Abstract
The development of biomaterials ensuring proper cell adhesion, polarization, migration and differentiation represents a true enabler for successful tissue-engineering applications. Surface nanostructuring was suggested as a promising method for improving cell-substrate interaction. Here, we study Wharton's Jelly human Mesenchymal Stem Cells (WJ-hMSC) interacting with nanogratings (NGs) having a controlled amount of nanotopographical noise (nTN). Our data demonstrate that unperturbed NGs induce cell polarization, alignment and migration along NG lines. The introduction of nTN dramatically modifies this behavior and leads to a marked loss of cell polarization and directional migration, even at low noise levels. High-resolution focal adhesions (FAs) imaging showed that this behavior is caused by the release of the geometrical vinculum imposed by the NGs to FA shaping and maturation. We argue that highly anisotropic nanopatterned scaffolds can be successfully exploited to drive stem cell migration in regenerative medicine protocols and discuss the impact of scaffold alterations or wear.
Cells are physiologically exposed to complex mechanical forces that induce cell morpho-functional responses, activate biochemical signal cascades and biophysical changes1. Mechanical stimuli modulate many aspects of cell physiology, including growth, differentiation, migration, gene expression, protein synthesis, and apoptosis2,3. The extracellular matrix (ECM) is the natural cell scaffold and the main extracellular tissue component. It provides most of these mechanical forces4. ECM is composed of proteins and polysaccharides5 whose aggregation and folding create a complex 3D architecture, which is characterized by a wide variety of geometries (topographies) and a physiological level of topographical noise at the micro/nanoscale6,7. Other examples of nanotopographical noise are cellular debris, protein aggregates, sclerotic plaques8,9,10, scar-tissue invasion11, and degradation of implantable prostheses12. Interaction with these noisy topographies is mediated by a process called mechanotransduction: physical cues are integrated and converted to intracellular biochemical responses, which in turn drive changes to cell morphology and function13.
Stem cells play an increasing role in tissue engineering and regenerative medicine. Bone-marrow human mesenchymal stem cells (BM-hMSCs) are the most commonly used cells in clinical applications and basic research. Owing to the rather invasive extraction procedure, however, in the last few years, MSCs have been actively sought for in other human tissues14. In this context neo-natal tissues are particularly promising15 because they can provide an easily-accessible and ethically non-controversial stem-cell source. Additionally, neo-natal hMSCs exhibit enhanced proliferation, lifespan, differentiation potential and reduced rejection after transplantation16,17. In particular, Wharton's Jelly hMSCs (WJ-hMSCs) are extracted from the part of the umbilical cord (UC) composed of a mucous-connective tissue matrix (named Wharton's Jelly) that is rich in stem cells, collagen fibers and proteoglycans18,19.
Many therapeutical approaches based on hMSCs have been proposed. Some of them take advantage of hybrid hMSC-biomaterial scaffolds that are implanted in the human body. When in situ, cells migrate off the scaffold and undergo terminal differentiation leading to device integration and regeneration of the damaged area. Beyond solubility, other factors can stimulate this process20,21,22 and improve the final transplantation outcome23,24 such as the physical properties of the scaffold (e.g. stiffness, density, topography, porosity, roughness).
The modulation of cell migration, invasion and terminal differentiation by artificial physical cues is of paramount importance for implementing effective stem cell based tissue engineering protocols for regenerative medicine. Although micro/nanomaterial driven differentiation has been studied by many groups25,26,27,28 the regulation of hMSC migration by contact interaction is still largely unknown. Mechanotransduced migration was studied for other cell types, however, primarily for the case of fibroblasts. For example, Lo et al. demonstrated that these cells tend to migrate towards rigid surfaces or local regions of high tension in an elastic polyacrylamide gel29. Jeon et al. investigated 3T3 morphology and motility on micron-scale cross and line patterns with different aspect ratios. They showed that cell alignment and migration directionality improved if the pattern size is smaller than the cell size, and that migration on shallow patterns (3 μm) was faster than on deep grooves (10 μm)30. Also, Kim et al. studied fibroblast response to gratings with constant ridge width and depth, but variable groove width (from 1 to 9.1 μm)31, and reported on cell-morphology variation in response to specific topographies. Cell adhesion, elongation and alignment were optimal on areas with small groove width, where migration directionality and speed also increased. Kaiser et al. correlated fibroblast cell migration with cell morphology and groove/ridge lateral size on Ti4Al6V patterned samples32: cell orientation, migration angle and migration velocity were different on structured surfaces compared to observations with plane surfaces, and shallow topographies with wide ridges supported more effectively cell migration. Using a different cell model we demonstrated that plastic nanogratings change PC12 cell-polarity state and lead to bipolar cells with aligned neurites33. Cells on flat substrates migrate randomly in all directions, while differentiated PC12 on nanogratings show slower migration with angular restriction and specific polarization of cell cytoskeleton. Other similar studies were based on the use of microstructured PDMS platforms. These substrates were exploited to obtain directed Schwann-cell or smooth-muscle-cell migration34,35. Interestingly, Yim et al. observed that smooth muscle cells on nanopatterned samples also elongate nuclei and align to the pattern, but with reduced proliferation.
In this paper we study WJ-hMSCs polarization and migration properties as a function of controlled directional stimuli. To this end, we exploited nanogratings (NGs, alternating lines of submicron ridges and grooves) with a controlled amount of topographical noise36. We measured mechanotransduction in WJ-hMSC by evaluating cell morphology and alignment, cytoskeleton polarization, focal-adhesion (FA) development and spatial distribution by fluorescence microscopy on fixed and living cells. Finally, WJ-hMSC functionality was assayed by studying single-cell migration driven by the contact interaction with noisy substrates.
Results
WJ-hMSC viability on noisy nanogratings
Six different topographies were produced by thermal NIL on 170-μm-thick cyclic olefin copolymer (COC) film (see Materials and Methods). Our fabrication method yielded substrates suitable for high-resolution microscopy with highly reproducible nanopatterns over macroscopic areas (16 mm2). For this study, two reference substrates were employed: the FLAT surface (with no nanopatterning), and the T1 nanograting (NG), i.e. the fully anisotropic condition (Fig. 1b). T1 is characterized by 1 μm period (50% duty cycle) and 350 nm depth and led to cell contacts restricted to ridge tops, driving early cell adhesion and spreading37. In order to progressively release this constraint and reduce substrate directionality, we added topographical noise to T1. This was obtained by adding random nanomodifications (NMs, bridges between ridges) with controlled density to the original layout36. In particular, four noisy NGs were tested (Fig. 1c–f) with a percentage of noise evenly spaced (20%, 40%, 60% and 80%), where T1 and FLAT correspond to control substrates.
Figure 1. Substrates morphological characterization.
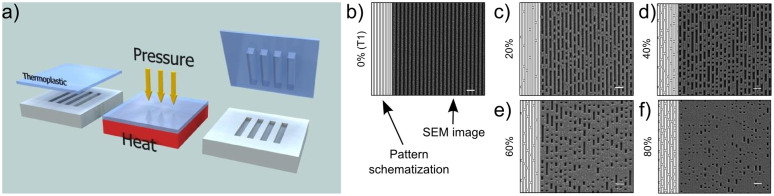
(a) Scheme of the nanoimprint process. (b–f) Replica scheme and scanning electron microscope images of noisy COC-NGs (500-nm ridge- and groove-width). In SEM images, dark and bright areas correspond to groove and ridge, respectively. Each substrate is defined by the percentage of noise and a cell-dependent directionality (δ). Negative noise on COC replicas consists of links between adjacent ridges. Scale bar = 2 μm.
As a preliminary test, Wharton's Jelly human Mesenchymal Stem Cells (WJ-hMSC) viability was assessed on the whole set of substrates. While COC biocompatibility was already reported38,39, nanopatterning by itself could in principle influence cell viability40 by altering cell morphological and functional properties. Experiments were performed at short and medium term: 24 hours and 1 week after seeding. In order to detect living and dead cells, we used different fluorescent dyes and three-channel epifluorescence microscopy. Measurements demonstrated that none of the six topographies affects cell viability significantly (Fig. S1). In particular, for both time points, the percentage of viable cells is (94 ± 2%) in line with typical values obtained for standard plastic substrates (e.g. TCPS). This result confirms the possibility to test our topographies in long-term experiments, such as cell proliferation, migration and differentiation assays.
WJ-hMSC morphology modulation by contact guidance
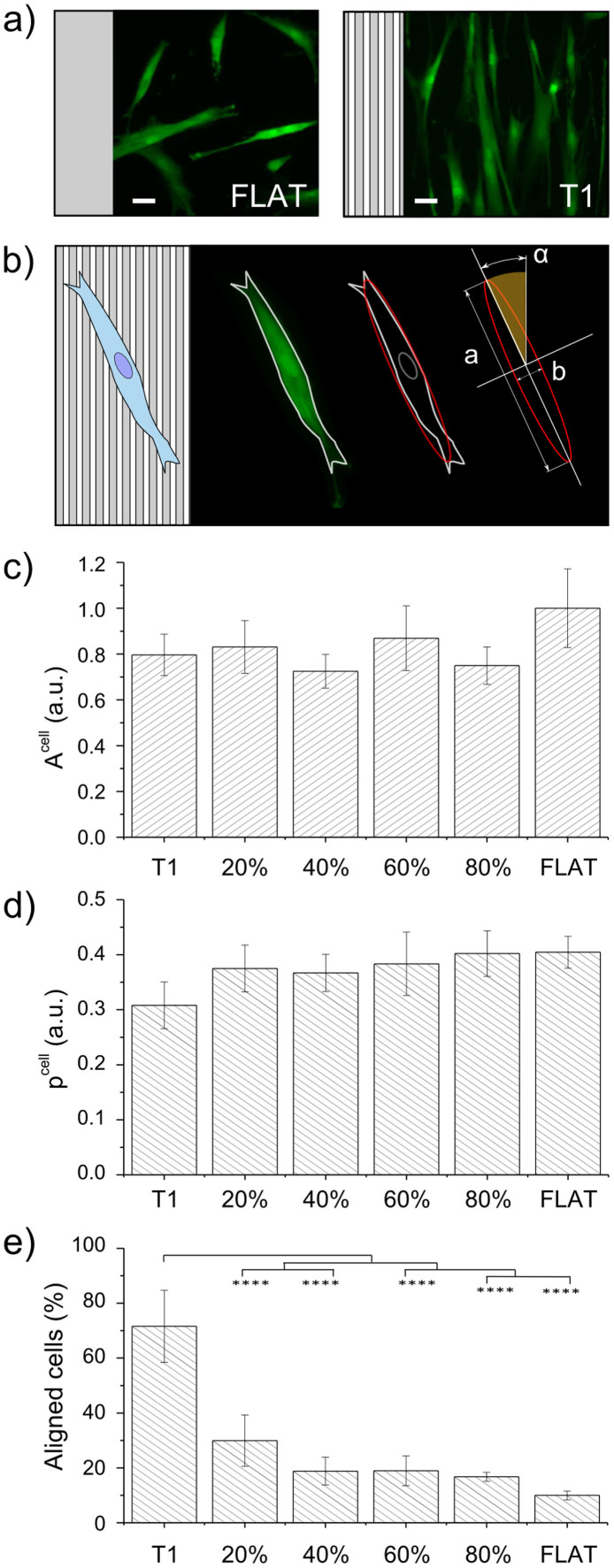
Substrate physical properties are known to influence cell morphotypes at different levels, from macroscopic shaping to molecular organization. In order to quantify cell area, polarization and alignment to the NGs, cells were stained with a vital, membrane-permeant fluorescent green dye (Calcein-AM) and images were acquired by epifluorescence microscopy (two representative fields are reported in Fig. 2a). Later, image binarization and automatic analysis were used to measure single-cell morphological parameters(Fig. 2b).
Figure 2. WJ-hMSC morphology.
(a) Representative cell images at t = 24 h after seeding on T1 and FLAT. Cells were stained with Calcein AM-green dye. Scale bar = 50 μm. (b) Schematic of the method used for cell morphology characterization. After fitting the cell contour with an ellipse, major (a) and minor (b) axes and orientation (αcell) were measured. (c) Cell area (Acell) vs noise. Data were normalized to the FLAT value. (d) Cell polarization (pcell)vs noise. pcell → 0 indicates full polarization, pcell → 1 correspond to round shaped cells. (e) Percentage of aligned cells (0° < αcell < 15°) vs noise. Four independent experiments were performed. (**** P < 0.0001).
Figure 2c shows the average cell area (Acell) measured with the different nanotopographies. Overall, patterned surfaces lead to about 20% cell-size reduction as compared to FLAT, though this difference is not statistically significant. Remarkably, noise does not affect Acell with respect to the no-noise condition (T1 substrate). Results on cell polarization (pcell) are shown in Fig. 2d. WJ-hMSCs are naturally polarized cells, with pcell = 0.4 ± 0.03 on FLAT. As expected, interaction with T1 leads to pcell increase of about the 25%. Contrary to what observed for cell area, pcell is restored to its FLAT value by nanotopographical noise, regardless of its density. Figure S2 reports the angular distribution of cell alignment (αcell) for each substrate and it shows that cells on all the noisy NGs and on FLAT were randomly oriented. Instead, as demonstrated in Fig. 2e, T1 induced a net (70%) and significant improvement of cell alignment with respect to all other conditions.
Analysis of cell cytoskeleton was performed by immunofluorescence techniques. Three fluorescent dyes were used to detect actin cytoskeleton, vinculin aggregation in focal adhesions (FAs), and cell nuclei (see materials and methods). Confocal microscopy was used to acquire high-resolution images. Figure 3a reports six representative images of actin cytoskeletons, acquired on the different substrates. WJ-hMSCs are characterized by a well-organized cytoskeleton, where actin stress fibers are evident. Fast Fourier transform functions (FFT) were exploited to study the spatial organization of actin fibers. Figure 3b shows that 85 ± 6% of cells grown on T1 present actin fibers well-oriented along the NG direction (αactin < 15°). Notably, this percentage was reduced by 42% in the presence of 20% noise. A significant difference in stress fiber alignment is also found between NGs with 20% noise and NGs with high percentage of noise (60% up to FLAT). Single-cell actin dispersion is finally reported in Fig. 3c. Also in this case T1 performs best, while all noisy NGs lead to a dispersion, which at least doubles with respect to that of cells cultured on T1. Interestingly, data relative to high noise percentage (60–80%) substrates indicate a greater dispersion with respect to data obtained on FLAT.
Figure 3. Actin cytoskeleton polarization.
(a) Representative fluorescence images of actin cytoskeleton acquired at t = 24 h after cell seeding. A schematic of the substrate geometry is shown for each panel. Scale bar = 20 μm. (b) Percentage of cells with aligned actin (0° < αactin < 15°) vs noise. (c) Single cell actin dispersion vs noise. Five independent experiments were performed. (*P < 0.05, ***P < 0.001, ****P < 0.0001).
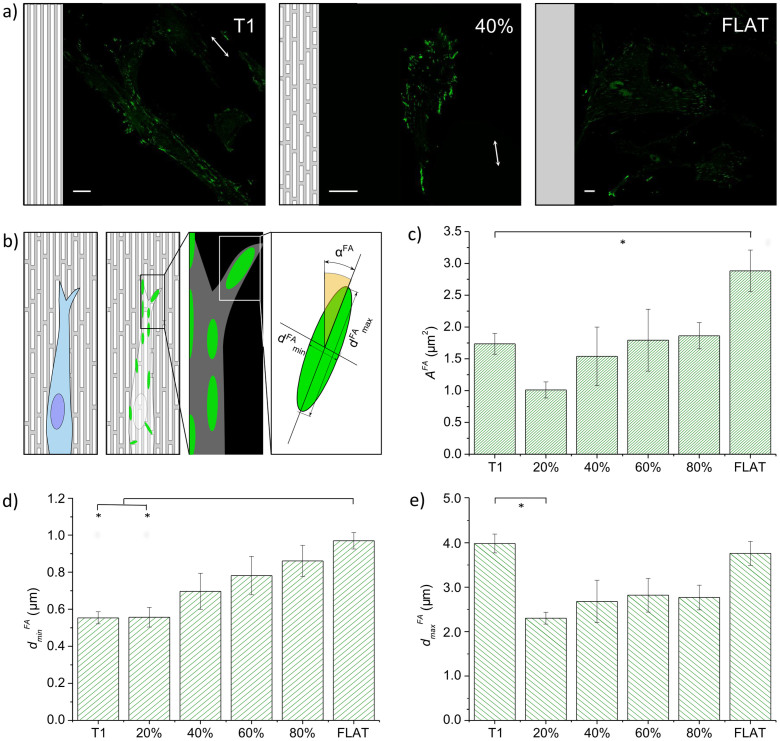
Focal adhesion morphology
As mentioned in the previous paragraph, FAs were detected as vinculin clusters by immunofluorescence techniques and confocal microscopy (representative images are shown in Fig. 4a). FA morphological parameters were semi-automatically measured from fluorescence images by image correlation methods41. FAs were modeled as ellipsoidal bodies (Fig. 4b) and described by the following parameters: area (AFA), axis length (dminFA and dmaxFA), elongation (pFA = dminFA/dmaxFA), and alignment to the pattern (αFA). We also quantified FA density (i.e. the average number of FAs per cell) and FA spatial distribution within single cells. Figure 4 indicates that NGs inhibit the full development of FAs. As shown in Fig. 4d, T1 and the NG with 20% noise led to dminFA ≈ 0.55 ± 0.03 μm, a value comparable to the substrate ridge width. Increased noise made a larger surface area available for cell adhesion and FA maturation. Indeed, dminFA reached values indistinguishable from those obtained on FLAT (≈1 μm) while dmaxFA displayed a different response (Fig. 4e). FAs could elongate freely on FLAT and T1 (dmaxFA = 3.99 ± 0.21 and 3.76 ± 0.27 μm, respectively), while noisy NGs reduced FA stretching by approximately 35%. Thus, the net result of these modifications affects both FA area and elongation, as shown in Figs. 4c and S3a. FAs were naturally ellipsoidal with pFA = 0.259 ± 0.007 on FLAT. Polarization almost halved (46%) on T1, while noise restored the conformation found on FLAT. Moreover, as shown in Fig. 4c, all the NGs —with or without noise— halved the FA area with respect to the FLAT value. The cells that were mainly affected by this effect were those grown on substrates with a low (20%–40%) percentage of noise, where we also observed an increased number of FAs (Fig. S4d).
Figure 4. Focal adhesions morphology.
(a) Representative fluorescence images of vinculin clusters on selected substrates (T1, 40% noise and FLAT). Scale bars = 20 μm. (b) Schematic of WJ-hMSC focal adhesion. FAs were modeled as ellipses, for which major (dFAmax) and minor axes (dFAmin), and directionality (αFA) were measured. (c) Area (AFA) (d) minor axis (dminFA) and (e) major axis (dmaxFA) vs noise. Three independent experiments were performed. (* P < 0.05).
FA positions within single cells were also modified by NGs (Fig. S4). Majority of FAs were aligned to the cell polarization direction on T1. FLAT and NGs with 80% noise maintained 60% of FAs aligned to the cell axis, whereas low and medium percentage of noise reduced this alignment slightly (Fig. S4c). As expected, the radial distribution of FAs was unaffected by any of the substrates: 60% of FAs were consolidated at the cell periphery (Fig. S4b) where they experienced the maximum traction applied by actin stress fibers.
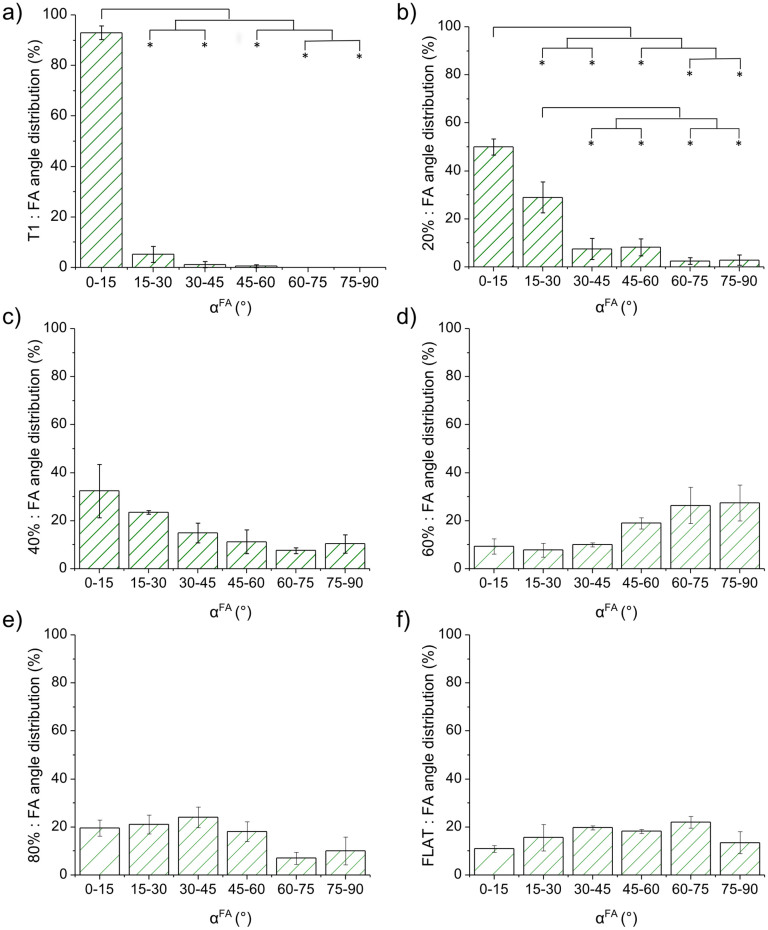
αFA was very dependent on the substrate under consideration (Fig. 5 and S3b), showing a behavior similar to that obtained for cell alignment. On T1, cells established almost all the FAs (>90%) parallel (0° < αFA < 15°) to the pattern, while no FAs were found with αFA > 60° (Fig. 5a). Noise broadened this distribution. On low-noise (20–40%) NGs, we measured a reduced number (40–60%) of parallel FAs and an overall rise of all the other bins of the angular distributions. Above 60% noise, the distribution flattened, showing a slight increase of misaligned (αFA > 45°) FAs for the 60% noise NG. Figure S3b focuses on fully aligned (0° < αFA < 15°) FAs demonstrating that T1 and 20% noise lead to a significantly greater number of aligned FAs with respect to all other substrates.
Figure 5. Focal adhesion alignment to nanogratings.
(a–f)FA angle distribution with respect to the NG direction for different noise levels. Bin size: 15°. (* P < 0.05).
Contact guidance in WJ-hMSC migration
WJ-hMSC migration was recorded by time-lapse microscopy after staining cell nuclei with a vital fluorescent dye (Syto16, Invitrogen, Italy). Frames were acquired every 15 minutes for 15 hours, which allowed information to be gathered at two time-scales.
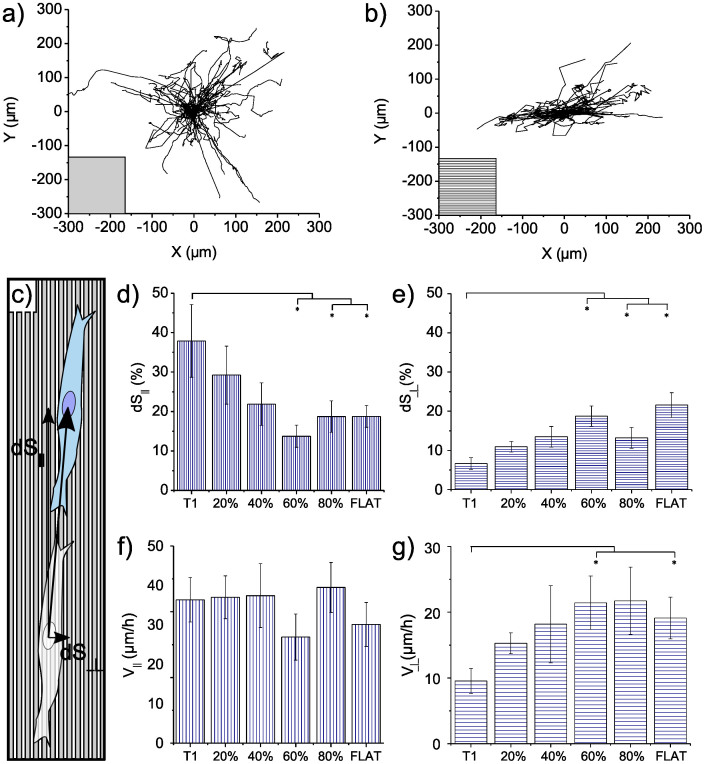
None of the NGs affected migration global parameters, namely cell displacement (R), total path (S), migration step (dS) and speed (V) (see Material and Methods). Specifically we measured R = 127.2 ± 3.5 μm, S = 324.4 ± 11.6 μm, dS = 5.2 ± 0.2 μm and V = 20.9 ± 0.8 μm/h. These quantities were calculated by averaging the values measured for each substrate. Nonetheless, migration directionality was driven by contact guidance. Figures 6a and 6b show representative tracks: cell random walks were observed on FLAT and are compared with T1-driven migration in which tracks follow NG lines.
Figure 6. Analysis of cell migration.
Cells were cultured for 24 h under standard conditions and then incubated with Syto 16 dye for nucleus staining. Migration was sampled every 15 minutes for 15 hours. (a–b) Representative tracks of cells moving on T1 and FLAT surfaces, respectively. (c) Measurement schematic. (d) Percentage of steps aligned (dS∥) to the pattern direction (0°–15°) and (f) relative speed (V∥). (e) Percentage of steps perpendicular (dS⊥) to the pattern directionality (75°–90°) and g) relative speed (V⊥). Four independent experiments were performed. (* P < 0.05).
Migration was then characterized along two directions: parallel (0°–15°) and perpendicular (75°–90°) to the NGs. For FLAT, one random direction was chosen together with its perpendicular. The percentage of parallel migration steps doubled on T1 with respect to FLAT and high-noise NGs (60% and 80%) (Fig. 6d). A reduction applied, though to a lesser extent, also in the case of 20% and 40% noise. On the contrary, NGs did not affect the parallel speed (V∥) (Fig. 6f) and results were not significantly modified. The same was obtained for the speed calculated along other directions with angles up to 75° with respect to the NG. Figure 6e reports the percentage of perpendicular steps on the different substrates. As expected, T1 led to a suppression of the migration across the ridges where increasing noise progressively reduced this constraint. Perpendicular movement was slower on T1, leading to a reduced perpendicular speed (V⊥) of 50% with respect to V⊥ on FLAT. Conversely to what was obtained for V∥, noise was found to affect V⊥ (Fig. 6g). The presence of bridges between ridges made more anchoring points available, allowing easier movement across the NGs.
Discussion
Cell-substratum interactions are central in many biological phenomena, such as cell adhesion, spreading, migration, alignment and proliferation. Moreover, they can play a significant role in regulating cytokine production, gene expression and cell differentiation42. Knowledge and control of these interactions is crucial to understand important biological questions, to design biomedical devices43,44, and to produce in vitro and in vivo optimized engineered tissues45,46.
In this context, nanoscale topographies have recently attracted much attention since they can mimic the extracellular-matrix (ECM) features47,48 and because they are promising to enhance the performance of biomedical devices49,50,51,52,53,54.
In this paper we analyzed Wharton's Jelly human Mesenchymal Stem Cells (WJ-hMSCs) contact guidance on nanostructured substrates as a function of nanotopographical noise. Significant examples of noise do occur both in vivo and in vitro. Yet, so far, very few studies addressed its role on cell/tissue function. By using Poly-methyl-methacrylate nanopatterned samples with randomly displaced nanopits Dalby et al. showed that nanoscale disorder can stimulate hMSCs to produce bone tissue without osteogenic supplements55. Other authors created a disordered network of nanoparticles functionalized with RGD peptides in a poly-ethylene-glycol background and found that disorder promotes the formation of thick actin bundles and, more importantly, cell adhesion and FA maturation56.
Here we exploited plastic substrates nanoimprinted with noisy nanogratings (NGs) and reported on the impact of WJ-hMSC-substrate interaction. Focal-adhesion (FA) and actin-polarization properties were measured as readout of the mechanotransduction efficiency. Vinculin staining was used to visualize FAs. This protein localizes between transmembrane integrins and actin fibers and was shown to be sensitive to the balance between intracellular and extracellular forces, modulating its shape and aggregation during cell migration57. With NGs FAs primarily assemble along cell periphery and exhibit a polarized shape defined by the pattern direction. FA size halved on T1 with respect to measurements on FLAT and was modulated by noise level. In particular, small FA clusters were favored by 20% noise, suggesting that this specific geometrical arrangement of imperfections may particularly influence FA maturation. This is supported by the presence of a larger number of FAs at this noise level that we argue are part of a feedback mechanism to successfully counteract the presence of weak anchor points and to maintain the correct internal mechanical stress balance58. FA spatial distribution drives the formation of actin stress fibers that in turn align2,59,60 reflecting FA angular distribution, and finally lead to cell remodeling and alignment to the pattern. We found a good correspondence between cell alignment (Fig. 2e) and actin alignment (Fig. 3b). Importantly, the actin reshaping does not significantly affect cell area, a condition that can promote pathological dysfunctions61.
It is known that cell migration is correlated with FA dynamics, i.e. cell motion direction is controlled by the FA spatial distribution and turn over. Cell migration is one of the important biological processes that are regulated by ECM-cell interaction35,62,63: it is fundamental for embryogenesis, development and for many other physiological processes such as tissue renewal or immune response. Migration is also pivotal for pathogenic processes (e.g. cancer), and tissue repair after injury. Controlled migration would be particularly desirable with stem cells in order to drive injected cells as rapidly as possible towards their target position. We showed that NGs do not strongly affect global migration parameters (path and speed), but can control movement direction. This property is consistent with previous studies with other cell types (e.g. fibroblasts) on similar anisotropic substrates32,64. We observed that on FLAT samples cells freely move in two dimensions, while on NGs their motion is confined along the pattern direction. This effect is tuned by pattern noise. For noise levels above 40%, the substrates are ineffective in directing cell motion.
Moreover we observed that noise interferes dramatically with NG topographical stimuli and does lead to loss of alignment. In order to better quantify the impact of noise on alignment and cell migration, we introduced a different parameter, δ based on the concept of substrate directionality (see Material and Methods, Eq. 1). δ measures the degree of directionality fed to cells, which is a function not only of surface geometrical properties, but also of the actual cell sensing area [δ = δ(A,p); where A and p are the average cell area and nanomodification (NM) density, respectively]. Note that δ is not an intrinsic substrate characteristic, but it depends on the biological system under consideration. Figure S6 shows δ = δ(AWJ-hMSC,p) calculated according to Eq. 1, with AWJ-hMSC = 3421 μm2 (i.e. the WJ-hMSC average area measured in our experiments) as a function of p. δ monotonically decreases with p, but with a marked non-linear behavior: δ reduces by 42% when the noise increases from 0 to 20%, but this effect rapidly saturates and further increase of p from 20% to 80% leads to a reduction in δ by only 8%. This shows that a significant variation in WJ-hMSC reaction to guiding substrates is already produced by a small amount of noise.
In conclusion, we investigated the influence of highly anisotropic and noisy substrates on Wharton's Jelly human Mesenchymal Stem Cells by focusing on cell morphotype, functionality and migration by means of biochemical assays and fluorescence/confocal microscopy. Our results show that migration on perfect nanogratings is highly directional. On the contrary, pattern degradation leads to a loss of cell directionality, even at very low noise levels. The same effect was observed also on cell morphology: while a slight cell area reduction was reported for all NGs, only the non-noisy NGs did induce cell polarization. Molecular analysis revealed that cell polarization corresponds to actin cytoskeleton organization, with ordered stress fibers following NG lines. Furthermore, this cytoskeletal structure correlates with focal-adhesion (FA) size and spatial arrangement. In the case of unperturbed NGs we observed FAs that developed at cell terminations and were aligned to the pattern. Finally, we observed that NGs, with or without noise, constrain and stretch FAs along ridges, reducing their area and inhibiting their maturation. Our results indicate that highly-anisotropic nanopatterned scaffolds might be successfully applied to drive stem-cell migration in new regenerative-medicine protocols. Importantly, we showed that pattern fidelity is crucial for proper translation of the topographical signal into the desired cell response: pattern degradation may indeed result in altered mechanotransduction at molecular (rearrangement of FA and actin fibers), morphological (cell area and polarization), and functional (cell migration) levels. This indicates that small scaffold alterations or its wear resulting from interaction with the host tissue (eg., deposition of extracellular matrix components, degradation of bioresorbable devices, cell invasion) need be carefully considered when evaluating the actual scaffold operation time for active regenerative therapies.
Methods
Mold fabrication
Nanopatterned silicon molds production protocol is described in36. Briefly, noisy geometries were generated by a Matlab program and transferred onto silicon molds by electron-beam lithography (EBL) and reactive-ion etching (RIE) before the imprinting process.
All noisy patterns were designed starting from an “ideal” NG with 500 nm ridge size, 500 nm groove size and 350 nm depth. A controlled density of randomly distributed nanomodifications (NMs) was then introduced into this “ideal”. In the present case, NMs are composed of bridges between adjacent ridges and will be named negative noise. Substrate directionality was calculated based on the Fourier transform (FT) of the actual nanopattern image (Fig. 1b–f). Directionality δ (in dB) is defined as the average ratio between the substrate periodic component (signal) and the low-frequency noise (noise) in the FT domain,
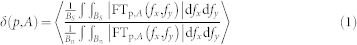 |
where p is the NMs density and A is the area on which the FT is calculated. We chose A as the average area occupied by a WJ-hMSC body, i.e. A = 3421 μm2. Images were generated by introducing a random variation in the value of each pixel, mimicking the impact of surface roughness on the topography. Roughness was modeled as noise with a constant probability density and amplitude of 6 nm.
COC-substrate fabrication
Nanostructured cell-culture substrates were fabricated by thermal nanoimprint lithography (NIL) (see Fig. 1a). Cyclic olefin copolymer (COC) foil (IBIDI; Martinsried, Germany) was selected as thermoplastic material for cell culture. The COC foils were imprinted37 using an Obducat Nanoimprint 24 system (Obducat, Sweden). Imprinted substrates were attached to the bottom of hollowed 35 mm Petri dishes with silicone glue (RS Components RS692-524). Substrate wettability (average water contact-angle value = 87 ± 7°, mean ± SD) was not significantly affected by the presence of the noise.
Before cell culturing the samples were sterilized with ethanol, rinsed with PBS twice, and then with complete medium once.
Wharton's jelly mesenchymal stem cell extraction and culture
Wharton's Jelly human Mesenchymal Stem Cells (WJ-hMSCs) were isolated from umbilical cords from full-term deliveries, which were collected at the Gynaecology Unit of the Azienda Ospedaliera Universitaria-Pisa Hospital. Wharton's jelly (WJ) was separated from the cord vessels and placed in 6-well dishes containing alpha-minimum essential medium (α-MEM; GIBCO) supplemented with 20% fetal bovine serum (FBS; Euroclone, Italy) and incubated at 37°C with 5% humidified CO2. Fresh medium was added twice a week up to 90% confluence. MSCs were then harvested with 0.25% trypsin and 1 mM EDTA solution (Cambrex, Italy) and re-plated at 8.000 cells/cm2. Successive passages were performed in DMEM medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 10 U/ml penicillin and 10 mg/ml streptomycin. The expanded cells were characterized after the primoculture (P0) by flow cytometric analysis (FACS Canto I, Becton Dickinson CA, USA) of specific surface antigens (CD14−, CD34−, CD20−, CD45−, CD73+, CD90+ and CD105+) according to the mesenchymal immunophenotype. Cells were used within the 8th passage. In order to perform single-cell experiments, WJ-hMSCs were seeded at the final concentration of 30 × 104 cells/cm2 and kept in humidified atmosphere until experiment time.
Cell viability and morphology analysis
Live and dead cells were counted by using 5 μM Calcein-AM (A-AC, C3100, Molecular Probes, Eugene, USA) and 8 μg/μl Propidium Iodide (PI, P35669, C3100, Molecular Probes, Eugene, USA), respectively. Nuclei were stained with 5 μg/μl Hoecst 33345 (H3570, Invitrogen, Italy) in order to measure the total cell number. Ten epifluorescence images for each sample (Fig. S1a) were acquired with a 20× air Nikon objective, N.A. 0.45, PlanFluor, using an Eclipse Ti inverted microscope (Nikon, Japan) equipped with an incubating chamber (Okolab, Italy), a CCD ORCA R2 (Hamamatzu, Japan), a mercury arc lamp, and 3 filter blocks (UV-2E/C, B-1°, G-2E/C, Nikon, Japan). Four independent experiments at short and medium term (1 and 7 days) were performed.
Green channel images (Fig. 2a) were used to evaluate cell area (Acell), polarization (pcell) and alignment (αcell). The software ImageJ was used for image analysis. Cell profile was obtained by image binarization (Fig. 2b). Acell was then calculated as number of pixels contained in cell profiles and normalized to the average value, measured on FLAT substrates (Fig. 2c). pcell was evaluated as mini-Feret/Feret-diameters (ImageJ analysis). pcell → 0 indicates fully polarized cells and pcell → 1 corresponds to round-shaped cells (Fig. 2d). αcell was calculated as the angle between cell Feret-diameter and the NG direction, as shown in Fig. 2e. The alignment distribution was plotted with a 15° bin-size. Cells were considered aligned if 0° < αcell < 15°.
Immunostaining protocol
24 hours after seeding, cells were fixed for 15 minutes in 4% paraformaldehyde, rinsed three times with phosphate buffered saline (PBS), and then processed for immunofluorescence analysis. Cells were permeabilized and stained at 4°C overnight with 0.165 μM AlexaFluor647phalloidin (Invitrogen, Italy) and the primary antibody 2.5 μg/ml, 0.5%Triton, 0.8 M NaCl, 30 mM Phosphate buffer). Cells were washed and incubated at room temperature for 45 minutes in 20 μg/ml secondary antibody AlexaFluor488 diluted in GDB solution. Nuclei were labeled with 5 μg/μl of Hoecst (Invitrogen, Italy) in PBS. Fixed and stained cells were mounted in Vectashield (VectorLaboratories, USA) and imaged using a confocal microscope (TCS SP5 AOBS Leica Confocal Microscopy, Germany) equipped with Ar (emission 488, detection 495–550 nm), He/Ne (emission 633, detection 650–800 nm) and UV (emission 405, detection 410–470 nm) lasers with a 40× oil Leica objective, 1.25 N.A. Images were acquired with four channels: green for FA, red for actin cytoskeleton, blue for nuclei and the transmission (DIC). The image size was 1024 × 1024 pixels.
Cytoskeleton characterization
At least ten one-channel confocal images per sample were used to analyze immunostained cell actin cytoskeleton. Representative images are shown in Fig. 3a. Actin fibers (αactin) alignment to NGs (Fig. 3b) in individual cells was measured using the directionality tool of Fiji free software. It calculates the image FFT and returns a directionality histogram for every analyzed image. The difference between histogram peaks and the angles of the underlying patterns yields the actin fibers alignment. The range of possible alignments varies between 0 and 90°. Average alignment angles of 0° and 45° indicate perfect and random alignment, respectively. Fibers were considered aligned if 0° < αactin < 15°. Actin-fiber dispersion (Fig. 3c) was returned by the same plugin. Data were calculated on five independent experiments.
Focal adhesion characterization
Green one-channel confocal microscopy images were used to analyze vinculin focal adhesions (FAs) of immunostained cells and transmission DIC images were used to extract cell contours. At least ten images were used for all samples. Representative images are shown in Fig. 4a. FAs were modeled as elliptic objects (Fig. 4b) and were semi-automatically processed by image correlation methods32. Adhesions were manually divided into groups according to their shape, size and orientation. Each group was manually selected using a free-hand selection tool, and a background threshold was set. These selections were spatially auto-correlated and the resulting correlation function was fitted using a 2D-Gaussian function, characterized by three parameters: two perpendicular standard deviations (dmin and dmax) (Fig. 4c,d) and the angle between the FA-major axis and the pattern direction (αFA). FAs were defined as aligned when 0° < αFA < 15° (Fig. S3b). A calibration curve was created to limit the artifacts due to non-punctual microscopy point spread function (PSF): a series of simulated images were produced and analyzed using the correlation method as described above. The calibration curve was validated using images of 1 μm and 4 μm fluorescent beads (Tetraspeck, Invitrogen, Italy). FA-shape was finally described by area (AFA) elongation (pFA) (Fig. 4b and S3a). pFA was calculated as the ratio between the FA long (dmaxFA) and short (dminFA) axis: pFA = dmaxFA/dminFA. This parameter spans from 0 (fully stretched, or linear, adhesions) to 1 (round shaped adhesions).
In order to evaluate FA radial distribution on single cells, the positions of the FA center and the cell edge were manually selected. The displacement of each FA with respect to the cells main center (Fig. S4a) (the cell body was divided in three regions: nuclear, perinuclear -or cytoplasmic- part and cell edge) and the angular distance (with 15° bin size) from the cell major axis (θFA) were measured (Fig. S4b). The distance from cell center was normalized to the cell radius in that radial direction, obtaining a distance range from 0 to 1. With this procedure, the FA density (i.e. the adhesions number/cell) for each cell was also measured (Fig. S4). Results were calculated on three independent experiments.
Migration experiments
500 μM Syto16 (Invitrogen, Italy) was used to stain living-cell nuclei. Syto 16 was administered 24 h after cell seeding (30 min incubation time at room temperature in culture medium without serum). Four independent time-lapse experiments were performed in epifluorescence by using a 20× air Nikon objective, N.A. 0.45, PlanFluor and an Eclipse Ti inverted microscope (Nikon, Japan) equipped with an incubating chamber (Okolab, Italy), a CCD ORCA R2 (Hamamatzu, Japan), a mercury arc lamp, and 3 blocks of filters (UV-2E/C, B-1°, G-2E/C, Nikon, Japan). Images were acquired for 15 hours, sampling every 15 minutes. Movies were analyzed with the ImageJ manual tracking plugin to extract the coordinates of single cells as a time function. Data were analyzed by a custom-made application written in Matlab. The following parameters were measured: cell displacement (R) (distance from the origin after 15 hours), total path covered in 15 hours (S), migration step (dS) and average speed (V) (calculated for intervals of 15 minutes). Directionality and speed of each step was measured and classified in two populations: parallel (dS∥, V∥) and perpendicular (dS⊥, V⊥) steps (Fig. 6c–g). dS was considered parallel if the angle between the step and the pattern was less than 15°, while it is considered perpendicular if this angle was between 75° and 90°.
Statistical analysis
All the experiments were independently repeated four times (unless stated) for each reported dataset. Data are reported as the mean value of the means of each experiment ± the standard error of the mean (mean ± SEM). Data were statistically analyzed using the commercial software OriginLab. For parametric data (after Shapiro-Wilk normality test), One-Way Anova with Tukey's post-test analysis was used; for non-parametric data, the Mann Whitney test was used. The statistical significance unless stated otherwise, is shown by *, **, ***, ****, which refers to P < 0.05, P < 0.01, P < 0.005, P < 0.001, respectively.
Author Contributions
E.J. and M.C. designed the experiments and wrote the main manuscript text. E.J. performed all the experiments. C.D.R. produced the correlation spectroscopy analysis. S.M. produced the silicon molds. F.N. extracted and characterized the Wharton Jelly mesenchymal stem cells. All authors reviewed the manuscript.
Supplementary Material
SUPPLMEMENTARY FIGURES
Acknowledgments
This work was supported in part by the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement no. NMP4-LA-2009-229289 NanoII and grant agreement no. NMP3-SL-2009-229294 NanoCARD. This work was also supported by the Regione Toscana through the projects “Biosensorbased assay for high-throughput quantitative screening of chloride transport” and NanoART.
References
- Guck J., Lautenschlager F., Paschke S. & Beil M. Critical review: cellular mecha- nobiology and amoeboid migration. Integr. Biol. (Camb). 2, 575–83 (2010). [DOI] [PubMed] [Google Scholar]
- Vogel V. & Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev. Mol. Cell Biol. 7, 265–75 (2006). [DOI] [PubMed] [Google Scholar]
- Patwari P. & Lee R. T. Mechanical control of tissue morphogenesis. Circ. Res. 103, 234–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. The extracellular matrix: not just pretty fibrils. Science 326, 1216–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. M. & George J. H. Exploring and engineering the cell surface interface. Science 310, 1135–8 (2005). [DOI] [PubMed] [Google Scholar]
- Nikkhah M., Edalat F., Manoucheri S. & Khademhosseini A. Engineering microscale topographies to control the cell-substrate interface. Biomat. 33, 5230–46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. L., Sims P. A. & Albrecht R. M. Three-dimensional extracellular matrix textured biomaterials. Biomat. 17, 2087–95 (1996). [DOI] [PubMed] [Google Scholar]
- Varga R. et al. Progressive vascular smooth muscle cell defects in a mouse model of HutchinsoneGilford progeria syndrome. Proc Natl Acad Sci U S A 103, 3250–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimon T. & Tabas I. Mechanisms and consequences of macrophage apoptosis in atherosclerosis. J Lipid Res 50, S382–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K., Plaks V. & Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. et al. Regenerative potential of silk conduits in repair of peripheral nerve injury in adult rats. Biomat. 33, 59–71 (2011). [DOI] [PubMed] [Google Scholar]
- Tabesh H. et al. The role of biodegradable engineered scaffolds seeded with Schwann cells for spinal cord regeneration. Neurochem In.t 54, 73–83 (2009). [DOI] [PubMed] [Google Scholar]
- DuFort C. C., Paszek M. J. & Weaver V. M. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorio O., Jacchetti E., Pacini S. & Cecchini M. Endothelial differentiation of mesenchymal stromal cells: when traditional biology meets mechanotransduction. Integr. Biol. (Camb). 5, 291–9 (2013). [DOI] [PubMed] [Google Scholar]
- Sarugaser R., Lickorish D., Baksh D., Hosseini M. M. & Davies J. E. Human umbilical cord perivascular (HUCPV) cells: a source of mesenchymal progenitors. Stem Cells. 23, 220–9 (2005). [DOI] [PubMed] [Google Scholar]
- Wang H. S. et al. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells. 22, 1330–7 (2004). [DOI] [PubMed] [Google Scholar]
- Fu Y. S., Shih Y. T., Cheng Y. C. & Min M. Y. Transformation of human umbilical mesenchymal cells into neurons in vitro. J Biomed Sci. 11, 652–60 (2004). [DOI] [PubMed] [Google Scholar]
- Tsagias N., Koliakos I., Karagiannis V., Eleftheriadou M. & Koliakos G. G. Isolation of mesenchymal stem cells using the total length of umbilical cord for transplantation purposes. Transfus Med. 21, 253–61 (2011). [DOI] [PubMed] [Google Scholar]
- Wang H. S. et al. Mesenchymal Stem Cells in the Wharton's Jelly of the Human Umbilical Cord. Stem Cells. 22, 1330–7 (2004). [DOI] [PubMed] [Google Scholar]
- Bulj Z. et al. Protein kinase B/AKT isoform 2 drives migration of human mesenchymal stem cells. Int J Oncol. 42, 118–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Marquez-Curtis L. A. & Janowska-Wieczorek A. Mesenchymal stromal cells derived from umbilical cord blood migrate in response to complement C1q. Cytotherapy. 14, 285–95 (2012). [DOI] [PubMed] [Google Scholar]
- Ponte A. L. et al. The in vitro migration capacity of human bone marrow mesenchymal stem cells: comparison of chemokine and growth factor chemotactic activities. Stem Cells. 25, 1737–45 (2007). [DOI] [PubMed] [Google Scholar]
- Martino S., D'Angelo F., Armentano I., Kenny J. M. & Orlacchio A. Stem cell-biomaterial interactions for regenerative medicine. Biotechnol Adv. 30, 338–51 (2012). [DOI] [PubMed] [Google Scholar]
- Buschmann J. et al. Tissue engineered bone grafts based on biomimetic nanocomposite PLGA/amorphous calcium phosphate scaffold and human adipose-derived stem cells. Injury. 43, 1689–97 (2012). [DOI] [PubMed] [Google Scholar]
- McMurray R. J. et al. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 10, 637–44 (2011). [DOI] [PubMed] [Google Scholar]
- McNamara L. E. et al. Nanotopographical control of stem cell differentiation. J Tissue Eng. 2010, 1–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay C. Y. et al. Bio-inspired icropatterned platform to steer stem cell differentiation. Small. 7, 1416–21 (2011). [DOI] [PubMed] [Google Scholar]
- Guilak F. et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 5, 17–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C. M., Wang H. B., Dembo M. & Wang Y. L. Cell movement is guided by the rigidity of the substrate. Biophys J. 79, 144–52 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H., Hidaim H., Hwang D. J., Healy K. E. & Grigoropoulos C. P. The effect of micronscale anisotropic cross patterns on fibroblast migration. Biomat. 31, 4286–95 (2010). [DOI] [PubMed] [Google Scholar]
- Kim D. H. et al. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomat. 30, 5433–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. P., Reinmann A. & Bruinink A. The effect of topographic characteristics on cell migration velocity. Biomat. 27, 5230–41 (2006). [DOI] [PubMed] [Google Scholar]
- Ferrari A., Cecchini M., Degli Innocenti R. & Beltram F. Directional PC12 cell migration along plastic nanotracks. IEEE Trans Biomed Eng. 56, 2692–6 (2009). [DOI] [PubMed] [Google Scholar]
- Mitchel J. A. & Hoffman-Kim D. Cellular scale anisotropic topography guides Schwann cell motility. PLoS One. 6, e24316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim E. K. et al. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomat. 26, 5405–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meucci S., Tonazzini I., Beltram F. & Cecchini M. Biocompatible noisy nanotopographies with specific directionality for controlled anisotropic cell cultures. Soft Matter 8, 1109–1119 (2012). [Google Scholar]
- Ferrari A. et al. Neuronal polarity selection by topography-induced focal adhesion control. Biomat. 31, 4682–94 (2010). [DOI] [PubMed] [Google Scholar]
- Franco D. et al. Control of initial endothelial spreading by topographic activation of focal adhesion kinase. Soft Matter. 7, 7313–24 (2011). [Google Scholar]
- Ferrari A. et al. Nanotopographic control of neuronal polarity. Nano Lett. 11, 505–11 (2011). [DOI] [PubMed] [Google Scholar]
- Di Rienzo C. et al. Unveiling LOX-1 receptor interplay with nanotopography: mechanotransduction and atherosclerosis onset. Sci Rep. 3, 1141–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. & George S. C. Theory and practical recommendations for autocorrelation-based image correlation spectroscopy. J Biomed Opt. 17, 080801–1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby M. J. Nanostructured surfaces: cell engineering and cell biology. Nanomed. (Lond). 4, 247–8 (2009). [DOI] [PubMed] [Google Scholar]
- Wang X., Ohlin C. A., Lu Q. & Hu J. Cell directional migration and oriented division on three-dimensional laser-induced periodic surface structures on polystyrene. Biomat. 29, 2049–59 (2008). [DOI] [PubMed] [Google Scholar]
- Wisniewski N. & Reichert M. Methods for reducing biosensor membrane biofouling. Colloids Surf B Biointerfaces. 18, 197–219 (2000). [DOI] [PubMed] [Google Scholar]
- Nerem R. M. Tissue engineering: confronting the transplantation crisis. Adv Exp Med Biol. 534, 1–9 (2003). [DOI] [PubMed] [Google Scholar]
- Patel N. et al. Spatially controlled cell engineering on biodegradable polymer surfaces. FASEB J. 12, 1447–54 (1998). [DOI] [PubMed] [Google Scholar]
- Matthews J. A., Wnek G. E., Simpson D. G. & Bowlin G. L. Electrospinning of collagen nanofibers. Biomacromolecules. 3, 232–8 (2002). [DOI] [PubMed] [Google Scholar]
- Chiquet M., Gelman L., Lutz R. & Maier S. From mechanotransduction to extracellular matrix gene expression in fibroblasts. Biochim Biophys Acta. 1793, 911–20 (2009). [DOI] [PubMed] [Google Scholar]
- Miyoshi H. et al. Characteristics of motility-based filtering of adherent cells on microgrooved surfaces. Biomat. 33, 395–401 (2012). [DOI] [PubMed] [Google Scholar]
- Yim E. K., Darling E. M., Kulangara K., Guilak F. & Leong K. W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomat. 31, 1299–306 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Lee K. B., Jeon H. S. & Park H. K. Effects of surface nano-topography on human osteoblast filopodia. Anal Sci. 27, 369 (2011). [DOI] [PubMed] [Google Scholar]
- Théry M., Pépin A., Dressaire E., Chen Y. & Bornens M. Cell distribution of stress fibres in response to the geometry of the adhesive environment. Cell Motil Cytoskeleton. 63, 341–55 (2006). [DOI] [PubMed] [Google Scholar]
- Curtis A. & Wilkinson C. Nantotechniques and approaches in biotechnology. Trends Biotechnol. 19, 97–101 (2001). [DOI] [PubMed] [Google Scholar]
- Flemming R. G., Murphy C. J., Abrams G. A., Goodman S. L. & Nealey P. F. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomat. 20, 573–88 (1999). [DOI] [PubMed] [Google Scholar]
- Dalby M. J. et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 6, 997–1003 (2007). [DOI] [PubMed] [Google Scholar]
- Huang J. et al. Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett. 9, 1111–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A. D. & Yamada K. M. Cell biology: Sensing tension. Nature. 466, 192–3 (2010). [DOI] [PubMed] [Google Scholar]
- Dalby M. J. Cellular response to low adhesion nanotopographies. Int J Nanomedicine. 2, 373–81 (2007). [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P. & Bershadsky A. D. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 10, 21–33 (2009). [DOI] [PubMed] [Google Scholar]
- Parsons J. T., Horwitz A. R. & Schwartz M. A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 11, 633–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby M. J. et al. Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp Cell Res. 276, 1–9 (2002). [DOI] [PubMed] [Google Scholar]
- Rørth P. Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev Cell. 20, 9–18 (2011). [DOI] [PubMed] [Google Scholar]
- Ridley A. J. et al. Cell migration: integrating signals from front to back. Science. 302, 1704–9 (2003). [DOI] [PubMed] [Google Scholar]
- Hamilton D. W., Oates C. J., Hasanzadeh A. & Mittler S. Migration of periodontal ligament fibroblasts on nanometric topographical patterns: influence of filopodia and focal adhesions on contact guidance. PLoS One. 5, e15129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLMEMENTARY FIGURES