Abstract
The biological actions of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) serve both to orchestrate calcium and phosphorus homeostasis in higher vertebrates and to regulate a diverse set of cellular functions unrelated to control of mineral metabolism. With regard to bone, mesenchymal lineage cells, including both early and late osteoblasts as well as osteocytes represent classic targets of the vitamin D hormone. Accordingly, much of the early information regarding our current understanding of the mechanism of action of 1,25(OH)2D3, of which gene regulation is central, derives from a broad array of studies in these cell types. Indeed, a gene that provided both the earliest and perhaps the most extensive information regarding this and additional mechanisms was that of osteoblast-specific osteocalcin. Subsequent work has provided much additional detail as to how 1,25(OH)2D3, through the vitamin D receptor (VDR), mediates the modulation of many bone cell genes. In recent years, however, a series of technical advances involving the coupling of chromatin immunoprecipitation (ChIP) to unbiased methodologies that involve next-generation DNA sequencing techniques (ChIP-seq) have opened new avenues in the study of gene regulation. In this review, we summarize early work and then focus on more recent studies that have used ChIP-seq analysis and other approaches to provide insight into not only the regulation of specific genes such as the VDR, TNFSF11 (RANKL), LRP5, CBS and CYP24a1, but overarching genome-wide principles of gene regulation as well. The results of these studies highlight the value of these new approaches and the increased insight that can be gained.
Introduction
Early studies of the biological activities of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) in the intestine, kidney and bone revealed that this hormone likely functioned following nuclear localization by regulating the expression of genes. This hypothesis was strengthened when it was discovered that 1,25(OH)2D3 interacted selectively with an intestinal binding protein, later termed the vitamin D receptor (VDR), and that this interaction prompted translocation of the protein to the nucleus.1,2,3 Subsequent studies demonstrated that the VDR could be found not only in the intestine, but also in the parathyroid gland,3 kidney4 and bone,5 and later in specific cell types in many if not most bodily tissues.6,7,8 Further characterization of the VDR as a DNA-binding protein supported its role as the primary mediator of vitamin D action in all tissues.9 This idea was eventually confirmed when the VDR was cloned and shown to belong to the nuclear receptor family of transcription factors10,11,12 and through the discovery that mutations in the VDR led to the human syndrome of hereditary 1,25(OH)2D3-resistant rickets,13,14 a phenotype replicated through genetic deletion of the VDR in mice.15,16 Interestingly, although the initial actions of vitamin D were found in the intestine, it was in bone cells and on bone cell-specific genes that the most striking molecular advances have been made.
Mechanistically, 1,25(OH)2D3 prompts VDR interaction directly with DNA sequences located within regulatory regions of target genes. This interaction requires heterodimer formation with either related steroid receptor family members RXRα or β17,18 and occurs with high affinity on selected sequences comprising two directly repeated AGGTCA hexameric half-sites separated generally by 3 bp.19,20,21 The formation of this protein–DNA complex, and in particular the exposure of unique binding sites on the surface of both the VDR and RXR, provides a platform for the recruitment of additional multi-protein co-regulatory complexes, a structure that has now been deduced from three-dimensional analysis.22 These complexes function directly through various epigenetic mechanisms to modulate regions of the chromatin environment surrounding the promoter of a target gene, to regulate the essential recruitment of RNA polymerase II and likely through additional mechanisms as well.23 Osteocalcin was the first gene for which a vitamin D response element or VDRE was identified,19,24 a finding followed thereafter by the discovery of a VDRE located in the SPP1 (osteopontin) gene20 and subsequently in the CYP24A1 gene.25,26,27 Although VDR/RXR DNA-binding sites that diverge from this overall organizational motif have emerged from time to time,28 the discovery of an osteocalcin/osteopontin type VDRE sequence in many subsequent genes firmly established this motif as a classic VDR/RXR-binding site for genes that are induced by 1,25(OH)2D3. Many additional features and details of this overall mechanism have been identified. Together, they have firmly established the molecular fundamentals of 1,25(OH)2D3 action at target genes, an overall process that is not unlike that of most steroid hormones and indeed most other transcription factors as well.
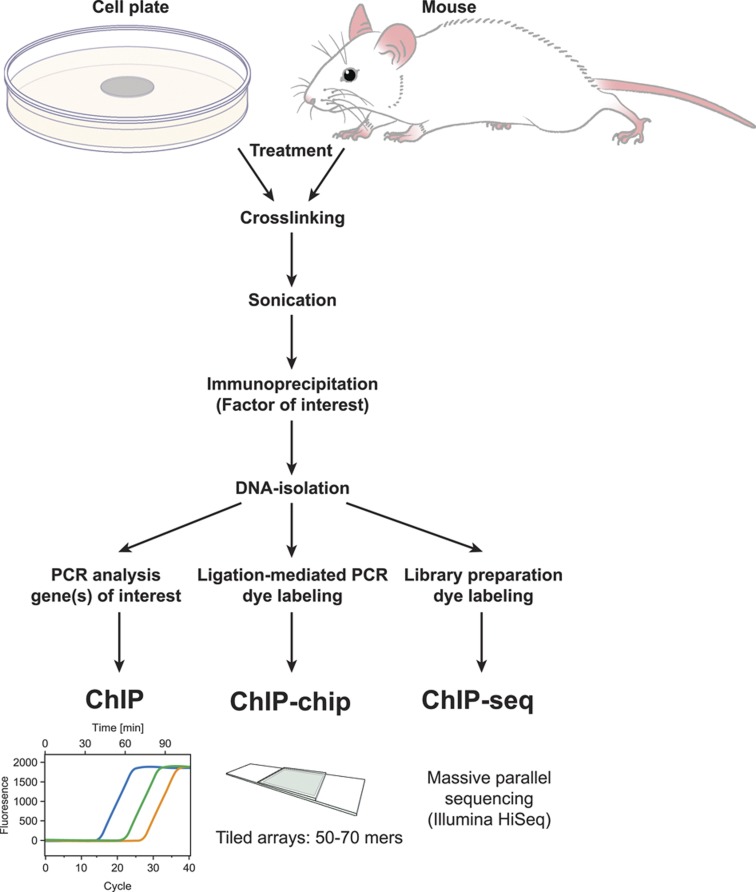
The development of chromatin immunoprecipitation (ChIP) methods coupled first to site-specific PCR analysis and shortly thereafter to methods capable of detecting in an unbiased manner the abundance of immunoprecipitated DNA segments on a genome-wide basis (tiled microarrays (ChIP-chip) and then massively parallel DNA sequencing (ChIP-seq)) are changing traditional molecular biological approaches to the study of transcriptional regulation (Figure 1). The primary advantage of the current ChIP-seq approach is the facile ability to detect proteins at endogenous sites on the genome in a largely unbiased and genome-wide manner. This approach has been exploited extensively over the past decade by many investigators, including those belonging to the ENCODE Consortium to not only explore specific pathways of gene regulation, but to also provide new annotation to the genome.29 A specific area of progress has also been a significant advance in our understanding of the genetics and epigenetics of cellular differentiation and reprogramming.30,31 In this review, we document advances that have been made using ChIP-chip and ChIP-seq approaches in understanding the regulation of bone cell gene expression by 1,25(OH)2D3.
Figure 1.
Site-specific and genome-wide methodologies associated with chromatin immunoprecipitation (ChIP) methods. ChIP-chip, ChIP linked to tiled microarray analysis; ChIP-seq, ChIP linked to DNA deep sequencing methods.
Observations
Although early mechanistic studies of the regulation of gene expression by 1,25(OH)2D3 defined sites of VDR/RXR interaction on genes such as osteocalcin and osteopontin, regulatory sites could not be detected on many genes whose RNAs were known to be regulated by the vitamin D hormone. They include the VDR gene itself as well as genes for receptor activator of NF-κB ligand (RANKL), low-density lipoprotein receptor-related protein 5 (LRP5), cystathionine β-synthase (CBS) and a number of others as well. Indeed, this feature still categorizes the majority of targets for 1,25(OH)2D3. The absence of sites suggests either that alternative mechanisms are at play or that the regions that control the expression of these genes are located outside those being explored. Although both are possible, the latter seemed the most likely as traditional methods are highly biased and limit investigative focus almost exclusively to regions near the transcriptional start sites (TSSs) of candidate genes. To begin to explore this question, we applied the unbiased technique of ChIP-chip analysis (confirmed via ChIP-seq analysis) to the study of 1,25(OH)2D3's regulation of the genes mentioned above. We also re-examined the CYP24A1 gene to determine if the regulatory mechanism previously defined for this gene was complete. In addition to these specific genomic loci, ChIP-seq analysis is inherently genome-wide, therefore we will also discuss the principles of 1,25(OH)2D3 regulation of the entire transcriptome in osteoblasts and other cell types.
The regulation of specific genes
Specific targets
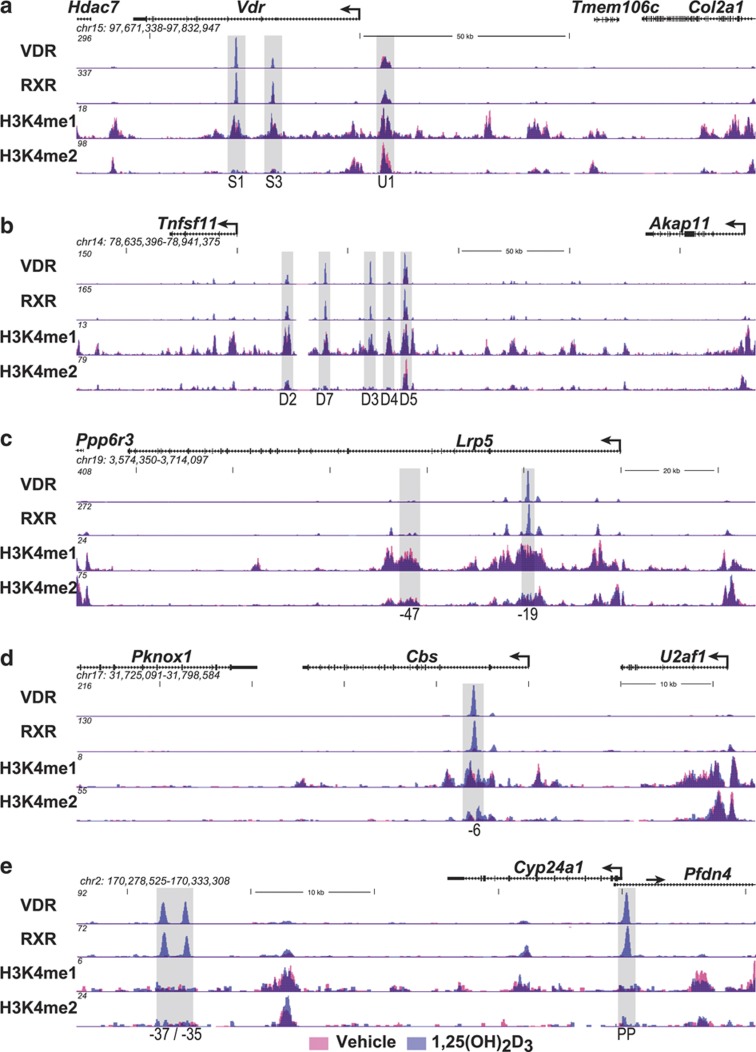
The VDR gene. The VDR gene is highly expressed in both early and late osteoblasts and terminally differentiated osteocytes32 and is believed to be synthesized in osteoclasts33 as well. Through the VDR, 1,25(OH)2D3 regulates a wide variety of genes which underlie both catabolic and anabolic cellular characteristics. Studies two decades earlier suggested that the VDR gene was transcriptionally regulated by several hormones including the glucocorticoids, estrogens and retinoic acid, and might be autoregulated by 1,25(OH)2D3 as well.34 More recent studies in vivo suggest that 1,25(OH)2D3 upregulates the VDR almost exclusively in bone cells (Meyer and Pike, unpublished). Despite this compelling evidence, studies using traditional approaches failed to identify binding sites for the VDR at regions surrounding the gene's TSS either in vitro or in vivo. As the mouse Vdr gene, like that of the human gene13,35 is complex, spanning some 54 kilobases (kb) on chromosome 15 (Figure 2a), we conducted in osteoblasts an unbiased ChIP-chip analysis across the Vdr gene locus that spanned several hundred kilobases upstream as well as downstream of the Vdr transcriptional unit.36,37 These initial studies as well as an additional unpublished investigation using ChIP-seq analysis (see Figure 2a) revealed the 1,25(OH)2D3-induced presence of both the VDR and RXR at two separate intronic sites approximately +19 and +29-kb downstream and at one intergenic site −6-kb upstream of the TSS. This group of regulatory sites was generally conserved in both the mouse and the human genes and their properties extensively characterized including identification of the VDREs contained within each region.36 Interestingly, retinoic acid receptor binding was induced by retinoic acid at the intronic +19-kb site while cAMP response element binding protein (CREB) binding was induced by protein kinase A activators (including PTH) at the upstream −6-kb site.37 A number of additional transcription factors including both runt-related transcription factor 2 (RUNX2) and CCAAT/enhancer-binding protein β (C/EBPβ) were also found at subsets of these sites as well. These results provided the first evidence that VDR-binding sites in genes could be located at remote sites distal to gene promoters and that multiple regions with unique regulatory properties could participate.
Figure 2.
ChIP-seq profiles at specific gene loci. Mouse MC3T3-E1 cells were treated for 3 h with either vehicle or 1,25(OH)2D3 (10−7 M) and then subjected to ChIP-seq analysis using antibodies to VDR, RXR, histone H3K4me1 or H3K4me2. ChIP-seq tag densities (normalized to 107 reads) were quantified and mapped to the mouse MM9 genome using MACS, HOMER and cistrome data analyses. The genomic loci (chromosome number and nucleotide interval are indicated) contain the Vdr (a), Tnfsf11 (b), Lrp5 (c), Cbs (d) and Cyp24a1 (e) genes and their respective neighbors. Each data track (scales are indicated on the Y axis) contains two mapped data sets derived from vehicle- and 1,25(OH)2D3-treated cells (red and blue, respectively). The TSS and direction of transcription for each gene is indicated by an arrow; exons are indicated by boxes. Shaded vertical columns highlight the locations and number/letter designation for each of the regulatory regions of the target genes. 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; CBS, cystathionine β-synthase; ChIP, chromatin immunoprecipitation; HOMER, hypergeometric optimization of motif enrichment; LPR5, lipoprotein receptor-related protein 5; MACS, model-based analysis for ChIP-seq; TSS, transcriptional start site; VDR, vitamin D receptor; PP, promoter proximal.
The TNFSF11 (RANKL) gene. Early studies by Suda et al.38 suggested that a factor derived from osteoblast lineage cells regulated osteoclast differentiation. Importantly, the expression of this factor was controlled by 1,25(OH)2D3 as well as other calciotropic agents such as PTH and the inflammatory cytokines. This factor was identified and cloned in 1999 and designated RANKL.39,40 RANKL is a membrane bound and sometimes soluble tumor necrosis factor (TNF)-like factor derived from the TNFSF11 gene that strongly induces osteoclast differentiation from hematopoietic precursors. Indeed, the signaling pathway that mediates activation of this complex differentiation pathway is now well described.41 Despite considerable effort, however, attempts using traditional methods to identify regions mediating the regulation of RANKL by 1,25(OH)2D3 as well as PTH and cytokines such as interleukin-6 and oncostatin M were largely unsuccessful. As the RANKL gene is located in a gene desert and bounded by intergenic segments of nearly 200 kb of DNA (Figure 2b), we explored this gene in osteoblasts for regulation by the vitamin D hormone by conducting a ChIP-chip analysis for both VDR and RXR spanning over 500 kb of DNA surrounding the mouse Tnfsf11 transcription unit.42 This initial study, confirmed through subsequent studies using ChIP-seq analysis (Figure 2b), was highly revealing. Although neither VDR nor RXR were present near the promoter region following administration of 1,25(OH)2D3, both were strikingly present at −16 (D1), −22 (D2), −60 (D3), −69 (D4) and −75 (D5) kb upstream of the Tnfsf11 gene promoter. Each of these regions, as well as a newly defined site at −40 kb (D7), was extensively characterized and the VDR-binding sites that mediated vitamin D hormone action were identified in a subset. Similar ChIP-chip studies of PTH action on RANKL expression through CREB were also carried out.43 As with the VDR, these studies revealed that PTH induced CREB binding at several but not all of these regulatory regions. Interestingly, O'Brien and Co-workers44 also identified one these upstream regions using an entirely independent approach. Importantly, this region was deleted in the mouse genome and shown to affect both the basal expression of RANKL at specific skeletal sites and to ameliorate but not eliminate overall response to both 1,25(OH)2D3 and PTH.45 We also showed that cytokine induction, on the other hand, was mediated through signal transducer and activator of transcription 3 induction at a novel region located at −88-kb upstream.46 Residual binding of both RUNX2 and C/EBPβ and other transcription factors were also observed.47 These studies provide a conclusion similar to that observed in the VDR gene; 1,25(OH)2D3 regulation of gene expression occurs through multiple regulatory regions that can be located many kilobases distal to the transcriptional start sites of genes and are frequently modular in their actions.
The LRP5 gene. The canonical Wnt signaling pathway has a crucial role in osteoblast proliferation and differentiation, and thus appears to exert a significant anabolic role in bone formation.48 It also has a central role in transducing mechanosensory inputs via the osteocyte.49,50 Wnt activity is mediated through an interaction between specific Wnt family members and the cell surface receptor Frizzled. This interaction triggers downstream events that lead ultimately to stabilization and nuclear translocation of β-catenin, a dominant coactivator of the transcription factor 7-like 2/lymphoid enhancer-binding factor 1 family of transcription factors often prebound at target genes. LRP5 and LRP6 serve as coreceptors of Frizzled, facilitating positively the process of β-catenin activation. That LRP5 and LRP6 represent anabolic potentiators of Wnt activation was strengthened initially by the observation that hypermorphic alleles of LRP5 were associated with high bone mass in humans.51,52 As preliminary studies in osteoblasts revealed that 1,25(OH)2D3 prompted an upregulation of LRP5 mRNA, we explored the mechanism through which this hormone might operate using an initial ChIP-chip analysis of VDR/RXR spanning the mouse Lrp5 locus similar to that conduced for the Vdr and Tnfsf11 genes.53 These studies revealed the presence of three potential regulatory regions, one located immediately downstream of the Lrp5 TSS and two additional regions also located within separate introns at +19 and +47 kb (see Figure 2c for updated ChIP-seq data). Further characterization of these regions confirmed the ability of the regions at +19 kb to mediate 1,25(OH)2D3 activity; additional unpublished studies revealed similar binding sites for the VDR near the Lrp5 promoter. These studies similarly demonstrate the enhanced frequency with which regulatory regions are located distal to gene promoters, although in this case two of these regions were accompanied by a regulatory region that was indeed located near the TSS. Perhaps more importantly, these studies provide support for the hypothesis that the actions of 1,25(OH)2D3 in bone cells may not be exclusively catabolic.
The CBS gene. Osteoporotic fracture risk is elevated during the aging process because of a multiplicity of factors that may include lifestyle, nutrition, and hormonal and genetic factors.54 One additional factor may be a raise in circulating levels of the sulfur-containing amino-acid homocysteine.55 Indeed, this increase in homocysteine can give rise to hyperhomocysteinemia, which negatively impacts collagen cross-linking and maturation thus affecting bone quality, bone strength and promotes bone resorption.56,57 A key step in the elimination of homocysteine is the condensation of this amino acid with serine, a reaction that is catalyzed by the enzyme cystathionine β-synthase and which culminates via several additional steps in the synthesis of cysteine.58 The importance of this enzyme in bone is highlighted by the observation that mice genetically deficient in CBS exhibit severe hyperhomocysteinemia, which is accompanied by elevated reactive oxygen species levels and a progressively aberrant skeletal phenotype.59,60 Initial studies in osteoblasts revealed that 1,25(OH)2D3 induced the synthesis of CBS mRNA, prompting further examination of the mechanism through which this induction might occur. We therefore assessed VDR and RXR binding across the CBS gene locus using ChIP-chip analysis.61 These and additional ChIP-seq analyses (Figure 2d) revealed the presence of a single potential regulatory region in the CBS gene located in the second intron approximately +6-kb downstream of the TSS. Examination of the DNA sequence within this downstream region led to the identification of a classic VDRE comprised of two hexameric half-sites separated by 3 bp. Further study revealed that this element was indeed capable of mediating 1,25(OH)2D3 response. These results provide additional support for the utility of this unbiased analysis in rapidly identifying regulatory sites of vitamin D hormone action. They also suggest that in addition to both anabolic and catabolic actions, 1,25(OH)2D3 is also capable of modulating internal pathways of amino-acid synthesis that are positive for bone structure and function.
The CYP24A1 gene. CYP24A1 encodes the 1,25-dihydroxyvitamin D3-24 hydroxylase, a p450 enzyme that catalyzes the initial steps that results in the inactivation of 1,25(OH)2D3.62 Accordingly, this enzyme represents a primary set point determinant of both the level and duration of response to 1,25(OH)2D3 that is achievable within all vitamin D target cells both in vitro and in vivo. Thus, global genetic deletion of CYP24A1 expression in mice or the production of mutant hydroxylases as observed in young children with idiopathic familial hypercalcemia results in exaggerated levels of 1,25(OH)2D3 that potentiate vitamin D hormone activity and can cause hypercalcemia.63 As might be anticipated, CYP24A1 expression is strongly regulated by 1,25(OH)2D3. Thus, traditional studies of this gene in 1993 quickly revealed the present of two highly conserved VDREs located within the first 300 bp of both the mouse and human homologs that were capable of binding both VDR and RXR and mediating strong induction by 1,25(OH)2D3.25,26,27 The presence of these two sites of 1,25(OH)2D3 action were confirmed following extensive ChIP-chip and ChIP-seq analyses of VDR and RXR binding in mouse bone cells and in many other cell types as well64 (Figure 2e). Interestingly, inspection of both VDR and RXR-binding profiles across the mouse Cyp24a1 locus following these analyses also revealed the presence of a cassette containing a cluster of intergenic VDR/RXR-binding sites spanning +35 to +37 kb in the mouse (Figure 2e) and +50 to +66 kb in the human genomes.64,65,66 Subsequently, detailed studies not only identified several of the multiple VDREs that were contained within these regulatory regions but linked their activity directly to enhancement of the Cyp24a1 expression by 1,25(OH)2D3. Thus, these unbiased approaches are capable of not only confirming the presence of previously characterized regulatory regions, but identifying new sites as well. It is reasonable to conclude that the traditional methods used to study the regulation of many genes has underestimated both in number and complexity the components of regulation regions actually present in many genes, and that the application of unbiased methods described above will be required to more fully understand the regulation of these genes by 1,25(OH)2D3 and other factors.
Derived principles of vitamin D hormone action
The targeted ChIP-chip and subsequent ChIP-seq analyses of the above candidate genes provide strong validation for many of the principles of gene regulation established for 1,25(OH)2D3 over the past several decades using more traditional methods of experimentation.65,66 Indeed, they reinforce, in part, the concepts that (1) VDR DNA binding involves an RXR partner, (2) classic binding sites are comprised of two directly repeated hexanucleotide half-sites separated by 3 bp, (3) VDRE-containing regulatory regions are modular, that is, contain adjacent binding sites for additional transcription factors and (4) diverse transcription factor occupancy at these regulatory regions correlates directly with the upregulation of the genes to which the corresponding cis elements are linked. Importantly, however, these data and additional studies67,68,69 also point to several new principles of gene regulation by 1,25(OH)2D3 as well including the idea that regulatory regions of genes may be less frequently located near promoters than within introns or within intergenic regions often 10's if not 100's of kilobases distal to transcriptional start sites and furthermore that many such regions are generally involved. These approaches have also illuminated an additional concept, that is, that regulatory regions of gene (enhancers) are strongly highlighted by a number of specific epigenetic histone marks, most predominant being histone H3K4 mono- and dimethylation (H3K4me1 and H3K4me2, respectively) and H3K27 acetylation (H3K27ac) (see Figures 2a–e for H3K4me1 and H3K4me2 marks at Vdr, Tnfsf11, Lrp5, Cbs and Cyp24a1).70,71,72 Unique histone modifications also mark other features of genomic loci as well. These dynamic regulatory marks are imposed upon the genome by a diverse set of chromatin regulatory factors.73 This epigenetic histone code for repeating features of the genome was identified and is supported by studies conducted through the ENCODE Consortium as well as other investigative groups.70 Importantly, features of this code make it possible to localize regulatory regions of many genes without knowing precisely which transcription factors might participate in their expression.
Genome-wide principles of gene regulation by 1,25(OH)2D3
As discussed above, ChIP-chip and now predominantly ChIP-seq analyses provide regulatory data for a diverse set of transcription factors on a genome-wide scale. This capability is particularly powerful because it can define thousands of sites of action for a specific factor, be used to understand the regulation of gene networks and enable the development of overarching principles of gene regulation.70 It has also facilitated deep exploration of individual components involved in genome structure and function as well as those that predispose to disease. Of particular importance, this approach can be applied equally to studies conducted both in vitro and in vivo, permitting detailed molecular studies in animals that have not heretofore been possible. Along with others, we have conducted genome-wide studies of VDR/RXR binding in a variety of cell types. The following summary of these studies reveal a series of overarching principles that both confirm and extend those that were highlighted by the candidate genes described above.
Identification and quantitation of the VDR cistrome in osteoblasts and colorectal cancer cells
Genome-wide analysis facilitates the identification and quantitation of all binding sites for a particular factor or feature on the genome of a specific target cell (termed a cistrome); it also facilitates detection of changes in that cistrome, which might occur as a function of an alteration in the cellular environment.74,75 Accordingly, we conducted a genome-wide ChIP-chip analysis as well as subsequent ChIP-seq analyses of the VDR in osteoblasts and the colorectal cancer cell line LS180 in the absence and presence of 1,25(OH)2D3, quantitating the number of binding sites identified on the genome and noting their relative locations.65,66 In both cell types, VDR binding was low in the absence and strongly elevated in the presence of 1,25(OH)2D3, reaching a limit in the presence of the hormone ranging from 2000 to 8000 sites per genome depending upon cell type. We also explored the locations of these individual binding sites, and, as seen for the five candidate genes described earlier, only a small percent were associated with transcriptional start sites; the vast majority (95%) was found either in introns or within intergenic regions adjacent to neighboring genes, often at significant linear distances. Both number and location were cell-type specific, a discovery not surprising in view of the fact that the regulation of genes by the vitamin D hormone is cell specific in nature.65,66,67 Finally, de novo motif analysis revealed the frequent presence of motifs comprised of two directly repeated half-sites separated by 3 bp. Thus, on virtually thousands of regions across numerous cell types, VDR binding to DNA likely involves classic VDREs. The cell-specific nature, locations and presence of VDREs confirm properties that were observed in the candidate genes described above.
Features of the VDR/RXR cistrome
Genome-wide analysis revealed numerous features of VDR-binding sites in additional to those above.65,66 ChIP-chip/ChIP-seq analysis of RXR revealed that although the RXR cistromes tend to be less sensitive to 1,25(OH)2D3 and larger than those of the VDR, likely reflecting the fact that RXR serves a complex role as heterodimer partner to a number of other nuclear receptors, the vast majority of the thousand or more VDR-binding sites per genome identified contain RXR. Genome-wide data reveal that VDR DNA binding is strongly influenced by the presence of 1,25(OH)2D3, despite many examples where VDR is prebound to DNA in the absence of ligand and either unaffected or displaced in its presence. These and other experiments suggest that there may be several modes of VDR/RXR DNA binding. Whether these modes contrast mechanisms of gene activation vs suppression, dual roles that the vitamin D hormone performs equally well, are unknown. Indeed, where the mechanism of gene suppression by 1,25(OH)2D3 has been investigated, both DNA-binding-independent (that is, CYP27B1 and PTH)76,77 and DNA-binding-dependent78,79,80,81 mechanisms have been identified. It should be noted as indicated above, however, that ChIP-seq analysis will be necessary to confirm these latter DNA-binding-dependent series of findings. Finally, an extensive analysis of the presence of epigenetic histone marks suggests that VDR/RXR binding occurs in regions where histone 3 is mono- and di-methylated at K4 and where histones 3 and 4 are extensively acetylated on several lysine residues including H3K27 (Meyer and Pike, unpublished). Moreover, the presence of the VDR at many of these sites upregulates the level of histone acetylation.82 These findings support the overarching idea that on a genome-wide scale, like that seen for candidate genes, VDR/RXR-binding sites are located in regions of the genome that contain significant epigenetic enhancer features that provide strong regulatory potential. Overarching principles of 1,25(OH)2D3 regulation, as determined on a genome-wide scale, are summarized in Table 1.
Table 1. Overarching principles of vitamin D-mediated gene regulation.
| VDR-binding sites: 2000–8000 1,25(OH)2D3-sensitive binding sites/genome; cell-type specific. |
| Binding site locations: dispersed in CRMs (CRMs or enhancers) across the genome; located in a cell-type-specific manner near promoters, but predominantly within introns and distal intergenic regions; frequently located in clusters of elements. |
| Active transcription unit: the VDR/RXR heterodimer. |
| VDR/RXR-binding site sequence (VDRE): induction mediated by classic hexameric half-sites (AGGTCA) separated by 3 bp; repression mediated by divergent sites. |
| Mode of DNA binding: predominantly, but not exclusively, ligand dependent. |
| Modular features: CRMs contain binding sites for multiple transcription factors that facilitate both independent or synergistic interaction. |
| CRM signatures: defined by the dynamic presence of distinct epigenetic histone H3 and H4 modification. |
Abbreviations: CRM, cis-regulatory module; VDR, vitamin D receptor; VDRE, vitamin D response element; 1,25(OH)2D3, 1,25-dihydroxyvitamin D3.
Linking regulatory regions to the genes they modulate
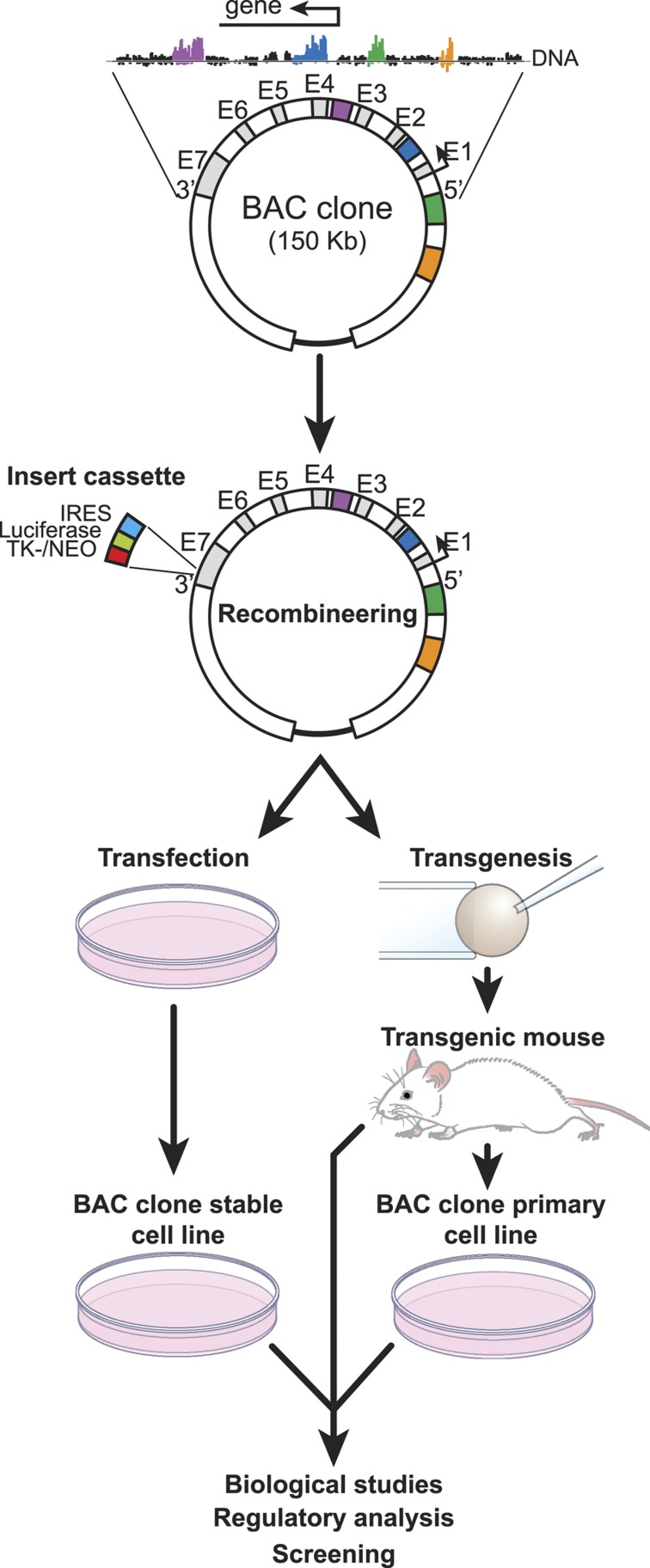
One consequence of the discovery that regulatory regions are frequently located large distances from the genes they control is that although their identification on the genome has become easier, identification of the genes they regulate has become much more difficult. The finding that chromatin structure is also exceptionally important also contributes to this difficulty. Although several ChIP-based approaches, including that of chromatin interaction analysis by paired-end tag sequencing, may ultimately provide detailed linkage at the genome-wide level, this highly bioinformatic-based linkage remains largely correlative.83,84 We have used several approaches to link the actions of regulatory regions directly to the genes they regulate. These include the use of bacterial artificial chromosomes, which contain a specific target gene surrounded by as much as 250 kb of endogenous linear DNA.37 The size of this segment of DNA frequently contains many candidate regulatory regions, which can be examined for their contribution to the internal gene of interest. A bacterial artificial chromosome clone can be recombineered (bacterial recombinant engineering) to contain a reporter function and then introduced in a stably integrated manner into the genomes of cells in culture and as transgenes into mice (Figure 3). Following characterization of the wild-type minigene in these backgrounds, enhancers can be removed and the consequence of this removal on activity assessed following reintroducing of the clone into the same cells or into the mouse genome. An additional approach has been to delete regulatory regions directly from the genome.45 Although this approach is similarly useful, it is both expensive and difficult to thoroughly investigate genes at the genomic level if they contain multiple regulatory regions. Recent advances in genome-editing methods likely will expedite this approach and facilitate simultaneous examination of more than one region.85 Along with others, we have used these methods to explore properties of both the VDR and TNFSF11 genes, studies that have been described in depth previously. Importantly, these studies confirm the functional role of many of the regulatory regions that were identified initially by ChIP-chip and ChIP-seq methodologies.
Figure 3.
Methodology associated with the preparation and use of bacterial artificial chromosomes (BACs). Chromatin immunoprecipitation (ChIP)-seq analysis is used to define the regulatory locus of a specific gene. A BAC clone containing this segment of DNA is modified through recombineering methods and then introduced via stable transfection into culture cells or through transgenic methods into mice. Orientation of the target transcription unit (5′ and 3′) together with representative exons (E) is indicated. The recombineering cassette contains an internal ribosome entry site (IRES)/luciferase and a thymidine kinase promoter-neomycin gene (TK-NEO) component. Direction of transcription is indicated by the arrow.
Conclusions
Early studies of a few selected target genes provided significant insight into how 1,25(OH)2D3 functions to regulate gene expression. The advent of ChIP, linked ultimately to unbiased methodologies that included the use of tiled microarrays and deep sequencing analysis, has revolutionized our approach to the study of gene regulation, and provided new insight into how 1,25(OH)2D3 as well as other systemic and local factors operate to control the expression of genes. These unbiased methods have revealed not only novel features of VDR and RXR binding, but novel features of the regions to which they bind and how these features, both genetic and epigenetic, contribute to the regulation of gene expression. As a side note, many of these more recent observations suggest that the more traditional molecular biological approaches used in the study of transcription over the past several decades can no longer be relied upon to provide a complete and representative picture of how 1,25(OH)2D3 functions. These findings do suggest, however, that much remains to be discovered in our ongoing efforts to understand how the vitamin D hormone functions to modulated the expression of genes and thus to direct the biological activities that are under its directive in vivo.
Acknowledgments
We thank members of the Pike Lab for their contributions to this review. This work was supported in part by research grants DK-72281, DK-73995 and DK-74993 from NIDDK and AR-45173 from NIAMS to JWP.
Footnotes
The authors declare no conflict of interest.
References
- Brumbaugh P, Haussler M. 1 Alpha,25-dihydroxycholecalciferol receptors in intestine. I. Association of 1 alpha,25-dihydroxycholecalciferol with intestinal mucosa chromatin. J Biol Chem 1974;249:1251–1257. [PubMed] [Google Scholar]
- Brumbaugh PF, Haussler MR. 1a,25-Dihydroxycholecalciferol receptors in intestine. II. Temperature-dependent transfer of the hormone to chromatin via a specific cytosol receptor. J Biol Chem 1974;249:1258–1262. [PubMed] [Google Scholar]
- Brumbaugh PF, Hughes MR, Haussler MR. Cytoplasmic and nuclear binding components for 1a25-dihydroxyvitamin D3 in chick parathyroid glands. Proc Natl Acad Sci USA 1975;72:4871–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Pike J, Haussler M. 1,25-Dihydroxyvitamin D3 receptors in rat kidney cytosol. Biochem Biophys Res Commun 1979;90:1057–1063. [DOI] [PubMed] [Google Scholar]
- Kream B, Jose M, Yamada S, DeLuca H. A specific high-affinity binding macromolecule for 1,25-dihydroxyvitamin D3 in fetal bone. Science 1977;197:1086–1088. [DOI] [PubMed] [Google Scholar]
- Stumpf WE, Sar M, Reid FA, Tanaka Y, DeLuca HF. Target cells for 1,25-dihydroxyvitamin D3 in intestinal tract, stomach, kidney, skin, pituitary, and parathyroid. Science 1979;206:1188–1190. [DOI] [PubMed] [Google Scholar]
- Stumpf WE, Sar M, Clark SA, DeLuca HF. Brain target sites for 1,25-dihydroxyvitamin D3. Science 1982;215:1403–1405. [DOI] [PubMed] [Google Scholar]
- Pike J, Goozé L, Haussler M. Biochemical evidence for 1,25-dihydroxyvitamin D receptor macromolecules in parathyroid, pancreatic, pituitary, and placental tissues. Life Sci 1980;26:407–414. [DOI] [PubMed] [Google Scholar]
- Pike JW, Haussler MR. Purification of chicken intestinal receptor for 1,25-dihydroxyvitamin D. Proc Natl Acad Sci USA 1979;76:5485–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Mangelsdorf DJ, Pike JW, Haussler MR, O'Malley BW. Molecular cloning of complementary DNA encoding the avian receptor for vitamin D. Science 1987;235:1214–1217. [DOI] [PubMed] [Google Scholar]
- Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR et al. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci USA 1988;85:3294–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester JK, Maeda N, DeLuca HF. Isolation and expression of rat 1,25-dihydroxyvitamin D3 receptor cDNA. Proc Natl Acad Sci USA 1988;85:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MR, Malloy PJ, Kieback DG, Kesterson RA, Pike JW, Feldman D et al. Point mutations in the human vitamin D receptor gene associated with hypocalcemic rickets. Science 1988;242:1702–1705. [DOI] [PubMed] [Google Scholar]
- Malloy P, Xu R, Peng L, Peleg S, Al-Ashwal A, Feldman D. Hereditary 1,25-dihydroxyvitamin D resistant rickets due to a mutation causing multiple defects in vitamin D receptor function. Endocrinology 2004;145:5106–5114. [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Handa Y, Uematsu Y, Takeda S, Sekine K, Yoshihara Y et al. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet 1997;16:391–396. [DOI] [PubMed] [Google Scholar]
- Li YC, Pirro AE, Amling M, Delling G, Baron R, Bronson R et al. Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proc Natl Acad Sci USA 1997;94:9831–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T, Ozono K, Pike JW. A 55-kilodalton accessory factor facilitates vitamin D receptor DNA binding. Mol Endocrinol 1991;5:1578–1586. [DOI] [PubMed] [Google Scholar]
- Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Näär AM et al. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell 1991;67:1251–1266. [DOI] [PubMed] [Google Scholar]
- Ozono K, Liao J, Kerner SA, Scott RA, Pike JW. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J Biol Chem 1990;265:21881–21888. [PubMed] [Google Scholar]
- Noda M, Vogel RL, Craig AM, Prahl J, DeLuca HF, Denhardt DT. Identification of a DNA sequence responsible for binding of the 1,25-dihydroxyvitamin D3 receptor and 1,25-dihydroxyvitamin D3 enhancement of mouse secreted phosphoprotein 1 (SPP-1 or osteopontin) gene expression. Proc Natl Acad Sci USA 1990;87:9995–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 1991;65:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd DR, Sutton AL, Zhang C, MacDonald PN. Comodulators of vitamin D receptor-mediated gene expression. In: Feldman D, Pike JW, Glorieux FH (eds).Vitamin D Vol 1, :second edn. Elsevier/Acadmeic Press: New York, 2005; 291–304. [Google Scholar]
- McKenna NJ, O'Malley BW. Minireview: nuclear receptor coactivators--an update. Endocrinology 2002;143:2461–2465. [DOI] [PubMed] [Google Scholar]
- Kerner SA, Scott RA, Pike JW. Sequence elements in the human osteocalcin gene confer basal activation and inducible response to hormonal vitamin D3. Proc Natl Acad Sci USA 1989;86:4455–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama Y, Ozono K, Uchida M, Shinki T, Kato S, Suda T et al. Identification of a vitamin D-responsive element in the 5'-flanking region of the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J Biol Chem 1994;269:10545–10550. [PubMed] [Google Scholar]
- Zierold C, Darwish HM, DeLuca HF. Identification of a vitamin D-response element in the rat calcidiol (25-hydroxyvitamin D3) 24-hydroxylase gene. Proc Natl Acad Sci USA 1994;91:900–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold C, Darwish HM, DeLuca HF. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. J Biol Chem 1995;270:1675–1678. [DOI] [PubMed] [Google Scholar]
- Carlberg C. Molecular basis of the selective activity of vitamin D analogues. J Cell Biochem 2003;88:274–281. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Iacovino M, Morrison CA, Mountoufaris G, Closser M et al. Embryonic stem cell-based mapping of developmental transcriptional programs. Nat Methods 2011;8:1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieben L, Carmeliet G. Vitamin D signaling in osteocytes: effects on bone and mineral homeostasis. Bone 2013;54:237–243. [DOI] [PubMed] [Google Scholar]
- Kogawa M, Findlay DM, Anderson PH, Ormsby R, Vincent C, Morris HA et al. Osteoclastic metabolism of 25(OH)-vitamin D3: a potential mechanism for optimization of bone resorption. Endocrinology 2010;151:4613–4625. [DOI] [PubMed] [Google Scholar]
- Krishnan AV, Feldman D. Regulation of Vitamin D receptor abundance. In: Feldman DGFPJW (ed)Vitamin D Academic Press: San Diego, 1997; 179–200. [Google Scholar]
- Miyamoto K, Kesterson RA, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol 1997;11:1165–1179. [DOI] [PubMed] [Google Scholar]
- Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol 2006;20:1231–1247. [DOI] [PubMed] [Google Scholar]
- Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, Pike JW. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol 2010;24:128–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie M, Martin T. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev 1999;20:345–357. [DOI] [PubMed] [Google Scholar]
- Jimi E, Akiyama S, Tsurukai T, Okahashi N, Kobayashi K, Udagawa N et al. Osteoclast differentiation factor acts as a multifunctional regulator in murine osteoclast differentiation and function. J Immunol 1999;163:434–442. [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999;397:315–323. [DOI] [PubMed] [Google Scholar]
- Takayanagi H. Mechanistic insight into osteoclast differentiation in osteoimmunology. J Mol Med 2005;83:170–179. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol 2006;26:6469–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Shevde NK, Pike JW. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol 2007;21:197–214. [DOI] [PubMed] [Google Scholar]
- Fu Q, Manolagas SC, O'Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol 2006;26:6453–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli C, Zella LA, Fretz JA, Fu Q, Pike JW, Weinstein RS et al. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology 2008;149:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KA, Meyer MB, Pike JW. A novel distal enhancer mediates cytokine induction of mouse RANKl gene expression. Mol Endocrinol 2009;23:2095–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone acetylation and regulated through distinct distal enhancers. J Cell Biochem 2011;112:2030–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med 2013;19:179–192. [DOI] [PubMed] [Google Scholar]
- Bonewald LF. Osteocytes as dynamic multifunctional cells. Ann NY Acad Sci 2007;1116:281–290. [DOI] [PubMed] [Google Scholar]
- Bonewald LF, Johnson ML. Osteocytes mechanosensing and Wnt signaling. Bone 2008;42:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet 2002;70:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001;107:513–523. [DOI] [PubMed] [Google Scholar]
- Fretz J, Zella L, Kim S, Shevde N, Pike J. 1,25-Dihydroxyvitamin D3 regulates the expression of low-density lipoprotein receptor-related protein 5 via deoxyribonucleic acid sequence elements located downstream of the start site of transcription. Mol Endocrinol 2006;20:2215–2230. [DOI] [PubMed] [Google Scholar]
- Hofbauer LC, Hamann C, Ebeling PR. Approach to the patient with secondary osteoporosis. Eur J Endocrinol 2010;162:1009–1020. [DOI] [PubMed] [Google Scholar]
- van Meurs JB, Dhonukshe-Rutten RA, Pluijm SM, van der Klift M, de Jonge R, Lindemans J et al. Homocysteine levels and the risk of osteoporotic fracture. N Engl J Med 2004;350:2033–2041. [DOI] [PubMed] [Google Scholar]
- Lubec B, Fang-Kircher S, Lubec T, Blom HJ, Boers GH. Evidence for McKusick's hypothesis of deficient collagen cross-linking in patients with homocystinuria. Biochim Biophys Acta 1996;1315:159–162. [DOI] [PubMed] [Google Scholar]
- Saito M, Marumo K. Collagen cross-links as a determinant of bone quality: a possible explanation for bone fragility in aging, osteoporosis, and diabetes mellitus. Osteoporos Int 2010;21:195–214. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Dominy JE, Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr 2006;136:(6 Suppl): 1652S–1659S. [DOI] [PubMed] [Google Scholar]
- Eberhardt RT, Forgione MA, Cap A, Leopold JA, Rudd MA, Trolliet M et al. Endothelial dysfunction in a murine model of mild hyperhomocyst(e)inemia. J Clin Invest 2000;106:483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert K, Maurin N, Vayssettes C, Siauve N, Janel N. Cystathionine beta synthase deficiency affects mouse endochondral ossification. Anat Rec A Discov Mol Cell Evol Biol 2005;282:1–7. [DOI] [PubMed] [Google Scholar]
- Kriebitzsch C, Verlinden L, Eelen G, van Schoor NM, Swart K, Lips P et al. 1,25-dihydroxyvitamin D(3) influences cellular homocysteine levels in murine pre-osteoblastic MC3T3-E1 cells by direct regulation of cystathionine β-synthase. J Bone Miner Res 2011;26:2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci 2004;29:664–673. [DOI] [PubMed] [Google Scholar]
- Schlingmann KP, Kaufmann M, Weber S, Irwin A, Goos C, John U et al. Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med 2011;365:410–421. [DOI] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem 2010;285:15599–15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. J Steroid Biochem Mol Biol 2010;121:136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol 2012;26:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1{alpha},25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res 2011;39:9181–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan S, Heger A, Berlanga A, Maugeri N, Lincoln M, Burrell A et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res 2010;20:1352–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 2013;153:601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 2010;28:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 2012;11:384–400. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005;122:33–43. [DOI] [PubMed] [Google Scholar]
- Carroll J, Meyer C, Song J, Li W, Geistlinger T, Eeckhoute J et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 2006;38:1289–1297. [DOI] [PubMed] [Google Scholar]
- Kato S, Kim MS, Yamaoka K, Fujiki R. Mechanisms of transcriptional repression by 1,25(OH)2 vitamin D. Curr Opin Nephrol Hypertens 2007;16:297–304. [DOI] [PubMed] [Google Scholar]
- Kim MS, Fujiki R, Murayama A, Kitagawa H, Yamaoka K, Yamamoto Y et al. 1Alpha,25(OH)2D3-induced transrepression by vitamin D receptor through E-box-type elements in the human parathyroid hormone gene promoter. Mol Endocrinol 2007;21:334–342. [DOI] [PubMed] [Google Scholar]
- Alroy I, Towers TL, Freedman LP. Transcriptional repression of the interleukin-2 gene by vitamin D3: direct inhibition of NFATp/AP-1 complex formation by a nuclear hormone receptor. Mol Cell Biol 1995;15:5789–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers TL, Freedman LP. Granulocyte-macrophage colony-stimulating factor gene transcription is directly repressed by the vitamin D3 receptor. Implications for allosteric influences on nuclear receptor structure and function by a DNA element. J Biol Chem 1998;273:10338–10348. [DOI] [PubMed] [Google Scholar]
- Cippitelli M, Santoni A. Vitamin D3: a transcriptional modulator of the interferon-gamma gene. Eur J Immunol 1998;28:3017–3030. [DOI] [PubMed] [Google Scholar]
- Joshi S, Pantalena LC, Liu XK, Gaffen SL, Liu H, Rohowsky-Kochan C et al. 1,25-dihydroxyvitamin D(3) ameliorates Th17 autoimmunity via transcriptional modulation of interleukin-17A. Mol Cell Biol 2011;31:3653–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res 2005;20:305–317. [DOI] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012;148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol 2013;25:387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes A, Lakshmipathy U. Advances in genetic modification of pluripotent stem cells. Biotechnol Adv 2013;31:994–1001. [DOI] [PubMed] [Google Scholar]