with black bars.
DISCUSSION
We isolated two genes from N. attenuata coding for PI precursors that belong to the potato PI-II family. One codes for a 455-amino acid protein with a seven-repeat TPI domain, whereas the other codes for a 396-amino acid protein with a six-repeat TPI domain, both having a N-terminal signal peptide (Nielsen et al., 1997) and a C-terminal putative vacuolar targeting sequence (Nielsen et al., 1996; Fig. 1; Supplemental Fig. 1). The N. attenuata PI precursor shares a conserved amino acid sequence with that of N. alata (93%; Atkinson et al., 1993) and Nicotiana glutinosa (84%; Choi et al., 2000) PIs, including the Ala-Lys cleavage site for signal peptide cleavage and the position of the Cys residues in the repeat domains for the formation of disulphide linkages. Previously two-, four-, six-, and eight-domain PI-II precursors have been identified in other Solanaceous plants (Graham et al., 1985; Atkinson et al., 1993; Choi et al., 2000; Miller et al., 2000), and the diversity of repeats is thought to have evolved by crossing-over events within the genomes (Barta et al., 2002). How the seven-domain repeat arose is unclear, but two three-domain PI-II family members have been identified in tomato (Lycopersicon esculentum) and tobacco (Taylor et al., 1993; Balandin et al., 1995), and single repeat members have been identified in other plant families (Barta et al., 2002). It is possible that unequal crossing over among even-numbered members, or fusions of odd- and even-numbered members produced the seven-repeat domain member.
Transgenic manipulation of the ability to produce TPIs allowed us to determine whether endogenous TPIs function defensively in response to attack from native, and presumably adapted, herbivores. We used the seven-repeat TPI gene to down-regulate (1.5-and 3.5-fold) and restore (50% and 2-fold of the uninduced WT) the expression of the pi gene in the WT and A genotypes of N. attenuata, respectively (Fig. 2). TPI activity followed the patterns of TPI mRNA (Figs. 2 and 3) and increased 2-fold, 4 d after caterpillar attack (van Dam et al., 2001), with highest activity values after 3 d (J.A. Zavala and I.T. Baldwin, unpublished data), and with the highest TPI mRNA expression after 24 h (Fig. 3). Because the transformed (AS–, AS-, C, S+, S++, and AC) and untransformed genotypes (A and WT) did not differ with respect to other defense traits, such as nicotine production (Zavala et al., 2004), this is an ideal system in which to examine defensive function of TPI expression. Hornworm larvae fed on genotypes with low or no TPI activity (A, AC, S+, AS–, and AS-) grew faster, had higher survivorship, and produced heavier pupae than those that fed on genotypes with high TPI activity (WT, C, and S++; Fig. 4). Similarly, mirids preferred genotypes with low or no TPI and CPI activity (AS–and A) to genotypes with high TPI and CPI levels (WT, C, and S++; Fig. 5). These results are qualitatively and quantitatively consistent with those from previous work on A and WT genotypes (Glawe et al., 2003) and they demonstrate that endogenous TPI functions defensively against native herbivores.
The defensive function of TPIs begins with their affinity for insect proteinases (Laing and McManus, 2002), which generates the expectation of a positive quantitative relationship between the level of TPI expression and the protection they afford. However, the relationship between TPI expression and defense is complicated by the numerous counter-responses that insects have evolved. Insects, for example, in response to ingestion of high TPI leaves, are known to increase their rate of leaf consumption (Cloutier et al., 2000; Winterer and Bergelson, 2001), the secretion of total proteolytic digestive enzymes (Broadway, 1997), or the secretion of particularly TPI-insensitive proteinases (Jongsma et al., 1995). If the insect responds by increasing its consumption rate, the net fitness effect of TPI expression may not favor the plant. TPI expression is frequently found to slow herbivore growth (Charity et al., 1999), and the fitness benefit of this direct defense can be greatly enhanced when simultaneously expressed with indirect defenses that increase the mortality rate of these slow-growing herbivores. In N. attenuata, TPI expression is coordinated with the release of volatile signals that attract the generalist predator Geocoris pallens to feeding larvae (Kessler and Baldwin, 2001). This voracious predator is size selective, preferentially attacking eggs and larvae in the first three instars. The up-regulation of TPIs by herbivore attack slows the growth of larvae and keeps them in stages that are more vulnerable to the predator. Interestingly, the volatile signals are elicited by the same signals that elicit TPI production (Halitschke et al., 2000, 2001; van Dam et al., 2001), underscoring the coordination of these direct and indirect defenses. TPIs affect not only herbivore performance, but can also affect the natural enemies of the herbivores. When the Colorado potato beetle (Leptinotarsa decemlineata) feeds on transgenic potato plants that express Cys PI, which increase the PI levels of the prey, these sequestered PIs do not influence the digestion of the predatory stinkbug (Perillus bioculatus) because the predator compensates by synthesizing de novo Ser-type proteinases (Bouchard et al., 2003a, 2003b).
TPI expression alone is known to increase the mortality rate of herbivores, particularly for neonate larvae (McManus and Burguess, 1995; Heath et al., 1997; Charity et al., 1999; McManus et al., 1999; Marchetti et al., 2000; De Leo and Gallerani, 2002). In this study, larvae that ingested plants with high TPI content had not only decreased growth rates, but also a lower survivorship (approximately 40% difference) than larvae fed on low TPI genotypes. The effects of constitutive differences in TPI expression between genotypes is clearly seen in the weight gain of caterpillars during the first 2 d of feeding: larval mass gain per day was 2-fold higher in AS-, AS–, and A than in WT and C genotypes (Fig. 4A) and in A, AC, and S+ than in S++ and WT genotypes (Fig. 4B). These early differences translated into significantly different pupal masses 22 d later, which in turn, is an accurate proxy for fecundity in Lepidoptera (Haukioja and Neuvonen, 1985; Awmack and Leather, 2002; De Leo and Gallerani, 2002; Klemola et al., 2003). For the high TPI-expressing lines (WT, C, and S++), survivorship decreased (Fig. 4, A and B) in tandem with the decreases in growth rate. Interestingly, for the larvae fed on the genotype with intermediate TPI levels (AS-: 0.94 nmol mg protein-1), the decrease in survivorship is correlated with inducible TPI expression, which attained maximum values 3 d after attack. Survivorship for these larvae during the first 3 d was similar to survivorship of those feeding on the low TPI (AS–) line, but after d 3 (TPI induction; 2.63 nmol mg protein-1), survivorship was similar to that of larvae fed on WT and C genotypes. These results suggest that a threshold amount of TPI expression is required to effect changes in larval survivorship and corroborate results from experiments with Helicoverpa armigera fed on artificial diets containing soybean (Glycine max) TPI and on transgenic tobacco plants expressing giant taro (Colocasia esculenta) TPI on (Johnston et al., 1993; Wu et al., 1997). In the AS-lines, TPI elicitation killed larvae with low mass, which increased the average mass of the survivors (Fig. 4A). The intermediate constitutive TPI levels in AS-plants may have allowed heavier larvae to adapt their digestive system to a diet rich in TPIs, whereas smaller larvae were unable to adapt. The process of adaptation may involve replacing the inhibited trypsin with the secretion of new trypsins, which are insensitive to the particular TPIs of the diet (Broadway, 1995; Jongsma et al., 1995). Our results suggest that the sensitivity threshold for TPI is likely to differ from insect to insect and that the developmental stage of the insect when first exposed to the inhibitor may also determine the defensive value of TPIs for plants.
In summary, this research demonstrates that despite the ongoing evolutionary interaction between N. attenuata and its herbivores, TPIs remain an effective defense against mirids and hornworm.
MATERIALS AND METHODS
Plant Material and Transformation
Nicotiana attenuata Torr. Ex Wats. (Solanaceae) used in this study was grown from seeds collected from Utah (Baldwin, 1998) or Arizona (Glawe et al., 2003) and was inbred 10 and four generations, respectively. To silence the expression of N. attenuata's pi gene in the genotype collected in Utah (WT), WT was transformed by an Agrobacterium tumefaciens-mediated transformation procedure with pNATPI1, which contains 175 bp of N. attenuata's seven-repeat domain pi gene in an AS orientation as described in Zavala et al. (2004). One homozygous T3 independently transformed line of WT plants (C) that had been transformed with an empty vector construct (lacking only the pi gene fragment) and whose TPI activity resembled that of WT plants was selected as a control for the bioassay experiments. Southern gel-blot analysis confirmed that all T3 lines were single-copy independent transformants (Zavala et al., 2004).
A genotype of N. attenuata collected from Arizona (A) has MeJA-inducible nicotine production identical to that found in WT plants, but completely lacked the ability to produce TPIs or accumulate TPI mRNA (Glawe et al., 2003). More recently, the mutation in the seven-domain repeat pi of A plants has been characterized and found to be located in the 5′ signal peptide, resulting in a premature stop codon (J. Wu and I.T. Baldwin, unpublished data). Because we never detected TPI activity with radial diffusion assay in A genotype, nor have we detected TPI mRNA transcripts with northern blots (van Dam et al., 2001; Glawe et al., 2003) or reverse transcriptase-PCR, we suggest that this transcript is rapidly silenced. Plants of the A genotype were transformed with a binary transformation vector pRESC2PIA2 containing the full-length seven-domain N. attenuata pi gene from the WT genotype in the sense orientation under control of a cauliflower mosaic virus 35S promotor (Zavala et al., 2004). Several T1 lines harboring a single copy of the transgene (Zavala et al., 2004) were screened for TPI activity; one of those lines (S) expressing low TPI activity (heterozygous; S+) or high TPI activity (homozygous; S++) at T1 was used in the experiments. S++ plants were bred to homozygosity in the T2 and were used again in the experiments, confirming the previous results. One homozygous T2 independently transformed line of A plants (AC: see supplemental information) that had been transformed with an empty vector construct (lacking only the pi gene fragment) and had TPI activity equivalent to that of A plants (not detectable) was selected as a control. All of these transformed and untransformed genotypes were used in the bioassay experiments.
Isolation of the TPI Gene
Based on the Nicotiana alata TPI sequence (Atkinson et al., 1993), primers PI2-FOR (5′-CTGATCCTAGAAATCCAAAGGC-3′) and PI2-REV (5′-GCATATTCAGATTCTCCTTCAC-3′) were designed and used in PCR of chromosomal DNA of tobacco. The product was cloned in pUC19 cut with SmaI (pUCPI2/14), sequenced, and used to screen a cDNA library (Hermsmeier et al., 2001) of hornworm (Manduca sexta)-attacked leaves of tobacco plants for isolating a full-length clone of the PI gene.
Feeding Bioassay Experiments
To determine the effect of down-regulation or restored expression of the pi gene in N. attenuata on caterpillar mass gain, survivorship, and pupal mass, two feeding experiments with different combinations of genotypes were performed with AS lines (AS–and AS-), untransformed genotypes (WT and A) and a transformed WT line with empty vector construct (C), or with A lines transformed to express the functional pi (S+ and S++), untransformed genotypes (WT and A), and a transformed A line with empty vector construct (C). Seeds were germinated in diluted liquid smoke solutions as described in Baldwin et al. (1994). Seedlings were transplanted in 1-L pots in a greenhouse in the conditions described in Glawe et al. (2003) with 800 to 900 μmol m-2 s-1 photosynthetic photon flux density supplied by 450-W Na-vapor HID bulbs. Eggs of hawkmoth (Lepidoptera: Sphingidae) were obtained from Carolina Biological Supply (Burlington, NC) and were placed in plastic containers (200 mL) on a moist tissue. The containers were kept in climate chambers at 28°C and 65% relative humidity with a 16:8-h light:dark photoperiod until the eggs hatched. Neonates were placed individually on the node +1 (one position older than the source-sink transition leaf; van Dam et al., 2001) leaf of 24 soil-grown rosette plants of each genotype. Larvae mass and number were determined daily for 7 d after hatching, and pupae were weighed 25 d after hatching. To ensure that caterpillars reached the pupal stage with sufficient leaf material, larvae between the third and fourth instars were transferred to new plants from the same genotype. Constitutive and TPI activity induced by caterpillar damage were determined from 20 replicates during rosette stage growth. Leaves growing at node +1 were harvested 3 d after the larvae started to feed, and protein concentrations and TPI activity were measured using a radial diffusion assay and are expressed as nanomoles per milligram as described in van Dam et al. (2001).
To determine whether N. attenuata TPI inhibits chymotrypsin, 10 rosette stage plants were unwounded or wounded with a pattern wheel over the source-sink transition leaf surface (W), and 20 μL of hornworm OS was applied to the fresh puncture wounds (W + OS; Halitschke et al., 2000). Source-sink transition leaf from unwounded and W + OS was harvested 3 d after induction for TPI and CPI analysis. The qualitative determination of TPI and CPI activities was measured using a radial diffusion assay as described in van Dam et al. (2001).
Colonization Experiments
To determine the colonization preference of mirids (Tupiocoris notatus; Hemiptera: Miridae), three pairs of genotypes with low or no TPI activity and high TPI levels (AS–versus WT, AS–versus C, and A versus S++) were placed in a greenhouse adjacent to N. attenuata plants infested with a mirids colony. We monitored the accumulation of the insects on the plants at 24 h and 3 d after the pairs of genotypes placement. These experiments were repeated twice.
TPI mRNA Expression Analysis
Leaves attacked by larvae during the first 24 h (CAT) and leaves from plants without larvae (CON) from the same position (+1) were harvested for northern-blot analysis of TPI mRNA accumulation as described in Winz and Baldwin (2001) and Glawe et al. (2003) in four replicate plants from each genotype and they were pooled. These plants were excluded from subsequent analysis.
For real-time PCR analysis, leaves growing at nodes +1 and -1 (one position younger than the source-sink transition leaf) from CAT and +1 from CON treatments were harvest from three replicate plants at 0, 6, 12, 24, and 48 h after the larvae started to feed. The relative expression of TPI mRNA was compared with that of undamaged WT. The isolated RNA was quantified spectrophotometrically and was diluted to 300 ng μL-1. The diluted RNA was reverse transcribed (Applied Biosystems, Foster City, CA), and 10 ng of the reverse-transcribed template was used in a 25-μL PCR reaction containing 1× universal mix (Eurogentec, Brussels, Belgium), 300 nm forward (5′-TCAGGAGATAGTAAATATGGCTGTTCA-3′) and reverse primers (5′-ATCTGCATGTTCCACATTGCTTA-3′), and 300 nm of FAM-(6-carboxyl-fluorescein) labeled Taqman probe (5′-TCCTTGCTCTCCTCCTCTTATTTGGAATGTCT-3′) with 18s RNA (Eurogentec) as internal standard. Thermal cycling and detection was performed on a sequence detector (ABI Prism 7700; Applied Biosystems).
Statistical Analysis
Data were analyzed with Stat View, version 5.0 (SAS, Cary, NC). The TPI, protein, and pupal mass were analyzed by ANOVA followed by Fisher's protected lsd post hoc comparisons in all experiments. For the survivorship analyses, we used the log-rank test for the global hypothesis of equality of survival distribution for hornworm, and performed the same test with a pairwise ranking of data using only two groups at a time (Zavala et al., 2001). Larval mass gain per day was calculated as (final larval mass–initial larval mass) and data were analyzed with repeated-measures ANOVA. Differences in colonization preference were analyzed by Wilcoxon signed-rank test.
Supplementary Material
Acknowledgments
We thank Michelle Lim for invaluable assistance in plant transformation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.035634.
This work was supported by the Max Planck Gesellschaft.
The online version of this article contains Web-only data.
References
- Antcheva N, Pintar A, Patthy A, Simonscits A, Barta A, Tchorbanov B, Pongor SN (2001) Proteins of circularly permuted sequence present within the same organism: the major serine proteinase inhibitor from Capsicum annuum seeds. Protein Sci 10: 2280-2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA (1993) Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5: 203-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awmack SC, Leather SR (2002) Host plant quality and fecundity in herbivorous insects. Annu Rev Entomol 47: 817-844 [DOI] [PubMed] [Google Scholar]
- Balandin T, Vanderdoes C, Albert JMB, Bol JF, Linthorst HJM (1995) Structure and induction-pattern of a novel proteinase-inhibitor class-I gene of tobacco. Plant Mol Biol 27: 1197-1204 [DOI] [PubMed] [Google Scholar]
- Baldwin IT (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95: 8113-8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT (2001) An ecology motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol 127: 1449-1458 [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Staszakkozinki L, Davidson R (1994) Up in smoke: smoke-derived germination cues for the post-fire annual Nicotiana attenuata Torr. Ex Watson. J Chem Ecol 20: 2345-2371 [DOI] [PubMed] [Google Scholar]
- Barta E, Pintar A, Pongor SN (2002) Repeats with variations: accelerated evolution of the Pin2 family of proteinase inhibitors. Trends Genet 18: 600-603 [DOI] [PubMed] [Google Scholar]
- Bergelson J, Purrington CB (1996) Surveying patterns in the cost of resistance in plants. Am Naturalist 148: 536-558 [Google Scholar]
- Bergey DR, Howe GA, Ryan CA (1996) Polypeptide signaling for plant defensive genes exhibit analogies to defense signaling in animals. Proc Natl Acad Sci USA 93: 12053-12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard E, Cloutier C, Michaud D (2003a) Oryzacystatin I expressed in transgenic potato induces digestive compensation in an insect natural predator via its herbivorous prey feeding on the plant. Mol Ecol 12: 2439-2446 [DOI] [PubMed] [Google Scholar]
- Bouchard E, Michaud D, Cloutier C (2003b) Molecular interactions between an insect predator and its herbivore prey on transgenic potato expressing a cysteine proteinase inhibitor from rice. Mol Ecol 12: 2429-2437 [DOI] [PubMed] [Google Scholar]
- Broadway RM (1995) Are insects resistant to plant proteinase inhibitors? J Insect Physiol 41: 107-116 [Google Scholar]
- Broadway RM (1996) Dietary proteinase inhibitors alter complement of midgut proteases. Arch Insect Biochm Physiol 32: 39-53 [Google Scholar]
- Broadway RM (1997) Dietary regulation of serine proteinases that are resistant to serine proteinase inhibitors. J Insect Physiol 43: 855-874 [DOI] [PubMed] [Google Scholar]
- Charity JA, Anderson MA, Bittisnich DJ, Whitecross M, Higgins TJV (1999) Transgenic tobacco and peas expressing a proteinase inhibitor from Nicotiana alata have increased insect resistance. Mol Breed 5: 357-365 [Google Scholar]
- Choi D, Park JA, Seo YS, Chun YJ, Kim WT (2000) Structure and stress-related expression of two cDNAs encoding proteinase inhibitor II of Nicotiana glutinosa L. Biochim Biophys Acta 1492: 211-215 [DOI] [PubMed] [Google Scholar]
- Cloutier C, Jean C, Fournier M, Yelle S, Michaud D (2000) Adult Colorado potato beetles, Leptinotarsa decemlineata, compensate for nutritional stress on Oryzacystatin I-transgenic potato plants by hypertrophic behavior and over-production of insensitive proteases. Arch Insect Biochm Physiol 44: 69-81 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Cooper P, East P (2002) cDNA cloning of a salivary chymotrypsin-like protease and the identification of six additional cD-NAs encoding putative digestive proteases from the green mirid, Creontiades dilitus (Hemiptera: Miridae). Insect Biochem Mol Biol 32: 1065-1075 [DOI] [PubMed] [Google Scholar]
- Colebatch G, East P, Cooper P (2001) Preliminary characterization of digestive proteases of the green mirid, Creontiades dilitus (Hemiptera: Miridae). Insect Biochem Mol Biol 31: 415-423 [DOI] [PubMed] [Google Scholar]
- De Leo F, Bonadé-Bottino M, Ceci LR, Gallerani R, Jouanin L (2001) Effects of mustard trypsin inhibitor expressed in different plants on three lepidopteran pests. Insect Biochem Mol Biol 31: 593-602 [DOI] [PubMed] [Google Scholar]
- De Leo F, Gallerani R (2002) The mustard trypsin inhibitor 2 affects the fertility of Spodoptera littoralis larvae fed on transgenic plants. Insect Biochem Mol Biol 32: 489-496 [DOI] [PubMed] [Google Scholar]
- Duan X, Li X, Xue Q, Abo-El-Saad M, Xu D, Wu R (1996) Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol 14: 494-498 [DOI] [PubMed] [Google Scholar]
- Gatehouse AMR, Norton E, Davison GM, Babbe SM, Newell CA, Gate-house JA (1999) Digestive proteolytic activity in larvae of tomato moth, Lacanobia oleraceae: effects of plant protease inhibitors in vitro and in vivo. J Insect Physiol 45: 545-558 [DOI] [PubMed] [Google Scholar]
- Glawe AG, Zavala JA, Kessler A, van Dam NM, Baldwin IT (2003) Ecological costs and benefits correlated with trypsin protease inhibitor production in Nicotiana attenuata. Ecology 84: 79-90 [Google Scholar]
- Graham JS, Pearce G, Merryweather F, Titani K, Ericsson LH, Ryan CA (1985) Wound-induced proteinase inhibitors from tomato leaves: the cDNA-deduced primary structure of the pre-inhibitor II. J Biol Chem 260: 6561-6564 [PubMed] [Google Scholar]
- Green TR, Ryan CA (1972) Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175: 776-777 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Kessler A, Kahl J, A. L, Baldwin IT (2000) Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia 124: 408-417 [DOI] [PubMed] [Google Scholar]
- Halitschke R, U. Schittko, G. Pohnert, W. Boland, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol 125: 711-717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukioja E, Neuvonen S (1985) The relationship between size and reproductive potential in male and female Epirritia autumnata (Lep., Geometridae). Ecol Entomol 10: 267-270 [Google Scholar]
- Heath RL, McDonald G, Christeller JT, Lee M, Bateman K, West J, van Heeswijck R, Anderson MA (1997) Proteinase inhibitors from Nicotiana alata enhance plant resistance to insect pest. J Insect Physiol 43: 833-842 [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol 125: 683-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilder VA, Gatehouse AMR, Sheerman SE, Barker RF, Boulter D (1987) A novel mechanism of insect resistance engineered in to tobacco. Nature 330: 160-163 [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA (1996) An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067-2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Narvaez J, An G, Ryan CA (1989) Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci USA 86: 9871-9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston KA, Gatehouse JA, Anstee JH (1993) Effects of soybean protease inhibitor on the growth and development of larval Helicoverpa armigera. J Insect Physiol 39: 657-664 [Google Scholar]
- Jongsma MA, Bakker PL, Peters J, Bosch D, Stiekema WJ (1995) Adaptation of Spodoptera exigua larvae to plant proteinase inhibitors by induction of gut proteinase activity insensitive to inhibition. Proc Natl Acad Sci USA 92: 8041-8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma MA, Bakker PL, Visser B, Stiekema WJ (1994) Trypsin inhibitor activity in mature tobacco and tomato plants is mainly induced locally in response to insect attack, wounding, and virus infection. Planta 195: 29-35 [Google Scholar]
- Jongsma MA, Bolter C (1997) The adaptation of insects to plant protease inhibitors. J Insect Physiol 43: 885-895 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141-2144 [DOI] [PubMed] [Google Scholar]
- Klemola T, Ruohomaki K, Tanhuanpaa M, P. K (2003) Performance of a spring-feeding moth in relation to time of oviposition and bud-burst phenology of different host species. Ecol Entomol 28: 319-327 [Google Scholar]
- Laing W, McManus MT (2002) Proteinase inhibitors. In MT McManus, WA Laing, AC Allan, eds, Protein-Protein Interactions in Plant Biology. CRC Press, Boca Raton, FL, pp 77-119
- Marchetti S, Delledonne M, Fogher C, Chiabá C, Chiesa F, Savazzini F, Giordano A (2000) Soybean Kunitz, C-II and PI-IV inhibitor genes confer different levels of insect resistance to tobacco and potato transgenic plants. Theor Appl Genet 101: 519-526 [Google Scholar]
- McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA (1992) Structure, expression, and antisense inhibition of the systemin precursor gene. Science 255: 1570-1573 [DOI] [PubMed] [Google Scholar]
- McManus MT, Burguess EPJ (1995) Effects of the soybean (Kunitz) trypsin inhibitor on growth and digestive proteases of larvae of Spodoptera litura. J Insect Physiol 41: 731-738 [Google Scholar]
- McManus MT, Burgess EPJ, Philip B, Watson LM, Laing WA, Voisey CR, White DWR (1999) Expression of the soybean (Kunitz) trypsin inhibitor in transgenic tobacco: effects on larval development of Spodoptera litura. Transgenic Res 8: 383-395 [Google Scholar]
- McManus MT, White DWR, McGregor PG (1994) Accumulation of the chymotrypsin inhibitor in transgenic tobacco can affect the growth of insect pests. Transgenic Res 3: 50-58 [Google Scholar]
- Miles PW (1972) The saliva of Hemiptera. Adv Insect Physiol 9: 183-255 [Google Scholar]
- Miller EA, Lee MCS, Atkinson AHO, Anderson MA (2000) Identification of a novel four-domain member of the proteinase inhibitor II family from the stigmas of Nicotiana alata. Plant Mol Biol 42: 329-333 [DOI] [PubMed] [Google Scholar]
- Moura DS, Ryan CA (2001) Wound-inducible proteinase inhibitors in pepper: differential regulation upon wounding, systemin, and methyl jasmonate. Plant Physiol 126: 289-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1-6 [DOI] [PubMed] [Google Scholar]
- Nielsen KJ, Hill JM, Anderson MA, Craik DJ (1996) Synthesis and structure determination by NMR of a putative vacuolar targeting peptide and model of a proteinase inhibitor for Nicotiana alata. Biochemistry 35: 369-378 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M, McGurl B, Ryan CA (1993) Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc Natl Acad Sci USA 90: 8273-8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A, Barillas-Mury C, Wells M (1994) Sequence of three cDNA encoding an alkaline midgut trypsin from Manduca sexta. Insect Biochem Mol Biol 24: 463-471 [DOI] [PubMed] [Google Scholar]
- Royo J, Leon J, Vancanneyt G, Albar JP, Rosahl S, Ortego F, Castañera P, Sánchez-Serrano JJ (1999) Antisense-mediated depletion of a potato lipooxygenase reduces wound induction of proteinase inhibitors and increases weight gain of insect pest. Proc Natl Acad Sci USA 96: 1146-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28: 425-449 [Google Scholar]
- Schuler TH, Poppy GM, Kerrry BR, Denholm I (1998) Insect-resistant transgenic plants. TIBTECH 16: 168-175 [DOI] [PubMed] [Google Scholar]
- Taylor BH, Young RJ, Scheuring CF (1993) Induction of a proteinase inhibitor II-class gene by auxin in tomato roots. Plant Mol Biol 23: 1005-1014 [DOI] [PubMed] [Google Scholar]
- van Dam NM, Baldwin IT (2001) Competition mediates costs of jasmonate-induced defenses, N acquisition and transgenerational plasticity in Nicotiana attenuata. Funct Ecol 15: 406-415 [Google Scholar]
- van Dam NM, Horn M, Mares M, Baldwin IT (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27: 547-568 [DOI] [PubMed] [Google Scholar]
- Winterer J (2002) The mixed success of protease inhibitors to combat insect pest in transgenic crops. AgBiotechNet 4: 1-7 [Google Scholar]
- Winterer J, Bergelson J (2001) Diamondback moth compensatory consumption of protease inhibitor-transformed plants. Mol Ecol 10: 1069-1074 [DOI] [PubMed] [Google Scholar]
- Winz RA, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: Insect-induced ethylene suppresses jasmonate-induced accumulation of nicotine biosynthesis transcripts. Plant Physiol 125: 2189-2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Llewellyn D, Mathews A, Dennis E (1997) Adaptation of Helicoverpa armigera (Lepidoptera: Noctuidae) to a proteinase inhibitor expressed in transgenic tobacco. Mol Breed 3: 371-380 [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004) Constitutive and inducible trypsin protease inhibitor production incurs large fitness cost in Nicotiana attenuata. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed]
- Zavala JA, Scopel AL, Ballaré CL (2001) Effects of ambient UV-B radiation on soybean crops: impact on leaf herbivory by Anticarsia gemmatalis. Plant Ecol 156
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.