Abstract
Lysophosphatidyl acyltransferase (LPAAT) is a pivotal enzyme controlling the metabolic flow of lysophosphatidic acid into different phosphatidic acids in diverse tissues. A search of the Arabidopsis genome database revealed five genes that could encode LPAAT-like proteins. We identified one of them, LPAAT1, to be the lone gene that encodes the plastid LPAAT. LPAAT1 could functionally complement a bacterial mutant that has defective LPAAT. Bacteria transformed with LPAAT1 produced LPAAT that had in vitro enzyme activity much higher on 16:0-coenzyme A than on 18:1-coenzyme A in the presence of 18:1-lysophosphatidic acid. LPAAT1 transcript was present in diverse organs, with the highest level in green leaves. A mutant having a T-DNA inserted into LPAAT1 was identified. The heterozygous mutant has no overt phenotype, and its leaf acyl composition is similar to that of the wild type. Selfing of a heterozygous mutant produced normal-sized and shrunken seeds in the Mendelian ratio of 3:1, and the shrunken seeds could not germinate. The shrunken seeds apparently were homozygous of the T-DNA-inserted LPAAT1, and development of the embryo within them was arrested at the heart-torpedo stage. This embryo lethality could be rescued by transformation of the heterozygous mutant with a 35S:LPAAT1 construct. The current findings of embryo death in the homozygous knockout mutant of the plastid LPAAT contrasts with earlier findings of a normal phenotype in the homozygous mutant deficient of the plastid glycerol-3-phosphate acyltransferase; both mutations block the synthesis of plastid phosphatidic acid. Reasons for the discrepancy between the contrasting phenotypes of the two mutants are discussed.
Glycerolipids are the most abundant lipids in higher plants (Somerville et al., 2000; Voelker and Kinney, 2001). They are synthesized in two subcellular compartments, which are denoted as the prokaryotic and eukaryotic systems. In the prokaryotic system of the plastids, glycerol-3-phosphate (GP) is acylated sequentially with acyl acyl-carrier protein (ACP) to lysophosphatidic acid (LPA) and phosphatidic acid (PA), which are catalyzed by GP acyltransferase (AT; EC 2.3.1.15) and lysophosphatidyl AT (LPAAT; EC 2.3.1.51), respectively. The PA is converted to different glyco- and sulfo-lipids for membrane synthesis within the plastids. In the eukaryotic system of mainly the endoplasmic reticulum (ER), GP is acylated sequentially with acyl-CoA to LPA and PA, which are also catalyzed by glycerol-3-phosphate acyltransferase (GPAT) and LPAAT, respectively. The PA is converted to phospholipids for incorporation into the ER membranes, which will eventually be distributed throughout the cell to become membranes of different eukaryotic cell components. Some of the cytoplasm-synthesized glycerolipids are channeled to the plastids to be converted to glycol- and sulfo-lipids. In maturing seeds, the cytoplasm-synthesized PA is also converted to triacylglycerols (TAGs) for storage. The prokaryotic and eukaryotic glycerolipid syntheses are similar in the two-enzymic conversion of GP to PA and different in the subsequent diversion of PA to special structural, storage, and signal lipids. Thus, the regulation of PA synthesis before the diversion is important in the early metabolic control of glycerolipid synthesis. Although the two-enzymic acylations of the prokaryotic and eukaryotic systems are very similar, they are catalyzed by enzymes unique to the respective system.
The genes encoding the plastid GPAT in Arabidopsis and several other species have been cloned (Ishizaki et al., 1988; Nishida et al., 1993). The plastid enzyme has a substrate preference for 18:1-ACP, and in some chilling-sensitive plants, it can utilize both 18:1- and 16:0-ACP (Murata and Tasaka, 1997). The plastid GPAT in Arabidopsis has been studied with use of mutants. An Arabidopsis homozygous mutant of a defective gene encoding the plastid GPAT has minimal prokaryotic GPAT activity but no apparent phenotype (Kunst et al., 1988). Analysis of the lipid compositions in the homozygous mutant has revealed that the need for glycerolipids in the plastids is met by shuffling glycerolipids synthesized in the ER to the plastids. This and other observations have led to the concept that shuffling glycerolipids between the two systems is readily operational whenever there is a need (Somerville et al., 2000). The in vitro activities of GPAT in the microsomes from leaves can utilize 18:1- and 16:0-CoA, whereas those from maturing seeds can also utilize 12:0-, 18:0-, and 22:1-CoA, depending on the plant species. The latter substrate preference is related to the acyl moieties at the sn-1 position of storage TAGs in the seeds of individual plant species (Voelker and Kinney, 2001). GPAT genes encoding the cytoplasmic enzymes have been studied only recently in Arabidopsis (Zheng et al., 2003). GPAT encoded by one of the seven potential GPAT genes, AtGPAT1, is active on 18:0-, 18:1-, and 16:0-CoA, and homozygous null mutation of AtGPAT1 leads to male sterility.
The gene encoding the plastid LPAAT in Brassica napus has been cloned, and its encoded enzyme in vitro has a preference for 16:0- over 18:1-CoA (Bourgis et al., 1999). Genes encoding the cytoplasmic LPAATs in maturing seeds from several species also have been cloned (Brown et al., 1994, 1995; Hanke et al., 1995; Knutzon et al., 1995). Some of the cytoplasmic LPAATs that have been studied biochemically exhibit a substrate preference for 18:1- over 16:0-CoA. In species in which the seed oils have unusual acyl moieties at the sn-2 position, additional seed-specific LPAATs are present that are reactive toward the respective unusual acyl-CoAs (Cao et al., 1990; Brown et al., 1995; Laurant and Huang, 1992; Frentzen, 1998).
Although LPAATs from several plant species have been cloned and studied, no detailed study of their genes in Arabidopsis has been reported. Thus, the known genome sequence and the mutation system of Arabidopsis have not been utilized to study the LPAAT reaction in glycerolipid synthesis. We searched the Arabidopsis genome for genes related to LPAAT. Five genes could encode putative LPAATs, and only one of them could encode the plastid LPAAT. We found a knockout mutant of this plastid LPAAT. Although the heterozygous mutant has no apparent phenotype, the embryo representing the homozygous mutant dies at an early stage during embryogenesis. Embryo death in the homozygous mutant of the plastid LPAAT contrasts with the normal phenotype of the homozygous mutant of the plastid GPAT (Kunst et al., 1988). The contrast is intriguing because the two enzymes work in tandem to produce PA, and the intermediate LPA is not known to be diverted to other metabolic reactions. In this report, we present our experimental findings and deduce possibilities that could explain the discrepancy between the contrasting phenotypes of the two mutants.
RESULTS
Arabidopsis Has Five Genes That Could Encode LPAATs
LPAATs and their genes in several species, including meadowfoam (Limnanthes douglasii; Brown et al., 1995; Hanke et al., 1995), maize (Zea mays; Brown et al., 1994), B. napus (Bourgis et al., 1999), almond (Prunus dulcis; GenBank accession no. AF213937), and coconut (Cocos nucifera; Knutzon et al., 1995) have been examined. No LPAAT or its gene in Arabidopsis has been documented. Also, a thorough search of the Arabidopsis database for genes that might encode LPAATs has not been described. A short report mentions 10 unspecified putative LPAAT genes in Arabidopsis (Maisonneuve et al., 2003). A Web site for Arabidopsis genes involved in lipid metabolism (Beisson et al., 2003; http://www.plantbiology.msu.edu/lipids/genesurvey) lists 11 putative LPAAT genes.
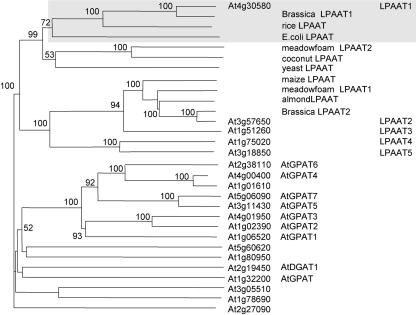
The BLAST algorithm was employed to search the Arabidopsis genome database for potential LPAAT genes. We used the maize cytoplasmic LPAAT (GenBank accession no. Z29518; Brown et al., 1994) and a B. napus plastid LPAAT (GenBank accession no. AF111161; Bourgis et al., 1999) as queries for Arabidopsis genes that encode proteins with similar amino acid sequences. From the results, we examined genes that encode proteins possessing the two conserved motifs (NHX4D and EGT). These two motifs are conserved in bacteria, yeast, and animal LPAATs (Heath and Rock, 1998; Lewin et al., 1999), and plant LPAATs (in species described in the preceding paragraph; our observation). NHX4D and EGT have been shown to be the catalytic site and GP-binding site, respectively (Heath and Rock, 1998; Lewin et al., 1999). Genes encoding proteins that lack either of these two motifs were eliminated. The retained genes, the 11 putative LPAAT genes suggested by the Web site of Beisson et al. (2003), and genes encoding studied ATs (GPAT and DGAT1) in the Kennedy pathway were subjected to amino acid sequence analyses to generate a phylogenetic tree (Fig. 1).
Figure 1.
A phylogenetic tree of Arabidopsis genes that encode proteins related to LPAAT constructed on the basis of their predicted amino acid sequences. It was inferred from the alignment using the neighbor-joining method with 1,000 bootstrap replicates. Only bootstrap values of over 50% are shown. Genes encoding these proteins were obtained after a BLAST search of the databases of The Arabidopsis Information Resource and National Center for Biotechnology Information with the use of the amino acid sequences of a maize cytoplasmic LPAAT (Z29518) and a B. napus plastid LPAAT1 (AF111161) as queries (for details, see “Results”). All of the preceding and following numbers of genes/proteins are from GenBank. Eleven putative LPAATs cited in a Web site (Beisson et al., 2003) and studied ATs (GPAT and DGAT1) of the Kennedy pathway are included. All reported LPAATs of other plant species (rice, AC068923; meadowfoam LPAAT2, S60477; coconut, U29657; meadowfoam LPAAT1, S60478; almond, AF213937; and B. napus LPAAT2, Z95637) and Escherichia coli (from plsC, M63491) and yeast (Saccharomyces cerevisiae) LPAAT (from slc1, L13282) are incorporated. The Arabidopsis genes are shown by their locus numbers, and the first five genes are also shown by their simplified protein names, as are those from other plants and microbes. Arabidopsis LPAAT1, B. napus plastid LPAAT1, a rice LPAAT (presumably in the plastids), and E. coli LPAAT are shaded.
Fifteen Arabidopsis genes encode proteins that have both NHX4D and EGT (in the upper portion of the phylogenetic tree, Fig. 1); none has been shown to encode LPAAT by experimentation. They can be divided into two groups on the basis of sequence similarities of their encoded proteins and other studied plant and microbial LPAATs. One group has five genes: one encodes the plastid LPAAT (LPAAT1) and four likely encode the cytoplasmic LPAATs (LPAAT2–5); their identifications will be described in the following section. The other group has 10 genes, whose encoded proteins are relatively dissimilar to those encoded by the first group; several of these genes (AtGPAT1–7 in Fig. 1) have been shown recently to encode putative cytoplasmic GPAT (Zheng et al., 2003).
There are five additional but quite dissimilar genes (in the lower portion of the tree, Fig. 1). Three of the five genes encode proteins containing the NHX4D motif but not the EGT motif; they include GPAT encoding the plastid GPAT (Nishida et al., 1993) and At3g05510 and At1g78690 (considered to be putative LPAAT genes by Beisson et al., 2003). Two of the five genes have neither of the motifs; they include diacylglycerol AT (DGAT1; Routaboul et al., 1999; Bouvier-Nave et al., 2000) and At2g27090 (considered to be a putative LPAAT gene by Beisson et al., 2003).
The above analyses suggest that there are only five genes (shown in the uppermost portion of the phylogenetic tree, Fig. 1) that could encode LPAATs. We analyzed these five genes further.
The meadowfoam (LPAAT2), coconut, and yeast LPAATs form a subgroup and do not have a closely related counterpart in Arabidopsis (Fig. 1). The meadowfoam (Hanke et al., 1995; Lassner et al., 1995) and coconut (Knutzon et al., 1995) enzymes are more active on the unusual 22:1-CoA and 12:0-CoA, respectively, than on 18:1-CoA; these activities are related to the presence of the unusual acyl moieties at the sn-2 position of the storage TAGs. The meadowfoam and coconut LPAATs might have diversified from the housekeeping LPAATs (e.g. meadowfoam LPAAT1) in individual species (Laurant and Huang, 1992); these housekeeping LPAATs, if studied, are more active on 18:1-CoA than on other acyl-CoAs. Arabidopsis apparently has only the housekeeping LPAATs, and its storage TAGs have the unusual 22:1 acyl moieties at the sn-1 and -3 positions but not at the sn-2 position (Voelker and Kinney, 2001).
Arabidopsis Has Only One Gene (LPAAT1) That Encodes the Plastid LPAAT
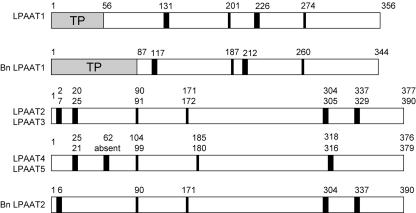
It is predicted that LPAAT1 (At4g30580) encodes the plastid LPAAT, and LPAAT2 to 5 encodes the cytoplasmic LPAATs for the following reasons. LPAAT1 is similar to the B. napus plastid LPAAT1 (84.6% amino acid sequence similarity; Bourgis et al., 1999) and the E. coli LPAAT (41.3%; Coleman, 1992), whereas LPAAT2 to 5 are relatively similar to the studied cytoplasmic LPAATs of other plants (Fig. 1). LPAAT1 is similar to the B. napus plastid LPAAT1 in having a putative N-terminal plastid transit peptide (Fig. 2), whereas LPAAT2 to 5 have putative ER retention sequences (data not shown). LPAAT1 has two putative trans-membrane segments located at positions similar to those of the B. napus plastid LPAAT1, whereas LPAAT2 to 5 have these segments located at the opposite ends of their sequences as in the B. napus cytoplasmic LPAAT2 (Fig. 2). LPAAT1 possesses in vitro LPAAT activity that is more active toward 16:0- than 18:1-CoA, a characteristic of the plastid enzyme (to be shown). The LPAAT1 transcript is present at a substantially higher level in leaves than in other organs, whereas the LPAAT2 transcript has a fairly even distribution among the various or- gans (to be shown). LPAAT1 can functionally complement a homozygous null lpaat1 mutant, whereas LPAAT1 having its sequence encoding the putative plastid targeting signal deleted cannot do so (to be shown). Thus, there is only one Arabidopsis gene (LPAAT1) encoding the plastid LPAAT. In B. napus, LPAAT encoding the plastid LPAAT has several copies (Bourgis et al., 1999). These multiple copies could have arisen via genome/gene duplication from a common ancestor shared by B. napus and Arabidopsis or could be because of the amphidipoidic nature of B. napus, or both.
Figure 2.
A comparison of the structures of the Arabidopsis LPAAT1 and putative cytoplasmic LPAAT(2–5) and those of the B. napus plastid LPAAT1 and cytoplasmic LPAAT2. The Arabidopsis (LPAAT1) and B. napus (Bn LPAAT1) plastid enzymes are similar in the locations of the putative plastid transit peptide (TP), the two trans-membrane segments (wide vertical bars), and the successive conserved motifs NHX4D and EGT (narrow vertical bars). LPAAT2 to 5 and the B. napus cytoplasmic LPAAT (Bn LPAAT2) share similar locations of the trans-membrane segments and the successive conserved motifs and an absence of a putative plastid transit peptide. The beginning residue numbers of the various parameters along the sequences are indicated.
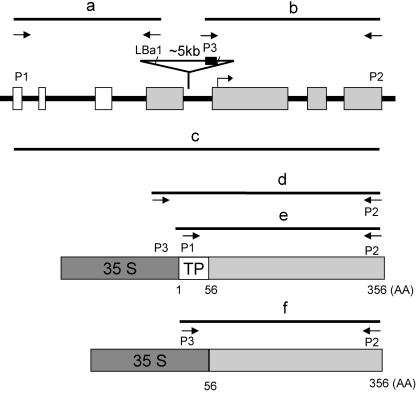
LPAAT1 has an open reading frame (ORF) consisting of seven exons (Fig. 3). Because of the length of the LPAAT1 transcript (to be described), the B. napus LPAAT1 sequence and the length of its transcript (Bourgis et al., 1999), and the annotated rice LPAAT sequence, we recognize an error in the Arabidopsis database in the prediction of the exons and introns in LPAAT1. This prediction omits the first four exons. The corrected exons and introns along the gene are shown in Figure 3.
Figure 3.
Structure of Arabidopsis LPAAT1 (At4g30580) encoding plastid LPAAT1, lpaat1, and the derived constructs used in the current studies. A 2-kb segment of the gene possessing seven exons, which encode the putative N-terminal plastid targeting signal (white boxes) and the mature protein (shaded boxes), is shown. The mutated gene, lpaat1, is interrupted by a T-DNA (indicated with an inserted triangle). The horizontal arrow along the ORF represents the starting point of the LPAAT1(234) segment, which was used to transform E. coli for testing functional complementation and enzyme activity. The structures of 35S:LPAAT1 and 35S:LPAAT1(-TP), which were used to transform Arabidopsis for testing functional complementation, are shown as horizontal boxes at the lower portion. The horizontal lines labeled a to f denote the predicted PCR fragments produced from the various primers. The primers correspond to the 5′ (P1) and 3′ (P2) termini of LPAAT1 or the left (LBa1) and right (P3) borders of the T-DNA. The 35S (heavily shaded box) also contains the P3 sequence.
LPAAT1 Functionally Complemented an E. coli LPAAT Mutant, and Its Encoded Protein Synthesized in Bacteria Had in Vitro LPAAT Enzyme Activity
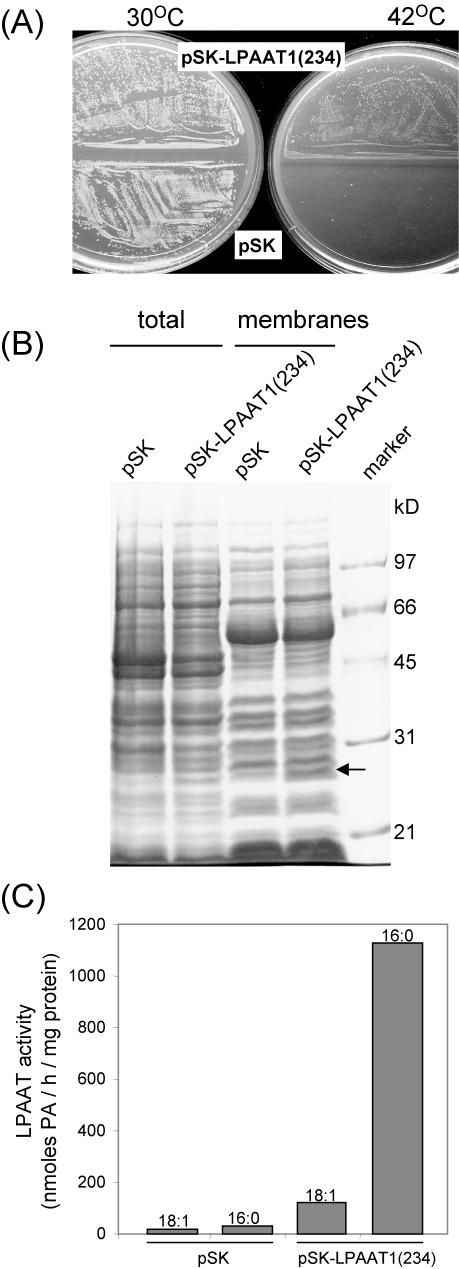
We tested whether LPAAT1 encodes LPAAT that is enzymically active in vivo and in vitro. The E. coli mutant JC201, which is a temperature-sensitive mutant of LPAAT (Coleman, 1990), was used. The mutant grows at 30°C but not at 42°C. We selected a segment of the LPAAT1 ORF [termed LPAAT1(234)] that encoded 234 residues instead of the full-length 356 residues (Fig. 3); this 234-residue polypeptide is most similar to the E. coli LPAAT in length and sequence. LPAAT1(234) was inserted into the expression vector pBluescript SK. The resulting pSK-LPAAT1(234) and the control pSK were transformed into E. coli JC201.
Figure 4A shows that E. coli JC201 harboring either of the two plasmids grew at 30°C but only that harboring pSK-LPAAT1(234) grew at 42°C. At 42°C, the colonies were smaller than those at 30°C. Thus, LPAAT1(234) was active in vivo and complemented the defective E. coli LPAAT.
Figure 4.
Expression of At LPAAT1 in E. coli strain JC201 temperature-sensitive mutant defective of LPAAT. A, Bacteria transformed with pSK or pSK-LPAAT1(234) were grown at 30°C or 42°C for 18 h and photographed. B, Total extracts and the membrane fractions of the two types of transformed bacteria grown at 30°C were analyzed for their protein constituents by SDS-PAGE. A protein of 28 kD (arrowed) was present in the total extract and the membrane fraction of bacteria transformed with pSK-LPAAT1(234) but absent in those of bacteria transformed with pSK. Mr markers are on the right lane. C, LPAAT activities in the membrane fractions from the two types of transformed bacteria assayed with the use of equal amounts of proteins (30 μg), LPA-18:1, and either 18:1- or 16:0-CoA are shown.
LPAAT1(234) synthesized in E. coli JC201 harboring pSK-LPAAT1(234) was detected by SDS-PAGE (Fig. 4B). The total extract and the membrane fraction derived from E. coli JC201 harboring pSK-LPAAT1(234) contained a 28-kD protein that was absent in corresponding fractions derived from E. coli JC201 harboring pSK. This molecular mass is that expected (28,172 Da) as deduced from the truncated ORF of LPAAT1(234).
Membrane fractions from the two E. coli samples were assayed for LPAAT enzymic activity with use of LPA-18:1 as the acyl acceptor and either 16:0- or 18:1-CoA as the acyl donor. The enzymic product PA was quantified (Fig. 4C). The membrane fraction from E. coli JC201 harboring pSK had minimal enzymic activity, which was higher with 16:0-CoA than with 18:1-CoA. The membrane fraction from E. coli JC201 harboring pSK-LPAAT1(234) had about 10 to 20 times higher activity; again, the activity was higher with 16:0-CoA than with 18:1-CoA.
Overall, our results show that LPAAT1(234) synthesized in E. coli JC201 had LPAAT enzymic activity, that this activity had acyl-CoA preference expected from the predominant 16:0 acyl moiety at the sn-2 position of plastid glycerolipids, and that the putative N-terminal plastid targeting transit peptide was not needed for enzymic activity.
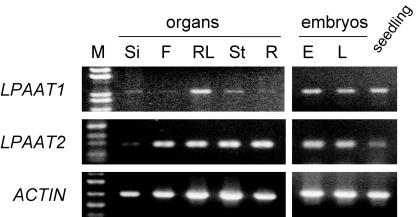
The Transcript of LPAAT1 Was Present in Diverse Organs
RNA-blot hybridization revealed that the transcript of LPAAT1 in leaves had approximately 1.2 kb (data not shown), which is slightly longer than the combined length of the exons (Fig. 3). The transcript, as detected by RT-PCR, was found in siliques, flowers, rosette leaves, stems, roots, and maturing embryos and seedlings (Fig. 5). The findings are consistent with, and expand from, the earlier report that in B. napus, the plastid LPAAT transcript was present in roots, stems, leaves, flowers, and embryos (Bourgis et al., 1999). In addition, the current findings show that the level of LPAAT1 transcript in leaves was substantially higher than those in other organs.
Figure 5.
RT-PCR analysis of the transcript from various organs with primers specific for LPAAT1 or LPAAT2. Approximately equal amounts of transcript of an Arabidopsis actin gene (ACTIN) were present in the various samples. Organs included maturing siliques (Si), maturing flowers (F), rosette leaves (RL), stems (St), and roots (R), as well as early (E) and late (L) maturing embryos and seedlings. Left lane, Markers of DNA length.
LPAAT2 encodes a putative cytoplasmic LPAAT, as judged from its close similarity in amino acid sequence with the cytoplasmic enzymes in other species, especially the B. napus LPAAT2 (Fig. 1). Its transcript was also ubiquitous (Fig. 5). However, unlike the LPAAT1 transcript, the LPAAT2 transcript was not present at a high level in leaves. The findings reinforce the idea that LPAAT1 and LPAAT2 encode the plastid and cytoplasmic enzymes, respectively.
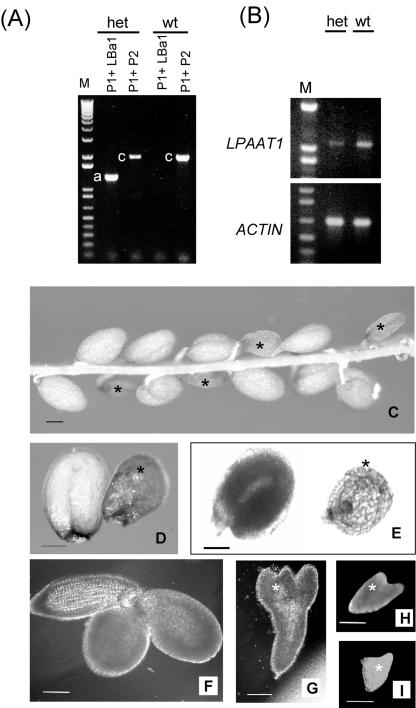
An LPAAT1/lpaat1 Heterozygous Mutant Had No Overt Phenotype
A homozygous mutant of Arabidopsis defective of the lone gene encoding plastid GPAT has little GPAT activity for plastid glycerolipid synthesis but no overt phenotype (Kunst et al., 1988). Analyses of its lipid composition in leaves indicate that cytoplasm-synthesized glycerolipids are transported to the plastids to compensate for the deficiency.
We examined an Arabidopsis mutant of LPAAT1 to test whether the mutant behaved the same as the above-mentioned plastid GPAT mutant. A mutant line containing T-DNA-inserted LPAAT1 (we termed it lpaat1) was available at the Salk Institute (http://signal.salk.edu/cgi-bin/tdnaexpress). Of the plants grown from the T3 seeds we received, one was heterozygous for lpaat1. Figure 6A shows that wild-type plants contained only LPAAT1, whereas the heterozygous mutant contained both LPAAT1 and lpaat1. In addition, the level of LPAAT1 transcript in the leaves of the heterozygous mutant was approximately one-half of that in the wild-type plants (Fig. 6B). The heterozygous mutant showed no apparent phenotype in its vegetative growth under our growth conditions. Its acyl composition of leaf lipids was similar to that of the wild-type plants (Table I). The lack of a difference in the vegetative growth phenotype and the leaf acyl composition in the heterozygous LPAAT1 mutant is similar to that in the heterozygous GPAT offspring from a cross between a homozygous GPAT mutant and a wild-type plant (Kunst et al., 1988).
Figure 6.
Characterization of the LPAAT1/lpaat1 heterozygous Arabidopsis mutant. A, PCR products of LPAAT1 and lpaat1 with the use of leaf DNA of wild type (wt) and a heterozygous LPAAT1/lpaat1 mutant (het). The primers representing the 5′ (P1) and 3′ (P2) termini of LPAAT1 (producing PCR fragment c) or P1 and the left border of the T-DNA (LBa1; producing PCR fragment a) are shown in Figure 3. Left lane, DNA size markers. B, RT-PCR products of LPAAT1 with the use of equal amounts of leaf RNA and the primers P1 and P2. The reaction detected LPAAT1 but not lpaat1 because of the exceedingly long, inserted T-DNA (approximately 5 kb). Approximately equal amounts of transcript of an Arabidopsis ACTIN gene were present in the two samples. DNA size markers are in the left lanes. C to I, Light microscopic images of the seeds and embryos produced by a LPAAT1/lpaat1 heterozygous mutant. C, Almost mature (approximately 15 d after flowering [DAF]) silique with its coat removed to reveal normal-sized and shrunken (asterisk) seeds. D, Enlarged view of normal-sized and a shrunken seeds taken from an almost mature (approximately 15 DAF) silique observed under a dissecting microscope. E, Same as D except under a transmission microscope. Both types of microscopy revealed that the normal-sized seed was completely filled with an upturned-U embryo, whereas the shrunken seed contained a very small embryo. F, Spread-out embryo from a normalsized seed of approximately 12 DAF. G to I, Spread-out embryos of different sizes from individual shrunken seeds of approximately 12 DAF. Bar in all images = 100 μm.
Table I.
Fatty acid composition of lipids extracted from leaves of wild-type and LPAAT1/lpaat1 mutant Each value represents the average of five individual plant samples. sds are shown in parentheses.
| Plants
|
Fatty Acid
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 | 16:2 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 | |
| mol % | ||||||||
| Wild type | 20.3 (3.3) | 4.9 (0.8) | 1.2 (0.7) | 10.3 (8.4) | 1.6 (0.4) | 1.2 (0.5) | 11.6 (3.2) | 49.0 (21.8) |
| LPAAT1/lpaat1 | 21.0 (4.4) | 5.8 (1.1) | 1.4 (0.5) | 8.6 (3.5) | 2.1 (0.5) | 2.3 (0.8) | 12.4 (3.6) | 46.2 (13.7) |
Homozygosity (lpaat1/lpaat1) Was Lethal during Embryogenesis
Selfing of the heterozygous mutant (LPAAT1/lpaat1) produced both normal-sized and shrunken seeds in mature siliques (Fig. 6C). One-fourth (155 of 450, P < 0.05) of the seeds were shrunken, whereas the remaining seeds were normal sized. The findings suggest that the shrunken-seed phenotype was associated with one recessive gene and that the normalsized seeds represented LPAAT1/lpaat1 heterozygous (two of four) and LPAAT1/LPAAT1 homozygous (one of four) individuals. None of the shrunken seeds but all of the normal-sized seeds germinated.
We tested further whether the phenotype of embryo death (shrunken seeds) was associated with the recessive lpaat1. The above-mentioned normal-sized seeds produced by selfing of a T3 heterozygous mutant should have a ratio of 2:1 for kanamycin resistance:kanamycin susceptibility because of the presence of KanR within the T-DNA, unless one or more additional T-DNA was inserted into other genes. We allowed 120 of these normal-sized seeds to germinate in a medium containing kanamycin, and 82 seedlings (two of three, P < 0.05) were resistant to kanamycin. The findings indicate that only one copy of T-DNA was inserted into LPAAT1 in the T3 heterozygous mutant. In addition, PCR analysis of 40 plants grown from the kanamycin-resistant T4 seedlings revealed all to be heterozygous for lpaat1. Furthermore, all of these 40 plants produced seeds, one-fourth of which were shrunken (data not shown). Thus, only one copy of T-DNA is in the heterozygous T3 plant, that this T-DNA is inserted into LPAAT1, and that lpaat1 is recessive for embryo death.
Development of the above normal-sized seeds and shrunken seeds was examined by light microscopy (Fig. 6, C–I). All the normal-sized seeds developed at a rate comparable with that of a wild-type plant (Mansfield and Briarty, 1991). In the shrunken seeds, the increase in embryo size was arrested at the heart or torpedo stage (Fig. 6, G–I). Thus, embryo death occurred at an early stage during embryogenesis.
Homozygous (lpaat1/lpaat1) Embryo Lethality Was Eliminated in Offspring of Heterozygous Plants That Had Been Transformed with cDNA Encoding LPAAT1
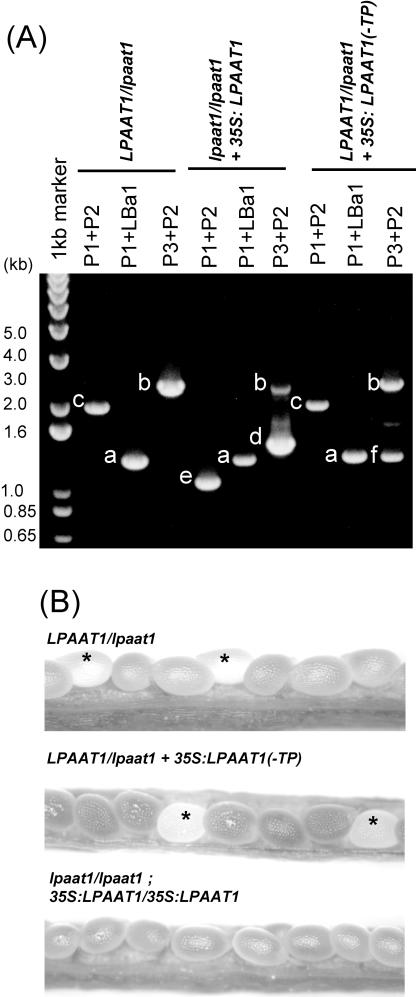
Kanamycin-resistant heterozygous (LPAAT1/lpaat1) plants were transformed with 35S:LPAAT1 (35S cauliflower mosaic virus [CaMV] promoter plus full-length LPAAT1 encoding the putative N-terminal plastid transit peptide and the mature protein) or 35S: LPAAT1(-TP) (35S CaMV promoter plus truncated LPAAT1 encoding only the mature protein) construct (Fig. 3). Many transformed plants (T1) survived the barstar selection. From each of the two transformations, 10 T1 individuals were randomly chosen and confirmed to contain the construct in their leaf DNA; they were further examined for the phenotypes of the maturing seeds by microscopy indicative of functional complementation.
Five T1 plants transformed with 35S:LPAAT1 were found to be heterozygous (LPAAT1/lpaat1) and possess 35S:LPAAT1. They had normal vegetative growth and produced seeds (T2) of normal and aborted phenotypes at ratios higher (in the range of 3:1–12:1, Table II) than the ratio of 3:1 in nontransformed heterozygous (LPAAT1/lpaat1) plants. Another five T1 plants transformed with 35S:LPAAT1 were found to be homozygous (lpaat1/lpaat1) and possess 35S:LPAAT1 (Fig. 7A). They had normal vegetative growth and produced maturing seeds (T2) of normal and aborted phenotypes at ratios of 2:1 to 3:1 (Table II). This result is indicative of a successful rescue of the homozygous (lpaat1/lpaat1) plants with 35S:LPAAT1. The 2:1 to 3:1 ratios are the consequence of uncertainties of the number of 35S:LPAAT1 construct per haploid genome in individual T1 plants and the location and positional effects of the construct on individual chromosomes. Homozygosity of 35S:LPAAT1 in a T2 lpaat1/lpaat1 plant produced seeds only of the normal phenotypes (Fig. 7B). The overall findings indicate that the heterozygous (LPAAT1/lpaat1) and homozygous (lpaat1/lpaat1) plants had been complemented functionally with 35S:LPAAT1.
Table II.
Phenotypic ratios of maturing seeds produced by homozygous (lpaat1/lpaat1) and heterozygous (LPAAT1/lpaat1) plants transformed with 35S:LPAAT1 or 35S:LPAAT1(-TP) The copy no. of 35S:LPAAT1 or 35S:LPAAT1(-TP) per haploid genome in each transformant is unknown. The nos. of normal seeds and abnormal seeds (white, transparent seeds to become shrunken seeds upon silique maturation) from five siliques of each plant line are shown.
| Plant Lines |
lpaat1/lpaat1 + 35S:LPAAT1
|
LPAAT1/lpaat1 + 35S:LPAAT1
|
LPAAT1/lpaat1a + 35S:LPAAT1(-TP)
|
|||
|---|---|---|---|---|---|---|
| Normal:abnormal | (Simplified ratio) | Normal:abnormal | (Simplified ratio) | Normal:abnormal | (Simplified ratio) | |
| 1 | 80:38 | (2:1) | 109:34 | (3:1) | 166:63 | (3:1) |
| 2 | 85:48 | (2:1) | 174:41 | (4:1) | 164:47 | (3.5:1) |
| 3 | 120:72 | (2:1) | 193:23 | (8:1) | 156:51 | (3:1) |
| 4 | 168:72 | (2:1) | 190:23 | (8:1) | 144:49 | (3:1) |
| 5 | 145:54 | (3:1) | 154:13 | (12:1) | 177:59 | (3:1) |
Five other transformants also produced normal/abnormal seeds in the ratio of 3:1; these data are not shown
Figure 7.
Functional complementation of Arabidopsis lpaat1 mutants with LPAAT1. A, PCR products from leaf DNA of heterozygous (LPAAT1/lpaat1) or homozygous lpaat1/lpaat1) mutants as offspring of heterozygous plants that have or have not been transformed with 35S:LPAAT1 or 35S:LPAAT1(-TP). The structures of these constructs and the expected PCR fragments a to f derived from the various primers are shown in Figure 3. Left lane, DNA size markers. B, Light microscopic images of the maturing (approximately 12 DAF) seeds in siliques in plants of the indicated genotypes. Stars indicate white, transparent maturing seeds, which would become shrunken upon silique maturation.
Corroborative evidence comes from the results of the analyses of the 10 T1 plants transformed with 35S:LPAAT1(-TP), all of which were found to be heterozygous (LPAAT1/lpaat1) and possess 35S: LPAAT1(-TP) (Fig. 7A). They had normal vegetative growth and produced seeds (T2) of normal and aborted phenotypes at a ratio of 3:1 (Fig. 7B; Table II). This ratio is similar to that expected 3:1 ratio of the heterozygous plants without transformation with 35S:LPAAT1(-TP). The findings indicate that only 35S:LPAAT1, but not 35S:LPAAT1(-TP), could functionally complement lpaat1.
The results confirm that the observed embryo lethality in homozygous offspring produced by heterozygous (LPAAT1/lpaat1) plants was caused by the loss of LPAAT1. In addition, they validate that LPAAT1 is a plastid enzyme. Without the transit peptide, LPAAT1(-TP) was unable to restore vitality in homozygous (lpaat1/lpaat1) plants, even though its shorter version [LPAAT1(234); Fig. 3] contained in vitro LPAAT activity (Fig. 4C) and functionally complemented the E. coli mutant JC201 (Fig. 4A).
DISCUSSION
Five Arabidopsis genes encode proteins that have sequences similar to those of LPAATs of other species and possess two conserved motifs. They include one gene for the plastid LPAAT and four genes for the cytoplasmic LPAATs. Both the plastid GPAT and LPAAT are each encoded by only one gene, and their synthesized glycerolipids are known to be used only for plastid membrane synthesis. In contrast, the cytoplasmic LPAATs apparently are encoded by several genes. This diversification may meet the needs of the enzymes and the synthesized glycerolipids in different compartments (e.g. ER, mitochondria, peroxisomes, etc.) for diverse purposes such as membrane formation, TAG storage, and signal transduction.
Embryo death in the homozygous LPAAT1 knockout mutant contrasts with the normal phenotype in the homozygous plastid GPAT mutant (Kunst et al., 1988). The latter mutant has little (3% or less of the wild type) plastid GPAT activity, and the lipid composition of its leaves suggests that cytoplasm-synthesized glycerolipids are elevated and transported to the plastids to compensate for the glycerolipid deficiency. In the plastids, GPAT and LPAAT work in tandem to convert GP to LPA and then PA, and, thus, a knockout mutation at either gene stops the production of PA for membrane biogenesis. If the loss of PA synthesis in the plastids were the only consequence of mutation at either enzymic step, and if replenishment by the cytoplasm-synthesized glycerolipids took place effectively, the LPAAT1 knockout mutant should have survived, as the GPAT mutant did. Yet, the LPAAT1 knockout mutant died at the embryo stage. We make the following speculations on the reasons behind this embryo death.
First, it is possible that in the GPAT mutant, the glycerolipids (possibly including LPA) reshuffled into the plastids require further action of the plastid LPAAT before utilization, especially in incorporating 16:0 at the sn-2 position. The GPAT mutant retained about one-half of the 16:0 moiety in the wild type, and the 16:0 presumably was present at the sn-2 position of the plastid glycerolipids as a consequence of the LPAAT1 catalysis. Plastid glycerolipids containing 16:0 moiety at the sn-2 position might be essential for the functioning of the plastids and presumably were not produced in the LPAAT1 knockout mutant. Second, the plastids may produce prokaryotic glycerolipids, not just for membrane synthesis but also for an unspecified and indispensable function that requires a minimal amount (e.g. a hormonal action). The GPAT mutant is not a knockout mutant, and the mutated enzyme still retains 3% or less activity. This 3% or less GPAT activity was that observed from an in vitro assay, and the percentage of retained in vivo GPAT activity to allow the metabolic flow could be higher. Therefore, the plastids in the GPAT mutant still would be able to produce a trace amount of prokaryotic PA (18:1, 16:0) to perform the hypothetical function. This trace amount of prokaryotic PA would not be produced in the LPAAT knockout mutant. Third, GP may act as a signal molecule, such that GP accumulated in the plastids or cytoplasm in the GPAT mutant, but not in the LPAAT mutant, triggers a compensatory response. Fourth, there may be an unknown GPAT or a GPAT-independent pathway for the synthesis of LPA in Arabidopsis plastids. Other possibilities include the following. In the LPAAT mutant, LPA may accumulate and act as a deleterious detergent, or the complete loss of LPAAT may alter a membrane enzyme complex resulting in disruption of metabolic flow or membrane permeability.
MATERIALS AND METHODS
Plant Materials
T3 seeds of Arabidopsis containing a T-DNA inserted in the At4g30580 locus (Salk_073445) were obtained from the Salk Institute (http://signal.salk.edu/cgi-bin/tdnaexpress) via the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Seeds containing T-DNA inserted in the genome were selected after allowing them to germinate on Murashige and Skoog medium supplemented with kanamycin at 50 μg mL-1. After 15 d, the plantlets were transferred to individual pots containing sterilized compost. These plantlets and those of the ecotype Columbia-0 were grown to flowering in a growth chamber maintained at 100 μE m-2 s-1 and 20°C under a 16-h-light/8-h-dark photoperiod.
For the studies of LPAAT1 and LPAAT2 transcripts, the following organs were collected. Unopened flowers (florets), siliques of mixed development stages, leaves, and stems were obtained from mature plants. Roots were collected from seedlings grown for 10 d on Murashige and Skoog medium. Developing embryos were dissected from seeds in siliques 10 (termed early maturation) and 18 (termed late maturation) DAF. Two-day-old seedlings were obtained from seeds grown on Murashige and Skoog medium.
For the studies of phenotypes of seeds produced by LPAAT1/lpaat1 plants, the siliques were cut open, and the numbers of normal and shrunken seeds were counted.
DNA Database Search and Sequence Analyses
We searched for putative LPAAT genes of Arabidopsis in The Arabidopsis Information Resource (http://www.Arabidopsis.org) using the amino acid sequences of a maize (Zea mays) cytoplasmic LPAAT (GenBank accession no. Z29518; Brown et al., 1994) and a Brassica napus plastid LPAAT (GenBank accession no. AF111161; Bourgis et al., 1999) as queries. The results gave genes that were then examined for the presence of the two conserved motifs, NHX4D and EGT, in their encoded proteins (Heath and Rock, 1998; Lewin et al., 1999). Genes encoding proteins that do not have either of these two motifs were eliminated. The retained genes, the 11 putative LPAAT genes present in the Web site for Arabidopsis genes involved in lipid metabolism (http://www.plantbiology.msu.edu/lipids/genesurvey; Beisson et al., 2003), and the genes encoding the reported ATs (GPAT and DGAT1) in the Kennedy pathway, were analyzed for similarities of the amino acid sequences of their encoded proteins.
Protein sequence alignments were conducted by the ClustalW algorithm (Thompson et al., 1994) with the use of the residue substitution matrix (blosum62mt2) of the AlignX application of Vector NTI Suite (InforMax, North Bethesda, MD). A phylogenetic tree of the aligned sequences was built with the use of the neighbor-joining method (Saitou and Nei, 1987). Bootstraps analyses were conducted with 1,000 replicates for the 30 LPAAT-like sequences.
The software programs TargetP (http://www.cbs.dtu.dk/services/TargetP; Emanuelsson et al., 2000) and PSORT (http://psort.ims.u-tokyo.ac.jp; Klein et al., 1985) were used to predict subcellular targeting motifs and to detect potential trans-membrane segments in protein sequences.
RNA and DNA Analyses
For gene expression studies and cDNA cloning, total RNAs were isolated from various organs by a phenol/SDS method (Verwoerd et al., 1989). Genomic DNAs were isolated from leaves with use of a Quick DNA Preparation procedure (http://www.dartmouth.edu/~tjack/TAILDNAprep.html).
For RT-PCR analysis, total RNA (1.5 μg) of each sample was treated with 30 units of RNase-free DNase I and then used to synthesize a first strand cDNA with SuperscriptII reverse transcriptase and oligo(dT)15 primer. The resulting cDNA was used as a template in the presence of a pair of genespecific primers for PCR amplification (Fig. 5). For LPAAT1, the primers represented sequences of the opposite ends of the ORF, 5′-ATGGATGTCGCTTCTGCTCG-3′ (P1) and 5′-TTAGAGATCCATTGATTCTGCAA-3′ (P2). For LPAAT2, the primers represented sequences close to the mid portion of the ORF and the 3′-untranslated region, 5′:-GCGTACTAACTCTTGGAGCAA-3′ and 5′-CAAAACTGACACGCGCTTCTT-3′, respectively. Primers for ACTIN gene (GenBank accession no. U37281) were those described earlier (Kim et al., 2002).
For PCR analysis of lpaat1, the primers were P1 and LBa1, which represents the left border of the T-DNA, 5′-TGGTTCACGTAGTGGGCCATCG-3′. The primers P1 and P2 were used for LPAAT1.
A cDNA containing the full-length ORF (1,071 bp) of LPAAT1 produced by RT-PCR using P1 and P2 as primers was cloned into pGEM-T vector, and its sequence was confirmed.
PCR amplification was performed for 30 or 40 cycles (for embryo tissue) of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. PCR fragments were analyzed by 1.2% (w/v) agarose gel electrophoresis.
Expression and Complementation of LPAAT1 in Escherichia coli JC201
An 884-bp fragment of the LPAAT1, termed LPAAT1(234), was obtained after digestion of the full-length cDNA with SacI/SacII. LPAAT1(234) encoded a polypeptide of 234 residues instead of the full-length 356 residues (Fig. 3); this polypeptide is most similar to the E. coli LPAAT in length and sequence. It was inserted into the pBluescript SK+ multicloning site. pSK-LPAAT1(234) and the control pSK were each transformed into JC201 (mutant defective in LPAAT; Coleman, 1990). Bacteria were grown on Luria-Bertani medium containing ampicilin (50 μg mL-1) and streptomycin (100 μg mL-1) at 30°C. In the test for functional complementation, bacteria were grown on Luria-Bertani agar at 30°C and 42°C.
The bacterial liquid cultures were used to obtain a total extract and a membrane fraction. A culture of 100 mL of JC201 cell harboring pSK-LPAAT1(234) or pSK was grown for 16 h at 30°C. Cells were pelleted by low-speed centrifugation, and the pellet was resuspended in 4 mL of 50 mm Tris-HCl (pH 8.0), 2 mm MgCl2, and 2 mm dithiothreitol. They were disrupted by sonication with a 40T probe in a Braun-Sonic 2000 ultrasonic generator (Freeport, IL) with a digital meter reading of 200. The total extract was centrifuged at 10,000g for 15 min at 4°C to remove unbroken bacteria and debris. The supernatant was centrifuged at 100,000g for 1.5 h at 4°C. The pellet containing the membranes was resuspended in 1 mL of the above buffer, and the resuspension in 50-μl aliquots was stored at -80°C.
The total extract and the membrane fraction were subjected to analysis of protein constituents by SDS-PAGE (Kim et al., 2002). The membrane fraction was also used to assay for LPAAT activity.
Construction of 35S:LPAAT1 and 35S:LPAAT1(-TP) and Their Transformation into Heterozygous (LPAAT1/lpaat1) Plants
Two expression constructs, pCL0011-35S:LPAAT1 (35S CaMV promoter plus full-length LPAAT1 encoding the putative N-terminal plastid transit peptide and the mature protein) and pCL0011-35S:LPAAT1(-TP) (truncated LPAAT1 encoding only the mature protein), were made. LPAAT1 cDNA (described above) was used as a template to produce RT-PCR fragments of modified LPAAT1 and LPAAT1(-TP) containing the appropriate restriction sites at the fragment ends. For modified LPAAT1, the primers BamHI-P1 (5′-GGATCCATTATGGATGTCGCTTCTGCTCGGAGC-3) and XbaI-P2 (5′-TCTAGAGATTTAGAGATCCATTGATTCTGCAAT-3′) were used. For modified LPAAT1(-TP), the primers BamHI-LP5-2 (5′-GATCCTTTATGGGCGAAACAAGACTGACTGGC-3′) and XbaI-P2 were used. Each of the two modified cDNA was inserted at the BamHI-XbaI sites of pCL0011, which was adapted from pCAMBIA3300 (http://www.cambia.org) to contain the 35S CaMV promoter for driving a foreign gene and a bar selection marker gene. The plasmid was transformed into heterozygous (LPAAT1/lpaat1) plants by the floral dip method with the use of Agrobacterium tumefaciens strain GV3101. Plants resistant to BASTA spray were examined by PCR for genotype and phenotype of seed appearance under a microscope. Native LPAAT1 and lpaat1 were detected as described in a preceding section. For detecting 35S:LPAAT1 or 35S:LPAAT1(-TP), the 5′primer (P3), 5′-GGGTAATATCCGGAAACCTCCTCGGAT-3′, representing a segment of the 35S, and the 3′ primer (P2), representing a segment of the LPAAT1 ORF, were used (Fig. 3).
LPAAT Enzyme Activity Assay
The activity was assayed according to a procedure established in our laboratory (Cao et al., 1990). The reaction mixture contained 50 mm Tris-HCl (pH 8.0), 1 mm MgCl2, 1 mm dithiothreitol, 20 μm 1-oleoyl-LPA, 20 μm [1-C14]acyl-CoA (either 18:1 or 16:0), and 30 μg of bacterial membrane proteins. The mixture was incubated for 2 and 4 min at 30°C, and the reaction was terminated with the use of chloroform and methanol. Lipids in the extract were subjected to thin-layer chromatography with the use of acetone:toluene:water (91:30:8 [v/v]). The PA spot was visualized by autoradiography, and the silica gel was scraped for scintillation counting. The proteins in the bacterial membranes were quantified by the Bradford method (Smith et al., 1985). The use of acyl CoA instead of acyl ACP followed a common practice in the studies of plastid ATs because of the unavailability of commercial radioactive acyl ACP. Apparently, the use of ACP and CoA derivatives can be interchanged in in vitro assays, and the results are consistent with their projected catalytic mechanism in vivo, such as acyl specificity.
Lipid Analysis
Rosette leaves were immediately frozen in liquid nitrogen after harvest, and the lipids were extracted according to the protocol of the Lipodomics Center at Kansas State University (http://www.ksu.edu/lipid/lipidomics/leaf-extraction.html). The acyl moieties of the lipids were saponified with alkaline in ethanol, and the acidified samples of free fatty acids were derivatized to methyl esters with boron trifluoride in methanol, which were subjected to gas-liquid chromatography analysis. Younger and older leaves had no substantial differences in their fatty acid compositions.
Microscopy
Developing seeds and embryos were viewed under a stereomicroscope (LEICAMZ125, Leica Microsystems, Wetzlar, Germany) or a light microscope (Nikon MICROPHOT-FXA, Nikon, Tokyo) attached to a spot digital camera. Seeds were dissected, and the removed embryos were placed in water and photographed similarly.
Acknowledgments
We sincerely thank Dr. Jack Coleman for the E. coli JC201 strain, Dr. Joseph Ecker of the Salk Institute (La Jolla, CA) and the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the T-DNA-inserted LPAAT1 Arabidopsis mutant, and the Lipidomics Center (Kansas State University, Manhattan) for lipid analysis.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.035832.
This work was supported by the National Science Foundation (grant no. MCB–0131358) and by the U.S. Department of Agriculture (National Research Initiative Competitive Grant no. 2000–01512).
References
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen J, Paddock T, Salas J, Savage L, Milcamps A et al. (2003) Arabidopsis thaliana genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of ESTs in organs and a Web-based database. Plant Physiol 132: 681-697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kader JC, Barret P, Renard M, Robinson D, Robinson C, Delseny M, Roscoe TJ (1999) A plastidal lysophosphatidic acid acyltransferase from oilseed rape. Plant Physiol 120: 913-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Nave P, Benveniste P, Oelkers P, Sturley SL, Schaller H (2000) Expression in yeast and tobacco of plant cDNAs encoding acyl CoA: diacylglycerol acyltransferase. Eur J Biochem 267: 85-96 [DOI] [PubMed] [Google Scholar]
- Brown AP, Brough CL, Kroon JTM, Slabas AR (1995) Identification of a cDNA that encodes a 1-acyl-sn-glycerol-3-phosphate acyltransferase from Limnanthes douglasii. Plant Mol Biol 29: 267-278 [DOI] [PubMed] [Google Scholar]
- Brown AP, Coleman J, Tommey AM, Watson MD, Slabas AR (1994) Isolation and characterization of a maize cDNA that complements a 1-acyl-sn-glycerol-3-phosphate acyltaransferase mutant of Escherichia coli and encodes a protein which similarities to other acyltransferases. Plant Mol Biol 26: 211-223 [DOI] [PubMed] [Google Scholar]
- Cao YZ, Oo KC, Huang AHC (1990) Lysophosphatidate acyltransferase in the microsomes from maturing seeds of meadowfoam (Limnanthes alba). Plant Physiol 94: 1199-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J (1990) Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J Biol Chem 265: 17215-17221 [PubMed] [Google Scholar]
- Coleman J (1992) Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC). Mol Gen Genet 232: 295-303 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Heijne Gv (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005-1016 [DOI] [PubMed] [Google Scholar]
- Frentzen M (1998) Acyltransferases from basic science to modified seed oils. Lipid-Fett 100: 161-166 [Google Scholar]
- Hanke C, Wolter FP, Coleman J, Peterek G, Frentzen M (1995) A plant acyltransferase involved in triacylglycerols biosynthesis complements an Escherichia coli sn-1-acylglycerol-3-phosphate acyltransferase mutant. Euro J Biochem 232: 806-810 [PubMed] [Google Scholar]
- Heath R, Rock CO (1998) A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol 180: 1425-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki O, Nishida I, Agata K, Eguchi G, Murata N (1988) Cloning and nucleotide sequence of cDNA for the plastid glycerol-3-phosphate acyltransferase from squash. FEBS Lett 238: 424-430 [DOI] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AHC (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277: 22677-22684 [DOI] [PubMed] [Google Scholar]
- Klein P, Kanehisa M, DeLisi C (1985) The detection and classification of membrane-spanning proteins. Biochim Biophys Acta 28: 468-476 [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Lardizabal KD, Nelsen JS, Bleibaum JL, Davies HM, Metz JC (1995) Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-glycerol-3-phosphate acyltransferase that accepts medium-chain-length substrates. Plant Physiol 109: 999-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst LJ, Browse J, Somerville C (1988) Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc Natl Acad Sci USA 85: 4143-4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassner MW, Levering CK, Davies HM, Knutzon DS (1995) Lysophosphatidic acid acyltransferase from meadowfoam mediates insertion of erucic acid at the sn-2position of triacylglycerols in transgenic rapeseed oil. Plant Physiol 109: 1389-1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurant P, Huang AHC (1992) Organ and development specific acyl CoA lysophosphatidate acyltransferase in palm and meadowfoam. Plant Physiol 99: 1711-1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin TM, Wang P, Coleman RA (1999) Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38: 5764-5771 [DOI] [PubMed] [Google Scholar]
- Maisonneuve S, Guyot R, Delseny M, Roscoe T (2003) A multigene family of lysophosphatidic acid transferases of Arabidopsis thaliana. In N Murata, M Yamada, I Nishida, H Okuyama, J Sekiya, W Hajime, eds, Advanced Research on Plant Lipids. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 183-186
- Mansfield SG, Briarty LG (1991) Early embryogenesis in Arabidopsis thaliana: II. The developing embryo. Can J Bot 69: 461-476 [Google Scholar]
- Murata N, Tasaka Y (1997) Glycerol-3-phosphate acyltransferase in plants. Biochim Biophys Acta 1348: 10-16 [DOI] [PubMed] [Google Scholar]
- Nishida I, Tasaka Y, Shiraishi H, Murata N (1993) The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate acyltransferase of Arabidopsis thaliana. Plant Mol Biol 21: 267-277 [DOI] [PubMed] [Google Scholar]
- Routaboul J-M, Benning C, Bechtold N, Caboche M, Lepiniec L (1999) The TAG1 locus of Arabidopsis encodes for a diacylglycerols acyltransferase. Plant Physiol Biochem 37: 831-840 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425 [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Malia AK, Gartner FH, Proverzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76-85 [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J, Jaworski JG, Ohlrogge JB (2000) Lipids. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 456-527
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52: 335-361 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872-1887 [DOI] [PMC free article] [PubMed] [Google Scholar]