Abstract
A distinct endogenous pararetrovirus (EPRV) family corresponding to a previously unknown virus has been identified in the genome of Nicotiana tomentosiformis, a diploid ancestor of allotetraploid tobacco (Nicotiana tabacum). The putative virus giving rise to N. tomentosiformis EPRVs (NtoEPRVs) is most similar to tobacco vein clearing virus, an episomal form of a normally silent EPRV family in Nicotiana glutinosa; it is also related to a putative virus giving rise to the NsEPRV family in Nicotiana sylvestris (the second diploid progenitor of tobacco) and in the N. sylvestris fraction of the tobacco genome. The copy number of NtoEPRVs is significantly higher in N. tomentosiformis than in tobacco. This suggests that after the polyploidization event, many copies were lost from the polyploid genome or were accumulated specifically in the diploid genome. By contrast, the copy number of NsEPRVs has remained constant in N. sylvestris and tobacco, indicating that changes have occurred preferentially in the NtoEPRV family during evolution of the three Nicotiana species. NtoEPRVs are often flanked by Gypsy retrotransposon-containing plant DNA. Although the mechanisms of NtoEPRV integration, accumulation, and/or elimination are unknown, these processes are possibly linked to retrotransposon activity.
Integrated (endogenous) viral sequences are increasingly recognized as common constituents of many plant genomes. Endogenous retroviruses have been detected recently in diverse plant species (Vicient et al., 2001; Wright and Voytas, 2001). In addition, sequences derived from the two types of plant DNA virus, the single-stranded DNA gemini-viruses (Bejarano et al., 1996; Ashby et al., 1997) and the double-stranded DNA pararetroviruses (Harper et al., 2002) have been identified in various plant genomes.
Retroviruses, which have an RNA genome, must integrate into host chromosomes by means of a retrovirus-encoded integrase activity to complete their replication cycle. By contrast, neither type of plant DNA virus encodes an integrase function and their replication normally proceeds without incorporation into the host genome. Thus, the mechanism by which DNA viral sequences integrate into plant chromosomes remains to be clarified. Together with retroviruses and retrotransposons, pararetroviruses are classified as retroelements because they use a virus-encoded reverse transcriptase (RT) to replicate their genome. However, the genome structure of pararetroviruses differs significantly from that of retroviruses or retrotransposons, and they are thought to have originated when a pre-existing virus captured an RT gene (Xiong and Eickbush, 1990).
Little is known about the potential pathogenicity of endogenous viral sequences or their impact on plant genome structure and function. Some endogenous pararetroviruses (EPRVs), such as those derived from banana streak virus (Harper et al., 1999; Ndowora et al., 1999) and tobacco vein clearing virus (TVCV; Lockhart et al., 2000), were initially noticed because they can be activated and cause symptoms of infection in hybrid plants. By contrast, the EPRV family in Nicotiana sylvestris (NsEPRV; formerly named TPVL [Jakowitsch et al., 1999] and TEPRV [Mette et al., 2002]) comprises mutated viral sequences that are presumably unable to reconstitute a functional virus. NsEPRVs were first detected during a routine characterization of plant DNA flanking transgene inserts in tobacco (Nicotiana tabacum; Jakowitsch et al., 1999). The conserved methylation pattern of NsEPRVs suggested that they might confer resistance to the exogenous form of the virus, which has yet to be detected, through an epigenetic gene silencing mechanism acting at the genome level (Mette et al., 2002).
Tobacco provides an interesting system for studying EPRVs and their contribution to plant genome evolution. Tobacco is an allotetraploid that was probably created by humans around 10,000 years ago (M. Chase, personal communication) from two wild diploid ancestors, N. sylvestris as maternal “S” genome donor and Nicotiana tomentosiformis as paternal “T” genome donor (Kenton et al., 1993; Lim et al., 2000a). Recent work analyzing the presence of geminiviral-related DNA sequences in different accessions of N. tomentosiformis pinpointed specifically N. tomentosiformis ac. NIC 479/84 as the paternal parent of tobacco (Murad et al., 2002). In principle, each diploid progenitor could have contributed one or more distinct EPRV families to the tetraploid tobacco genome. DNA-blot hybridization experiments indicated that the aforementioned NsEPRV family accumulated to approximately 1,000 copies in the N. sylvestris genome before polyploid formation (Jakowitsch et al., 1999). The copy number and epigenetic state of NsEPRVs have remained essentially unchanged in N. sylvestris and in tobacco since polyploidization. When DNA isolated from diploid N. tomentosiformis was hybridized to a probe derived from NsEPRV sequences, a unique pattern comprising DNA fragments not visible in tobacco or N. sylvestris was observed (Mette et al., 2002). This finding suggested that N. tomentosiformis harbors a related but distinct family of EPRVs that is enriched in, or exclusive to, the diploid genome.
To examine this idea further, we have isolated and sequenced a number of genomic λ clones containing portions of N. tomentosiformis EPRV (NtoEPRV) family members. We report here a characterization of this dispersed repetitive sequence family, a consensus sequence for the putative virus giving rise to Nto- EPRVs, and we discuss the differential accumulation of the two EPRV families during evolution of the three Nicotiana species.
RESULTS
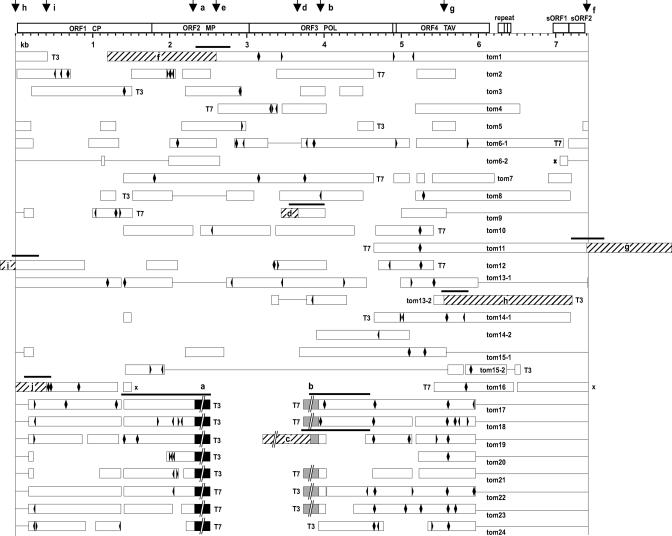
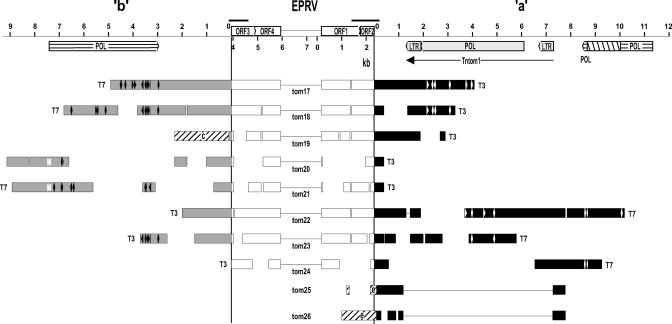
To isolate NtoEPRVs, genomic λ libraries were prepared using DNA isolated from N. tomentosiformis ac. NIC 479/84. These libraries were hybridized to a 5.5-kb probe containing the region of NsEPRV ranging from approximately 2 to 7.5 kb (Jakowitsch et al., 1999). Twenty-four positive clones were chosen for further analysis (Fig. 1). Partial sequence analysis of 16 clones that contained primarily viral sequences (Fig. 1, clones tom1 through tom16; viral sequences indicated by white bars) identified overlapping regions, which could be used to assemble a consensus sequence of a putative viral genome that is approximately 7.4 kb in length (Fig. 1, top). From these 16 clones, six unique virus DNA-plant DNA junctions were identified (Fig. 1: clones tom1-f, tom9-d, tom11-g, tom12-I, tom13-2-h, and tom16-j; plant DNA regions lettered and diagonally hatched). The eight remaining clones comprised a subclass consisting of a truncated (5.96–0.17 kb missing) and internally deleted (2.32–3.93 kb removed) copy of the putative viral genome (termed here ΔEPRV) flanked by identical plant DNA sequences designated “a” and “b” (Fig. 1, clones tom17 through tom24; redrawn in Fig. 2).
Figure 1.
Structure of NtoEPRV sequences. Twenty-four independent clones from genomic λ libraries made from N. tomentosiformis DNA were partially sequenced. White bars represent viral sequences; black bars represent plant “a” sequences; shaded bars represent plant “b” sequences; and diagonally hatched bars represent other plant sequences identified by lowercase letters within the hatched regions. The genome of the putative virus giving rise to NtoEPRV is shown at the top and is bounded by two vertical lines beginning with nucleotide 1 (tRNA-binding site) at the left and ending at approximately 7.4. kb on the right. Accession numbers for overlapping clones used to assemble the consensus sequence of the putative viral genome are AJ431198, AJ431199, AJ431200, AJ431201, AJ431202, AJ431203, AJ431204, and AJ431205. The consensus sequence of the putative virus genome can be obtained from our website (http://gmi.oeaw.ac.at). Within the NtoEPRV clones, frame shifts are denoted by arrowheads; stop codons by diamonds; crosses indicate sequence inversions. The lines connecting white bars indicate gaps compared with the consensus sequence; spaces between unconnected white bars represent unsequenced regions. T3 and T7 indicate the ends of the λ clones. Because these are arranged according to a linear projection (top) of the putative circular virus genome, ends of clones—depending on their position—can appear to be located internally. Vertical arrows at the top point out the position of junctions between viral sequences and plant DNA. Black bars above plant-virus junctions show PCR products analyzed in Figure 5. Detailed information on elements identified in the plant sequences can be found in supplemental data. CP, Coat protein; MP, movement protein; POL, polyprotein; TAV, transactivation protein. Two short ORFS reside in the intergenomic region between 7 and 7.4 kb. Accession numbers of plant sequences: a, AJ517511; b, AJ517512; c, AJ517513; d, AJ517514; f, AJ551256; g, AJ551257; h, AJ551258; I, AJ551259; and j, AJ551260.
Figure 2.
Structure of a-ΔEPRV-b subclass of NtoEPRV insertions. This figure is redrawn from Figure 1 (which illustrates the NtoEPRV clones arranged according to a linear projection of the circular genome of the putative virus) to show the actual arrangement in the plant genome. The truncated and internally deleted viral sequence (ΔEPRV) is flanked by plant sequences “a” (black bars) to the right, and “b” (gray bars) to the left. Black bars above plant-virus junctions show PCR products analyzed in Figure 5. The horizontally hatched bars represent sequences related to the polyproteins of gypsy-like retroelement remnants. The right-diagonally hatched region in the retroelement remnant in a represents an approximately 1.4-kb insertion that is similar to the NPR18 repeat in N. plumbaginifolia (Kovtun et al., 1993). The “a” sequences contain an intact gypsy-like retroelement, Tntom1 (accession no. AJ508603), with long terminal repeats. In clone tom19, “b” sequences are fused to plant sequence “c” (left-diagonally hatched). Clones tom25 and tom26 were obtained using only “a” sequence as a probe. These do not contain Tntom1 and are joined directly to a new plant sequence “e” (accession no. AJ517515; left-diagonally hatched). Detailed information on the plant sequences can be found in supplemental data. The white M in the “b” sequence of tom20 and tom21 indicates the putative start codon of the polyprotein. Other abbreviations are defined in the legend to Figure 1.
The NtoEPRV sequences contained various point mutations and small deletions, indicating they are independent clones derived from different integrated copies. The positions of the plant-virus junctions are distributed throughout the putative virus genome (Fig. 1, top: short vertical arrows), suggesting there are no preferential sites in viral DNA for recombination into plant chromosomes. Clones comprising virus-plant junctions are discussed in more detail below.
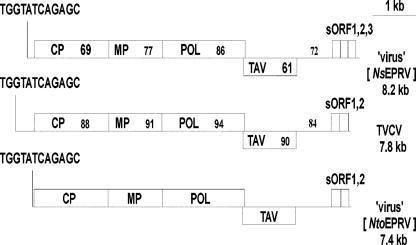
The putative pararetrovirus giving rise to Nto- EPRVs, which has not been described previously, is highly similar to TVCV (Lockhart et al., 2000), with amino acid identities in the four major open reading frames (ORFs) ranging from 88% to 94% (Fig. 3). The putative N. tomentosiformis pararetrovirus is also related to the putative virus giving rise to the NsEPRV family in N. sylvestris and the S subgenome of tobacco (Jakowitsch et al., 1999; Mette et al., 2002), although the amino acid identities in the four major ORFs are lower (range of 61%–87%; Fig. 3). The organization of the four ORFs is identical among the three viruses. In all cases, several short ORFs follow a region containing several tandem repeats and the putative enhancer-promoter. A distinguishing feature of the putative N. tomentosiformis pararetrovirus is the unusually short 5′ leader, placing the tRNA-binding site immediately upstream of the first ORF (Fig. 3). The significance of this is unknown.
Figure 3.
Comparison of the genomic organization of the putative viruses giving rise to NtoEPRV (http://www.gmi.oeaw.ac.at), NsEPRV (accession no. AJ238747; Jakowitsch et al., 1999) and TVCV (accession no. AF190123; Lockhart et al., 2000). In each case, the tRNA-binding site (TGGTATCAGAGC) is followed by four major ORFs (CP, Coat protein; MP, movement protein; POL, polyprotein; TAV, transactivation protein); a putative promoter-enhancer region (black line); and then two to three short ORFs that are putative enhancers of translational initiation and reinitiation of the polycistronic viral mRNA (Pooggin et al., 2001). The percentage of identity at the amino acid level of the putative virus giving rise to NtoEPRV and either TVCV or the putative virus giving rise to NsEPRV is shown in the respective ORFs of the latter two viruses. The percentage of DNA sequence identity is similarly indicated for the putative promoterenhancer regions. Amino acid sequence identities between TVCV and the putative virus giving rise to NsEPRV are not shown but are as follows: ORF1, 72%; ORF2, 78%; ORF3, 87%; and ORF4, 61%; and nucleotide sequence identity in the putative enhancer-promoter region is 72%.
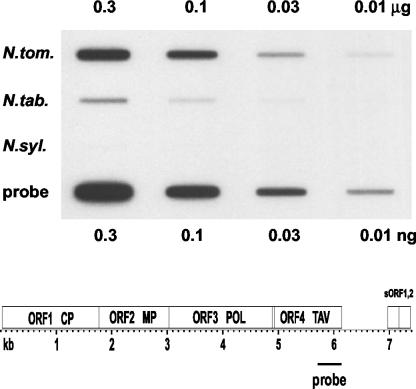
To determine the copy numbers of NtoEPRVs in the genomes of N. tomentosiformis and tobacco, a slot-blot analysis was performed. The availability of the Nto- EPRV sequence allowed us to identify a region in ORF4 of the putative virus that showed relatively low DNA sequence identity (69%) to the NsEPRV and could therefore be used as an NtoEPRV-specific probe. From the slot-blot analysis, we estimate that the genome of N. tomentosiformis contains approximately 4,000 copies of NtoEPRV, whereas the genome of tobacco DNA harbors around 600 copies (Fig. 4). Thus, the genome of the diploid species contains approximately seven times more copies of NtoEPRV than the polyploid species.
Figure 4.
Slot-blot analysis to determine NtoEPRV copy number. Plant DNA amounts are shown at the top; plant species to the left. Amounts of control probe DNA (identical to the hybridization probe) are shown at the bottom. The probe is specific for NtoEPRV under the hybridization conditions used and is derived from a region in ORF4 extending from approximately nucleotide 5,700 to 6,200. From scans of the blot, the approximate copy numbers were determined as described in “Materials and Methods.”
The difference in NtoEPRV copy number is not consistent with a simple additive model in which the polyploid tobacco genome comprises an unaltered combination of the two diploid progenitor genomes. The lower copy number in tobacco could be due to preferential loss from the polyploid genome or to accumulation specifically in the diploid N. tomentosiformis genome after polyploid formation. For each scenario, it should be possible to identify NtoEPRV inserts that are common to the tobacco and the N. tomentosiformis genomes, and inserts that are present only in the diploid species. This point is considered first for the “a-b” subclass.
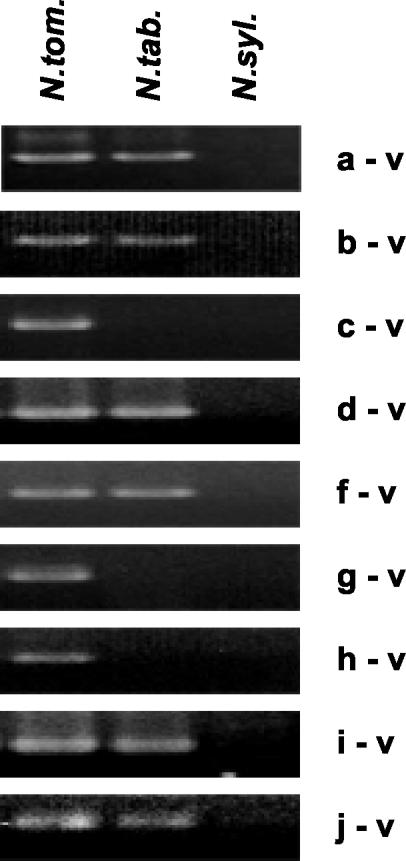
The cloning frequency of the a-b subclass in the λ libraries prepared from N. tomentosiformis DNA suggested that it comprises around 33% of the total NtoEPRV population in the diploid species. To determine whether any members of the a-b subclass are present in the tobacco genome, we performed a PCR analysis across the two plant-virus junctions using primer pairs anchored in viral DNA and a or b plant DNA sequence (Fig. 2, top). Products of the expected size were amplified from N. tomentosiformis DNA, but also from tobacco (Fig. 5, a-v and b-v). Using the “a” and “b” primer pair, the entire 4-kb ΔEPRV fragment was isolated from tobacco. Sequencing this fragment confirmed that the virus-plant junctions as well as the internal deletion in ΔEPRV are identical to those identified in the λ clones from N. tomentosiformis (data not shown). The presence of these fragments in tobacco DNA suggests that at least some members of the a-b subclass are present in tobacco despite the overall lower copy number of NtoEPRVs in this species.
Figure 5.
PCR analyses across plant DNA-virus (v) DNA junctions. Plant sequences are labeled a through j (plant sequence e, present in clones tom25 and tom 26 [Fig. 2] is not included) and are present in the following clones (Fig. 1 and Fig. 2): a and b, tom17 through tom24; c, tom19; d, tom9; f, tom1; g, tom11; h, tom13-2; I, tom12; and j, tom16. Primers used are listed in “Materials and Methods.” Details about the plant sequences including accession numbers can be found in supplemental data. As expected, none of the junction fragments are in N. sylvestris, confirming that they were contributed to the tobacco genome by N. tomentosiformis.
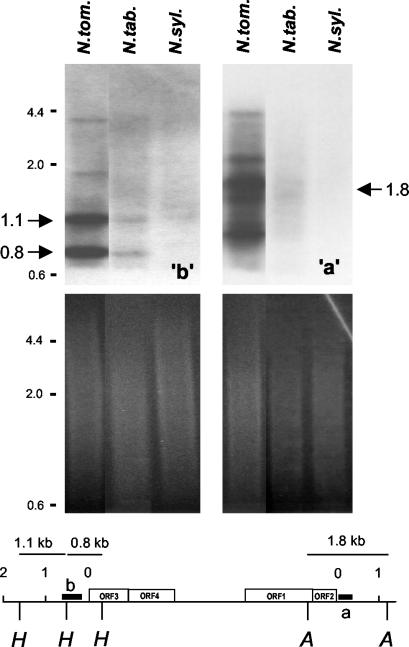
The PCR analysis we performed was not quantitative and could not be used to estimate the relative copy numbers of the a-b subclass in the diploid and polyploid genomes. Therefore, this question was examined by Southern-blot analysis. The identical “a” and “b” flanking plant DNA sequences in approximately 33% of the cloned NtoEPRV sequences suggest that these plant sequences coamplified with the ΔEPRV derivative as part of a larger self-amplifying unit. If true, the “a” and “b” plant DNA sequences should have a higher copy number in N. tomentosiformis than in tobacco, similar to the NtoEPRV population as a whole. To test this, plant DNA was cut with restriction enzymes recognizing sites on both sides of the plant-virus junctions, blotted onto nitrocellulose, and hybridized to probes specific for the “a” or “b” plant sequences. This strategy should detect selectively a-v and b-v fragments. Fragments of the expected size were observed for “a” (1.8 kb) and “b” (1.1 and 0.8 kb) probes in N. tomentosiformis (Fig. 6). Scanning these blots revealed that the “b” signals were approximately seven times stronger for the diploid species than for tobacco. A substantial enhancement of the “a” signal was also observed in N. tomentosiformis compared with tobacco, but the increase in strength is difficult to quantify owing to the presence of many extra bands (discussed further below).
Figure 6.
DNA-blot analysis of a-v and b-v junction fragments. DNA isolated from the species shown at the top was cut with HindIII (“b” blot, left) or AseI (“a” blot, right) and probed with respective plant DNA sequences (heavy black bars). Expected junction fragments from these digests and probes are 1.1 and 0.8 kb for “b” and 1.8 kb for “a” (large arrows). Autoradiograms of Southern blots are shown at the top; ethidium bromide-stained gels for loading controls are shown at the bottom. Size markers are shown in small numbers to the left. Scans of the blots were performed to quantify the differences in the signals in the tobacco lanes (N. tab.) as compared with the N. tomentosiformis (N. tom) lanes. N. sylvestris (N. syl.) DNA is included as a control.
These results indicate that the a-v and b-v sequence junctions are less abundant in tobacco than in N. tomentosiformis, which parallels the overall lower copy number of NtoEPRVs in the polyploid species. The data also support the idea that the “a” and “b” plant sequences, together with the ΔEPRV, comprise a large, self-amplifying unit (referred to hereafter as a-ΔEPRV-b) that is at least 25 kb in length (Fig. 2).
Although it is not yet possible to describe the exact nature of this postulated self-amplifying unit, it is tempting to invoke duplicative transposition. The “a” and “b” plant DNA sequences contain retrotransposon remnants (Fig. 2, horizontally hatched bars). In addition, “a” contains an intact Gypsy-like long terminal repeat retrotransposon, Tntom1, described here for the first time (Fig. 2; Tntom1, accession no. AJ508603). The retrotransposon remnants in “a” and “b” do not appear to be parts of the same element because they are in opposite orientation relative to each other. The “a” and “b” sequences show amino acid sequence similarity to the polyprotein regions of Gypsy-like retrotransposons (“a”) and Athila retroelements (“b”), which are also in the Gypsy group (Wright and Voytas, 2001; supplemental data, available in the online version of this article at http://www.plantphysiol.org).
Although the “b” hybridization pattern consists mainly of two discrete bands of the anticipated size (1.1 and 0.8 kb), the expected “a” fragment (1.8 kb) is accompanied by extra bands (Fig. 6), indicating considerable heterogeneity in sequences hybridizing to the a probe. This suggests that at least part of the “a” sequence makes up a separate repeat family containing members that amplify independently of ΔEPRV sequences. “a” sequences that are not associated directly with ΔEPRV but joined to a new plant sequence, “e”, were recovered when the N. tomentosiformis λ library was probed with an “a”-specific probe that lacked viral sequences (Fig. 2, clones tom25 and tom26). In addition, the “a” sequences in these two clones were deficient in Tntom1, which is consistent with “a” being a polymorphic repetitive element.
Independent from the subclass of a-ΔEPRV-b insertions, six unique plant-virus junctions were cloned (Fig. 1, clones tom1-f, tom9-d, tom11-g, tom12-I, tom13-2-h, and tom16-j; plant DNA lettered and diagonally hatched). To determine whether any of the unique plant-virus junctions are common to tobacco and N. tomentosiformis (corresponding to ancestral copies that have been retained in both species) or specific to the N. tomentosiformis genome (representing copies that had been lost from the polyploid genome or gained in the diploid genome since polyploid formation), PCR analyses spanning the junctions were performed (Fig. 1, black bars above plant-virus junctions). Two plant-virus junctions (g-v and h-v) were found only in N. tomentosiformis (Fig. 5). The “g” and “h” regions consist exclusively of internal Gypsy-like retroelement sequences that run directly into viral sequences (supplemental data). Whether these are parts of intact retrotransposons cannot be determined from the sequences recovered in the λ clones.
The four remaining unique plant-virus junctions were found in tobacco and in the diploid species, consistent with maintenance in both genomes (Fig. 5, d-v, f-v, i-v, and j-v). In contrast to “g” and “h”, the flanking plant DNAs “f” through “j” lack retroelement sequences; only “d” is partially composed of retroelement-related DNA (supplemental data).
Another type of junction that was found only in N. tomentosiformis comprised a new plant DNA-plant DNA fusion in a clone containing the a-ΔEPRV-b region. This junction brought together a short portion of “b” and a new plant sequence “c” (Figs. 1 and 2, clone tom4; and 5, c-v). The b-c fusion might reflect instability of the a-ΔEPRV-b region as a result of frequent amplification in N. tomentosiformis.
DISCUSSION
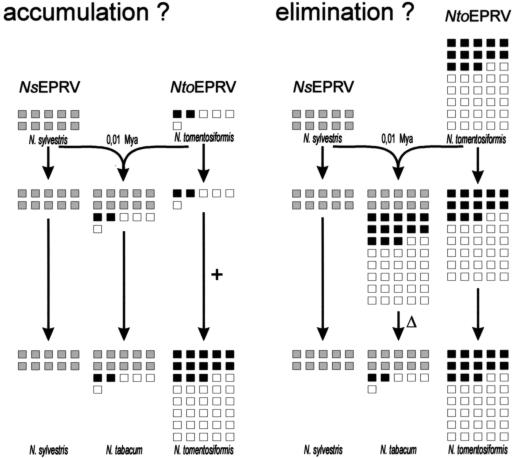
We have identified and characterized a distinct endogenous pararetrovirus family, NtoEPRV, in the genome of N. tomentosiformis. It is the second EPRV family to be associated with polyploid tobacco (N. tabacum), an allotetraploid derived from two diploid progenitors: N. tomentosiformis ac. NIC 479/84 and N. sylvestris. The first tobacco EPRV family to be identified, NsEPRV (Jakowitsch et al., 1999), appears to have integrated into the N. sylvestris genome before polyploid formation and to have survived virtually unchanged with respect to copy number, arrangement, and pattern of DNA methylation in N. sylvestris and the S subgenome of tobacco since the polyploidization event (Mette et al., 2002). The stability of the NsEPRV family is consistent with the idea that it has been selected as a virus resistance determinant (Mette et al., 2002). By contrast, as shown here, the copy number of NtoEPRVs is 7-fold higher in N. tomentosiformis than in tobacco. This difference could be due to preferential elimination from the tobacco genome or expansion of the Nto- EPRV family in the diploid species after polyploid formation (Fig. 7). Although the elimination of species-specific repeats is emerging as a common theme in allopolyploid genome evolution (Volkov et al., 1999; Shakad et al., 2001; Kashkush et al., 2002), the latter “accumulation” hypothesis cannot yet be ruled out. Further work to analyze the presence of NtoEPRV-like sequences in closely related Nicotiana species is required to decide between these alternatives.
Figure 7.
Possible models for the differential accumulation of two EPRV families during the evolution of N. tabacum and its diploid progenitors, N. tomentosiformis and N. sylvestris. Each box represents 100 copies of the respective EPRV family. The ancestral copy number of the NsEPRV family (gray boxes) is presumed to correspond to the present copy number because DNA isolated from N. sylvestris and tobacco appears identical on blots probed with an NsEPRV sequence (Mette et al., 2002). By contrast, the NtoEPRV family (black boxes, the putative chimeric retrotransposon a-ΔEPRV-b; white boxes, unique inserts) is seven times larger in N. tomentosiformis than in tobacco. This could reflect either a low ancestral copy number and accumulation (+) exclusively in the diploid species (left) or a high ancestral copy number and preferential elimination (Δ) from the polyploid species (right) after the polyploidization event approximately 10,000 years ago (0.01 Mya). Although the 7-fold change in NtoEPRV copy number in tobacco involves both unique inserts (Fig. 5) and members of the a-ΔEPRV-b subclass (Fig. 6), it is not yet known whether each class is affected equally as shown here.
The reason for the apparent lability of NtoEPRVs is not known, but it is intriguing to consider a role for retrotransposons. Strikingly, plant DNA that flanks NtoEPRVs often consists of Gypsy retrotransposon-containing sequences, including a new Gypsy retro-transposon, Tntom1, identified in this study. No bias toward these sequences was observed in an analysis of a comparable number of clones containing members of the NsEPRV family (Jakowitsch et al., 1999), which might account for the relative stability of this family. Of the two unique NtoEPRV inserts identified so far that are not present in tobacco (i.e. those corresponding to ones that were eliminated from the polyploid genome or accumulated in the diploid genome subsequent to polyploid formation), both are joined directly to Gypsy retrotransposon sequences (plant DNA “g” and “h”). In addition, a chimeric element containing Gypsy retrotransposon sequences was possibly involved in the amplification of the a-ΔEPRV-b subclass in N. tomentosiformis. The transduction of cellular genes by plant retroelements, including Athila in Arabidopsis (Pélissier et al., 1995), has been documented previously (Bureau et al., 1994; Jin and Bennetzen, 1994; Palmgren, 1994). It is not yet known whether the ΔEPRV sequence was captured by a replicating retrotransposon in a template switch during reverse transcription, or whether pararetroviral sequences integrated by chance into retrotransposon DNA and subsequently became part of the transposing unit.
Similarly to the putative virus giving rise to NsEPRV, the putative virus giving rise to NtoEPRV corresponds to a previously unknown pararetrovirus. Nevertheless, the two putative viruses are distinct from each other, sharing overall less than 80% amino acid sequence identity. The N. tomentosiformis “virus” is most similar to TVCV (the overall amino acid sequence identity is approximately 90%), the episomal infectious form of a normally silent family of EPRVs in N. glutinosa (Lockhart et al., 2000). Even though NtoEPRVs are a distinct repetitive sequence family in the N. tomenosiformis genome, the putative virus from which they originated might have been a strain of TVCV. In that case, the amino acid variation we observed may be due to differences in selective pressure on the endogenous copies within the host genomes. Activation of endogenous copies of TVCV occurs in Nicotiana edwardsonii, a hexaploid hybrid formed between diploid Nicotiana glutinosa and tetraploid Nicotiana clevelandii, which is devoid of EPRVs. Given the high sequence similarity between the putative N. tomentosiformis virus and TVCV, it will be interesting to see whether NtoEPRV can be reactivated to produce symptoms of virus infection in hybrids produced by crossing N. tomentosiformis and N. clevelandii. The fact that N. tomentosiformis does not show obvious symptoms of virus infection under normal growth conditions indicates that the NtoEPRVs are normally kept under control in this species, perhaps through a gene silencing mechanism. A recent study has indicated that endogenous petunia vein clearing virus is repressed by DNA methylation in host plants grown under standard conditions (Richert-Pöggeler et al., 2003).
In previous work on the NsEPRV family, virus-plant junctions were found to cluster at two regions of the putative viral genome, which possibly corresponded to recombinogenic gaps in the open circular form of the viral DNA (Jakowitsch et al., 1999). By contrast, the plant-viral junctions in the NtoEPRV family are distributed throughout the putative virus genome. Therefore, even though illegitimate recombination is still the most plausible mechanism for pararetroviral DNA integration into host chromosomes, we cannot conclude in the case of NtoEPRVs that recombination has occurred preferentially at gaps in the open circular form of the putative viral genome.
The two EPRV families we have identified in tobacco and its diploid progenitors illustrate the contribution of viral sequences to plant genome structure and diversity. The Nicotiana EPRV families offer a good system for studying further the differential behavior of repetitive sequences in polyploid genomes, the role of EPRVs in viral pathogenicity, and potential interactions of EPRVs with other retroelements. As additional EPRVs are identified and characterized (Harper et al., 2002), their role as intermediaries in the dynamic interplay between free viruses, retrotransposons, and plant genomes will be clarified.
MATERIALS AND METHODS
Plant Material and Plant DNA Isolation
The following plant material was used in this analysis: Nicotiana tabacum cv SR1, Nicotiana tomentosiformis ac. NIC 479/84, and Nicotiana sylvestris. Total DNA from the various Nicotiana species was extracted from fresh leaves with the DNeasy Plant Maxi kit (Qiagen, Hilden, Germany) following the manufacturer's instructions.
λ Cloning and Sequencing
λ libraries were constructed using the λ FixII kit (Stratagene, Vienna) according to the protocols provided by the supplier. The libraries were screened with a subcloned 5.5-kb NotI-HindIII fragment of the NsEPRV (formerly TPVL) clone V6 (Jakowitsch et al., 1999), corresponding to approximate NsEPRV coordinates 2 to 7.5 kb. Clones tom25 and tom26 were selected by hybridizing with probe a (see below), not with the 5.5-kb probe. λ DNA was isolated using the Lambda Midi kit (Qiagen) and was sequenced as described previously (Jakowitsch et al., 1999).
Amino Acid Alignments
To determine the percentage of amino acid identities of the four major ORFs of TVCV (accession no. AF190123), and the putative “viruses” giving rise to the NtoEPRV (http://gmi.oeaw.ac.at) and NsEPRV (accession no. AJ238747) repetitive sequence families, as well as the DNA sequence identities in the promoter-enhancer regions, multiple alignments using ClustalW were carried out.
Hybridization Techniques
For λ screening, and Southern and slot blots, Protran BA nitrocellulose membrane (Schleicher & Schuell, VWR, Vienna) was used. DNA probes (25–50 ng) were labeled using the Megaprime DNA labeling system (Amersham Pharmacia, Vienna) and 32P-dATP, and were then purified on a 1-mL Sephadex G50 column. The probes were hybridized at 64°C in 3× SSC (Thomashow et al., 1980) omitting EDTA and prehybridization. Blots were washed twice for 10 min at 64°C with 3× SSC (Thomashow et al., 1980). For Southern blots, 1 μg of total plant DNA was digested overnight, precipitated, dissolved in water, and loaded on a 1.5% (w/v) agarose gel. For slot blots, total plant DNA was precipitated and washed with 70% (w/v) ethanol to remove free nucleotides, quantified with a GeneQuant photometer (Amersham Pharmacia), and blotted with a Bio-Dot SF apparatus (Bio-Rad, Vienna) according to the manufacturer's instructions. For quantification, the developed X-Omat AR films (Kodak, Vienna) were scanned on a Sharp scanner JX 330 (Amersham Pharmacia Biotech Europe, Vienna) and band intensities proportional to the dilution factors were used. To estimate the NtoEPRV copy number, the following 1C values were used: N. tomentosiformis (2.83 pg); N. tabacum (5.85 pg); and N. sylvestris (2.88 pg; Bennett and Leitch, 2003). These 1C values were converted to Giga basepairs (Gbp) using the conversion factor 1 pg = 0.965 Gbp (Arumuganathan and Earle, 1991). This resulted in the following values for the actual genome sizes: N. tomentosiformis (5.46 Gbp/2C = 2x) and N. tabacum (11.29 Gbp/2C = 4x). NtoEPRV copy numbers were calculated as (ngpr/ngge) × (kbpge/kbppr), where ngpr and ngge are the amounts in nanograms of probe and genomic DNA, respectively, which correspond to equal intensities on the blot (only intensities proportional to the dilution factors were used); and kbpge/kbppr are the DNA sizes in kilobase pairs (kbp) of the plant genome (per 2C) and the probe, respectively. 1 Gbp = 1 × 106 kbp.
PCR Analysis and Oligonucleotides
PCR was performed using 1 ng of total plant DNA and Takara Ex Taq polymerase (BioWhittaker, Verviers, Belgium). The PCR primers are as follows (note that in the list below, the single letters—a, b, c, etc.—denote a specific plant sequence). The letter v, together with a plant sequence letter, denotes the viral sequence primer used for the corresponding plant DNA-virus DNA junction (va, vb, vc, etc.). These are indicated simply as v in the text and figures because they are always referred to together with the corresponding plant DNA primer (i.e. a-v, b-v, c-v, etc.).
PCR primers: a, 5′-AAAGGGAAATACACAATTTCCACTCACG-3′; va, 5′-CAGCACCACAATTTGGATGTAC-3′; b, 5′-TCCGTTGAGGTGGACCATG-3′; c, 5′-CAAGTTGTGGTGCGTATATAAAGC-3′; vbc, 5′-CTCCTCCATAACTATGATTACTCG-3′; d, 5′-TGCATATCTGGACAACTCACTAAAC-3′; vd, 5′-CTTGTGGTACTGTAAATGCTGTTAG-3′; f, 5′-AACTTGTGATGATCTTTGCCATC-3′; vf, 5′-GGTAATTCAGGGCATAGTTGTTC-3′; g, 5′-TTTCGTCATCCTCTTCATCCATC-3′; vg 5′-CTACAATACTGGCAACAAACTACAG-3′; h, 5′-TCTTAATCTCCATAGCTGAAGTGGG-3′; vh, 5′-CCACCTGCTACAATTATGGATTAC-3′; i, 5′-ATGAAACCAATTAACAACGAAAGGG-3′; vi, 5′-CCAATTGTTGTATTTCTTTGCTTGC-3′; j, 5′-CACTTGCAACGGCAACATC-3′; vj, 5′-TTCTGGGAATTTCTTATGGTTG-3′ using annealing temperatures of 63°C (a/va, b/vbc, c/vbc, f/vf, and g/vg), 61°C (d/vd, h/vh, and i/vi), and 59°C (j/vj). Probes a and b and the probe used for the slot blot were amplified from total DNA as PCR fragments using primer pairs 5′-CTTACAAGAGAAATGCTCATACCAG-3′ and 5′-CGTTTAGTTTGTCGTCTTCTTCTCG-3′; 5′-GTCCTTCTTCAACAGCATTTCTTC-3′ and 5′TGATTTAGAATCGAAGCACAAG-3′; 5′-AGGTTTGTGAAATCAAGGTCAATGC-3′ and 5′-CAGTGCCAAACATTTCTTCTGCAC-3′, and annealing temperatures of 50°C, 60°C, and 60°C, respectively.
Supplementary Material
Acknowledgments
We thank Andrew Leitch, Ilya Leitch, Marie-Angele Grandbastien, Werner Aufsatz, Tatsuo Kanno, and members of the European Union Consortium PARADIGM for helpful discussions. We also thank Ales Kovarik for communicating unpublished results, Johannes van der Winden for tech nical assistance, and the IPK Gatersleben for supplying seeds of N. tomen tosiformis Goodspeed accession no. NIC 479/84.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.031112.
This work was supported by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung (grant no. Z21–MED) and by the European Union (contract no. QLK3–CT–2002–02098).
The online version of this article contains Web-only data.
References
- Arumuganathan K, Earle E (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 208-218 [Google Scholar]
- Ashby A, Warry A, Bejarano E, Khashoggi A, Burrell M, Lichtenstein C (1997) Analysis of multiple copies of geminiviral DNA in the genome of four closely related Nicotiana species suggest a unique integration event. Plant Mol Biol 35: 313-321 [DOI] [PubMed] [Google Scholar]
- Bejarano E, Khashoggi A, Witty M, Lichtenstein C (1996) Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc Natl Acad Sci USA 93: 759-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ (2003) Plant DNA C-values database. http://www.rbgkew.org.uk/cval/homepage.html
- Bureau TE, White S, Wessler SR (1994) Transduction of a cellular gene by a plant retroelement. Cell 77: 479-480 [DOI] [PubMed] [Google Scholar]
- Harper G, Hull R, Lockhart B, Olszewski N (2002) Viral sequences integrated into plant genomes. Annu Rev Phytopathol 40: 119-136 [DOI] [PubMed] [Google Scholar]
- Harper G, Osuji J, Heslop-Harrison JS, Hull R (1999) Integration of banana streak badnavirus into the Musa genome: molecular and cytogenetic evidence. Virology 255: 207-213 [DOI] [PubMed] [Google Scholar]
- Jakowitsch J, Mette MF, van der Winden J, Matzke M, Matzke AJM (1999) Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc Natl Acad Sci USA 96: 13241-13246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YK, Bennetzen J (1994) Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6: 1177-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy A (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651-1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenton A, Parokonny A, Gleba Y, Bennett M (1993) Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol Gen Genet 240: 159-169 [DOI] [PubMed] [Google Scholar]
- Kovtun YV, Komarnitsky I, Gleba Y (1993) A new middle repetitive sequence of Nicotiana plumbaginifolia genome. Plant Mol Biol 23: 435-438 [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyásek R, Lichtenstein C, Leitch A (2000a) Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma 109: 245-258 [DOI] [PubMed] [Google Scholar]
- Lockhart B, Menke J, Dahal G, Olszewski N (2000) Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J Gen Virol 81: 1579-1585 [DOI] [PubMed] [Google Scholar]
- Mette MF, Kanno T, Aufsatz W, Jakowitsch J, van der Winden J, Matzke M, Matzke AJM (2002) Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J 21: 461-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad L, Lim KY, Christopodulou V, Matyasek R, Lichtenstein C, Kovarik A, Leitch AR (2002) The origin of tobacco's T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanaceae). Am J Bot 89: 921-928 [DOI] [PubMed] [Google Scholar]
- Ndowora T, Dahal G, LaFleur D, Harper G, Hull R, Olszewski N, Lockhart B (1999) Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology 255: 214-220 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (1994) Capturing of host DNA by a plant retroelement: Bs1 encodes plasma membrane H+-ATPase domains. Plant Mol Biol 25: 137-140 [DOI] [PubMed] [Google Scholar]
- Pélissier T, Tutois S, Deragon J, Tourmente S, Genestier S, Picard G (1995) Athila, a new retroelement from Arabidopsis thaliana. Plant Mol Biol 29: 441-452 [DOI] [PubMed] [Google Scholar]
- Pooggin M, Futterer J, Skryabin K, Hohn T (2001) Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc Natl Acad Sci USA 98: 886-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert-Pöggeler K, Noreen F, Schwarzacher T, Harper G, Hohn T (2003) Induction of infectious petunia vein clearing (pararetro)virus from endogenous provirus in petunia. EMBO J 22: 4836-4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakad H, Kashkush K, Ozkan H, Feldman M, Levy A (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749-1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M, Nutter R, Postle K, Chilton MD, Blattner F, Powell A, Gordon M, Nester E (1980) Recombination between higher plant DNA and the Ti plasmid of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 77: 6448-6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient C, Kalendar R, Schulman A (2001) Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 11: 2041-2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov R, Borisjuk N, Panchuk I, Schweizer D, Hemleben V (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol 16: 311-320 [DOI] [PubMed] [Google Scholar]
- Wright D, Voytas D (2001) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retrovirus. Genome Res 12: 122-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush T (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9: 3353-3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.