Abstract
Ewing sarcoma provides an important model for transcription-factor mediated oncogenic transformation because of its reliance on the ETS-type fusion oncoprotein EWS/FLI. EWS/FLI functions as a transcriptional activator and transcriptional activation is required for its oncogenic activity. Here we demonstrate that a previously less-well characterized transcriptional repressive function of the EWS/FLI fusion is also required for the transformed phenotype of Ewing sarcoma. Through comparison of EWS/FLI transcriptional profiling and genome-wide localization data, we define the complement of EWS/FLI direct downregulated target genes. We demonstrate that LOX is a previously undescribed EWS/FLI-repressed target that inhibits the transformed phenotype of Ewing sarcoma cells. Mechanistic studies demonstrate that the NuRD co-repressor complex interacts with EWS/FLI, and that its associated histone deacetylase and LSD1 activities contribute to the repressive function. Taken together, these data reveal a previously unknown molecular function for EWS/FLI, demonstrate a more highly coordinated oncogenic transcriptional hierarchy mediated by EWS/FLI than previously suspected, and implicate a new paradigm for therapeutic intervention aimed at controlling NuRD activity in Ewing sarcoma tumors.
Keywords: Ewing sarcoma, EWS/FLI, NuRD, transcription, repression
INTRODUCTION
Ewing sarcoma provides an attractive model to understand the role of transcriptional regulation in tumor development because of its reliance on a single oncogenic transcription factor, EWS/FLI, created by the t(11;22)(q24;q12) chromosomal translocation (1). Although the function of wild-type EWS is uncertain, the portion of EWS contained in the fusion contributes a strong transcriptional activation domain (2). FLI is an ETS-family transcription factor, and contributes an ETS-type DNA binding domain to the fusion (3).
Early studies suggested that EWS/FLI functions as a transcriptional activator to mediate oncogenic transformation. In support of this model were data demonstrating that the EWS portion of the fusion functions as a strong activation domain relative to the amino-terminal portion of FLI lost in the fusion protein (2), that EWS/FLI binds the RNA polymerase II core subunit, hsRBP7 (4) and co-activators CBP/p300 (5), and that replacement of the EWS portion of the fusion with strong transcriptional activation domains (such as the VP16 transcriptional activation domain) resulted in heterologous fusions that retained full oncogenic activity in NIH3T3 murine fibroblasts (6). Thus, in the NIH3T3 model, EWS/FLI functions mainly as a transcriptional activator, and transcriptional activation was thought to be solely required for its oncogenic function (2, 6).
Subsequent analyses with patient-derived Ewing sarcoma cell lines suggested a possible role for transcriptional repression by EWS/FLI in oncogenic transformation. For example, gene expression profiling after modulation of EWS/FLI levels in Ewing sarcoma cells revealed that many more genes were downregulated by EWS/FLI than upregulated (7, 8). A part of this downregulated signature is due to upregulation of transcriptional repressors, such as NKX2.2 and NR0B1, and is therefore indirectly regulated by EWS/FLI (9, 10). However, other studies suggested that some targets, such as TGFBR2 and IGFBP3, may be directly-repressed by EWS/FLI (8, 11). These data suggested that the dogmatic model of EWS/FLI functioning as a transcriptional activator may be incomplete. However, the full complement of directly-repressed EWS/FLI target genes, the importance of transcriptional repression for the oncogenic function of EWS/FLI, and the mechanistic basis of this repression, remain unknown. We therefore performed experiments to address these questions.
RESULTS
Identification of directly-downregulated targets of EWS/FLI
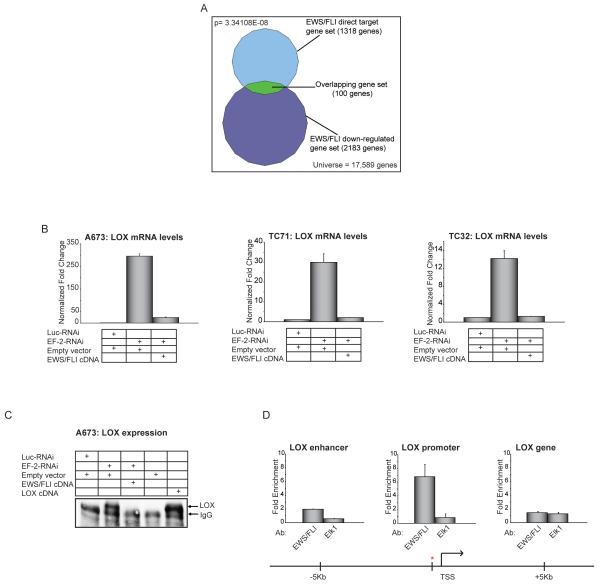
To identify the complement of genes downregulated through direct binding of EWS/FLI, we compared previously published EWS/FLI gene expression data and genome-wide EWS/FLI chromatin immunoprecipitation on microarray (ChIP-Chip) data (9, 12) and found 100 genes that overlapped between the two datasets as potential EWS/FLI direct-downregulated target genes (Figure 1A; Table S1). We validated a subset of these genes with quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) following knock-down of EWS/FLI using the EF-2-RNAi construct (Figure S1A). This gene set included Lysyl Oxidase (LOX), which has been shown to function as a tumor suppressor in a number of cancers, and had not been previously studied in Ewing sarcoma. We therefore analyzed this gene in greater detail, and used Transforming Growth Factor β Receptor II (TGFBR2) as a positive control as it had been previously identified as a direct downregulated target of EWS/FLI (11). Downregulation of LOX and TGFBR2 were not off-target RNAi effects because the gene expression changes mediated by EWS/FLI knockdown were reversed by re-expression of an RNAi-resistant EWS/FLI cDNA in multiple Ewing sarcoma cell lines (Figure 1B; S1B). Changes in LOX mRNA levels were well correlated with LOX protein levels (Figure 1C).
Figure 1. Identification of direct downregulated targets of EWS/FLI.
(A) Venn diagram representation of the overlap between EWS/FLI downregulated genes (by transcriptional profiling) and EWS/FLI direct targets (by ChIP-chip) in A673 Ewing sarcoma cells. The Chi square-determined p-value is indicated.
(B) qRT-PCR validation of EWS/FLI mediated transcriptional repression of LOX in A673, TC71 and TC32 Ewing sarcoma cells following retroviral knockdown of endogenous EWS/FLI (EF-2-RNAi, versus the negative-control Luc-RNAi) and rescue with an RNAi-resistant EWS/FLI cDNA (versus an empty vector control). Error bars indicate standard deviations. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control Luc-RNAi condition, with the data in each condition normalized to an internal housekeeping control gene GAPDH.
(C) Immunoprecipitation-Western blot analysis of LOX protein from A673 cells expressing the indicated RNAi constructs (Luc-RNAi negative-control versus EWS/FLI knockdown with EF-2-RNAi), and the indicated expression vectors (empty vector negative control, EWS/FLI cDNA resistant to the RNAi construct, or a LOX cDNA). The same antibody was used to perform the immunoprecipitation and the Western blotting. The IgG band refers to the heavy chain.
(D) ChIP of EWS/FLI at the LOX locus using antibodies against FLI (which recognizes only EWS/FLI) or ELK1 (negative control). The ChIP experiments were performed in A673 Ewing sarcoma cells which express EWS/FLI but do not have any detectable expression of wild-type FLI (7), and so the anti-FLI antibody used only detects the fusion protein. Thus, there is no competition between the two transcription factors for the same binding sites. The red asterisk indicates the ChIP-Chip identified EWS/FLI binding site at the LOX promoter, and the transcriptional start site (TSS) is indicated. The level of enrichment for EWS/FLI or ELK1 are plotted as fold enrichment compared to the average enrichment of EWS/FLI or ELK1 at two negative control housekeeping genes ALB and BCL2L1 used as normalization controls. Elk1 immunoprecipitation is used as a negative control for the ChIP experiment. Enrichment of EWS/FLI or Elk1 at regions 5 kb upstream and downstream of the ChIP-Chip identified binding site were used as negative controls to further demonstrate binding specificity for EWS/FLI at the LOX promoter. The error bars indicate standard error of the means of five independent experiments.
To determine if repression of LOX and TGFBR2 are directly mediated by EWS/FLI, we performed chromatin immunoprecipitation experiments, and found that EWS/FLI bound both the LOX and TGFBR2 promoters in vivo (Figures 1D; S1C). These data suggest that LOX is a directly-downregulated target of EWS/FLI in Ewing sarcoma and provide independent validation of TGFBR2 as an appropriate control gene for our study.
LOX functions as a tumor suppressor in Ewing sarcoma
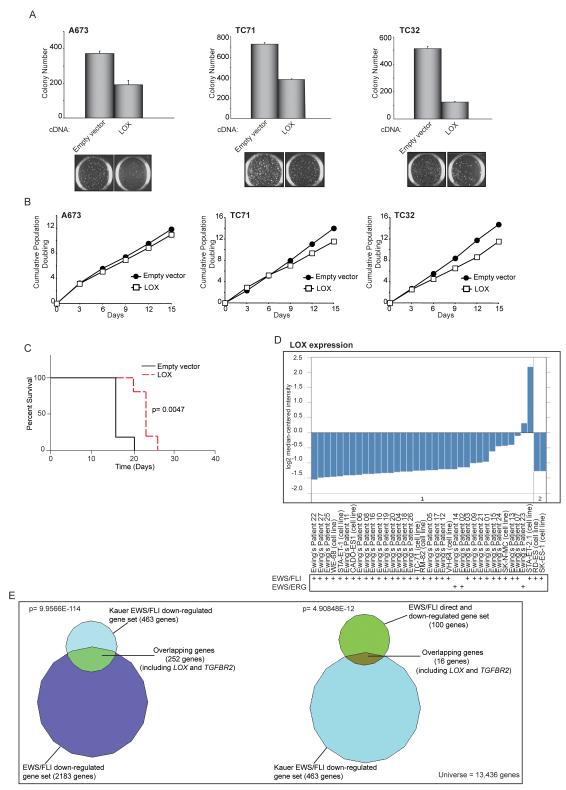
Since LOX was directly bound and downregulated by EWS/FLI, we asked whether it functions as a tumor suppressor in Ewing sarcoma. Forced expression of LOX (or TGFBR2) impaired colony growth in soft agar and in vivo xenograft tumor formation without affecting monolayer growth (Figures 2A-C; S2A-E). In the tumors that did form, LOX or TGFBR2 were still expressed, indicating that the tumors may have adapted to overcome the suppressive effect of each protein (Figure S2F).
Figure 2. LOX functions as a tumor suppressor in Ewing sarcoma.
(A) Quantification of colonies formed in soft agar by A673, TC71 and TC32 cells expressing a 3X-FLAG LOX cDNA construct in comparison to an empty vector control. Error bars indicate standard deviations of duplicate assays. Representative images of soft agar colonies are included.
(B) Growth assays (3T5) for A673, TC71 and TC32 cells described in (A).
(C) Survival curves for immunodeficient mice injected with TC32 cells expressing 3X-FLAG LOX or an empty vector construct. Five mice underwent bilateral subcutaneous injection for each condition, and each animal was sacrificed once one of their tumors reached a 2 cm endpoint. Percent survival was plotted as a Kaplan-Meier survival curve using GraphPad Prism. The log-rank (Mantel-Cox Test) p-value is indicated.
(D) Graphical representation of LOX expression levels in 27 primary Ewing sarcoma patient-derived tumors and 10 Ewing sarcoma cell lines in the Schaefer et al. dataset. The EWS/FLI or EWS/ERG translocation fusion status of each sample is indicated. The reason for the higher levels of LOX expression observed in the STA-ET-2.1 cell line is uncertain, but possibilities include that this cell line may harbor a mutant LOX allele, or perhaps has activated an adaptive “bypass” pathway that allows for growth in the presence of high levels of LOX expression (such as the effect seen in our own xenograft experiments with Ewing sarcoma cells expressing the LOX cDNA (Figures 2C and S2F).
(E) Venn diagram overlaps of EWS/FLI downregulated or EWS/FLI direct downregulated-genes datasets with the Kauer et al. EWS/FLI downregulated-genes dataset. In the Kauer et al. dataset, a molecular function map of Ewing sarcoma was constructed based on an integrative analysis of gene expression profiling experiments following EWS/FLI knock-down in a panel of five Ewing sarcoma cell lines, and 59 primary Ewing sarcoma tumors using mesenchymal progenitor cells (MPC, a suggested cell-of-origin of Ewing sarcoma) as the reference tissue. The Chi-square determined p-values are indicated.
If LOX and TGFBR2 are indeed important EWS/FLI-repressed target genes, then their expression should be relatively low in primary Ewing sarcoma tumor samples. To test this, we analyzed the expression pattern of LOX and TGFBR2 in a published microarray dataset (13). This dataset includes 37 Ewing sarcoma samples consisting of 27 primary tumors and 10 cell lines. Analysis of this dataset revealed low levels of expression of both LOX and TGFBR2 in nearly all of the primary tumor samples analyzed (Figure 2D; Figure S2G). We also evaluated a second published dataset containing 59 Ewing sarcoma primary tumors and five different Ewing sarcoma cell lines (in which endogenous EWS/FLI was knocked-down using an RNAi approach), compared to mesenchymal progenitor cells as the reference tissue (a suggested cell-of-origin of Ewing sarcoma). This dataset also demonstrated low levels of LOX and TGFBR2 in primary Ewing sarcoma tumors (Figure 2E) (14). These data sustain the assertion that repression of LOX and TGFBR2 is important for the development of Ewing sarcoma.
Mapping of the EWS/FLI transcriptional repressive domains
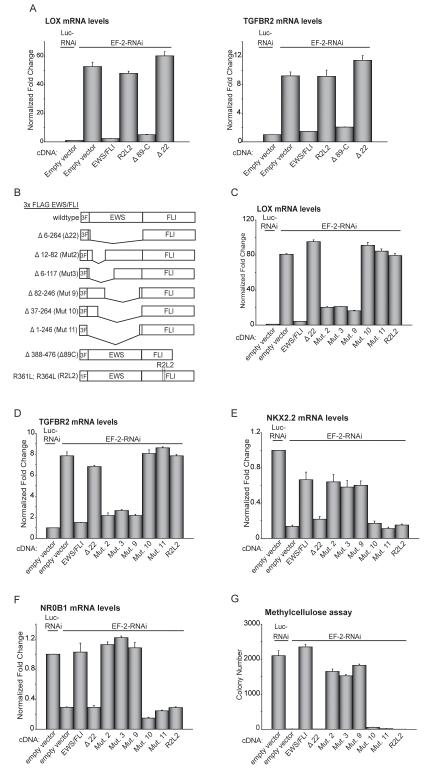
We next sought to identify the key domains required for transcriptional repression mediated by EWS/FLI by knocking-down endogenous EWS/FLI and re-introducing mutant forms of the protein (Figures S3A-S3E), followed by evaluation of endogenous LOX and TGFBR2 expression. We found that the DNA binding mutant R2L2 (15) failed to repress LOX or TGFBR2, but the mutant Δ89-C, lacking the carboxyl-terminal 89 amino acids of EWS/FLI (16) retained full repressive capability at these loci (Figures 3A, 3B, S3A-S3C). In contrast, the Δ22 mutant (17), lacking nearly all of the EWS portion of EWS/FLI, did not repress LOX or TGFBR2 when introduced in place of full-length EWS/FLI (Figures 3A, 3B, S3A-S3C). This indicates that the repressive activity is localized to the EWS portion of EWS/FLI and that DNA binding is required for transcriptional repression at these loci.
Figure 3. Structure-function analysis of EWS/FLI mediated repression.
(A) qRT-PCR analysis of LOX and TGFBR2 expression in A673 cells following knockdown of EWS/FLI (with the EF-2-RNAi construct) and rescue with 3X-FLAG wild-type EWS/FLI, 3X-FLAG Δ22, 1X-FLAG R2L2, 3X-FLAG Δ89C, or an empty vector control. Luc-RNAi is a negative control vector. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control Luc-RNAi condition RNAi, with the data in each condition normalized to an internal housekeeping control gene GAPDH.
(B) Schematic representation of 3X-FLAG (3F) EWS/FLI wild-type and mutant constructs and 1X-FLAG (1F) R2L2 mutant construct. Amino acids deleted/mutated in the EWS and FLI1 domains are indicated.
(C, D) Repression of LOX and TGFBR2, by wild-type EWS/FLI, or mutants, as analyzed by qRT-PCR. EWS/FLI was knocked-down in A673 cells and rescued with the indicated constructs. Error bars indicate standard deviations.
(E, F) Activation of NKX2.2 and NR0B1, by wild-type EWS/FLI, or mutants, as analyzed by qRT-PCR. EWS/FLI was knocked-down in A673 cells and rescued with the indicated constructs. Error bars indicate standard deviations.
(G) Quantification of colonies formed in methylcellulose by A673 cells infected with the indicated RNAi and cDNA constructs. Error bars indicate standard deviations of duplicate assays.
We then used a previously-described panel of deletion mutants to further refine the location of the repression domain within EWS/FLI (Figures 3B; S3C-S3E) (6). Mutants 2, 3, and 9 retained full transcriptional repressive activity, but mutants 10 and 11 failed to repress target genes (Figures 3C, 3D). All deletion mutants tested were appropriately localized to the nucleus and bound DNA in vitro (Figure S3F and data not shown).
These findings suggest the presence of two general regions in the EWS portion of EWS/FLI that mediate repression: an amino-terminal region (amino acids 1-82), and a distal region (amino acids 118-264). This underlying organization was reminiscent of the organization previously found for transcriptional activation in EWS/FLI (6). Consistent with this observation, EWS/FLI mutants that retained transcriptional repressive function also retained the ability to activate the critical EWS/FLI upregulated target genes NKX2.2 and NR0B1, while mutants deficient in transcriptional repression also failed to transactivate these genes (Figures 3E, 3F). Importantly, the mutants that mediate both transcriptional repression and activation also rescue oncogenic transformation of Ewing sarcoma cells following EWS/FLI knockdown, while those that are inactive in repression and activation do not (Figure 3G).
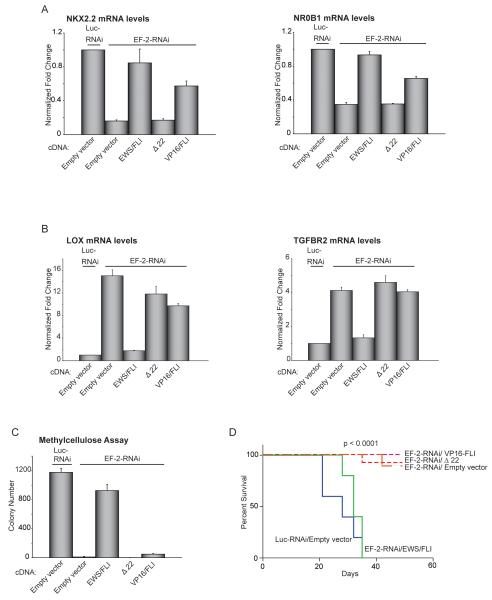
We next wanted to test the contribution of transcriptional repression to EWS/FLI-mediated oncogenesis directly. However, we were unable to identify mutants that separate transcriptional repression and activation functions in the fusion protein. As an alternate approach, we replaced EWS/FLI with a previously described engineered construct, VP16/FLI, that contains two copies of the strong transcriptional activation domain of the VP16 protein fused to the carboxyl-terminal FLI protein (6) (Figure S4A). This construct was previously shown to effectively transactivate a reporter construct and induce oncogenic transformation of NIH3T3 cells (6). VP16/FLI rescued activation of NKX2.2 and NR0B1 nearly as well as EWS/FLI itself (Figure 4A), but was unable to transcriptionally repress LOX or TGFBR2 (Figure 4B). Thus, VP16/FLI functions as a transcriptional activator in Ewing sarcoma cells.
Figure 4. EWS/FLI-mediated transcriptional activation and transcriptional repression are required for oncogenic transformation in Ewing sarcoma cells.
(A, B) qRT-PCR analysis of NKX2.2 and NR0B1, or LOX and TGFBR2, following knock-down of EWS/FLI (with the EF-2-RNAi construct) and rescue with the indicated cDNAs or an empty vector control. Luc-RNAi is a negative control. Error bars indicate standard deviations. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control Luc-RNAi condition, with the data in each condition normalized to an internal housekeeping control gene GAPDH.
(C) Quantification of colonies formed in methylcellulose by A673 cells infected with the indicated RNAi and cDNA constructs. Error bars indicate standard deviations of duplicate assays.
(D) Survival curves for immunodeficient mice injected with control knock-down A673 cells re-expressing empty vector or EWS/FLI knock-down A673 cells re-expressing empty vector, EWS/FLI cDNA, Δ22 or 2x VP16/FLI cDNA constructs. Five mice underwent bilateral subcutaneous injection for each condition, and each animal was sacrificed once one of their tumors reached a 2 cm endpoint. Percent survival was plotted as a Kaplan-Meier survival curve using GraphPad Prism. The log-rank (Mantel-Cox Test) p-value is indicated.
We found that Ewing sarcoma cells expressing VP16/FLI (in lieu of EWS/FLI) could not form in vitro colonies under anchorage-independent conditions, and that in vivo xenograft formation was also reduced, as compared to Ewing sarcoma cells expressing EWS/FLI (Figures 4C, 4D, S4B). Thus, transcriptional activation without transcriptional repression, in the context of a FLI-based DNA binding domain, is insufficient to rescue oncogenic transformation of Ewing sarcoma cells. Thus, transcriptional repression is required for the oncogenic function of EWS/FLI (6, 18-21).
Transcriptional repression by EWS/FLI requires HDAC activity
Given the importance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma oncogenesis, we next sought to determine the mechanistic basis for this function. We previously demonstrated that two critical EWS/FLI target genes, NKX2.2 and NR0B1, act as transcriptional repressors in Ewing sarcoma (9, 10). Neither of these are required for repression of LOX or TGFBR2 as shown by knockdown experiments (Figure S5A).
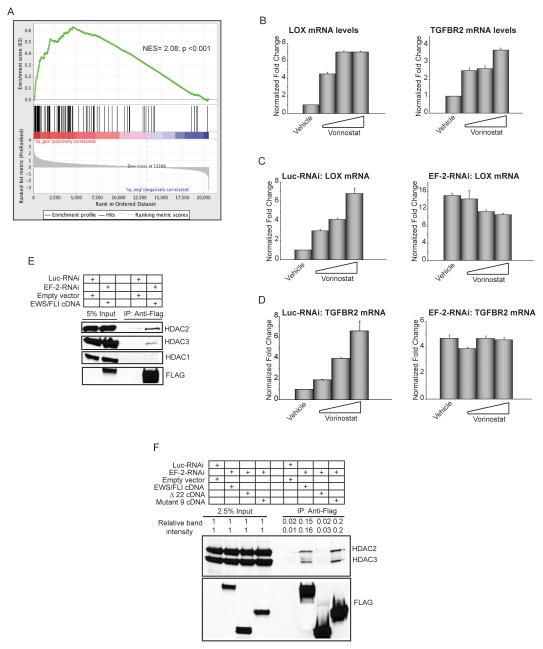
Histone deacetylases (HDACs) play an important role in transcriptional repression mediated by NKX2.2 in Ewing sarcoma (9). To test if HDACs are also involved in EWS/FLI-mediated transcriptional repression, we used gene set enrichment analysis (GSEA) to compare the transcriptional profile of A673 cells treated with the HDAC inhibitor vorinostat (9) to a gene set consisting of the 100 EWS/FLI-bound and downregulated genes (Figure 1A; Table S1). We found that these 100 genes clustered strongly with those genes upregulated by vorinostat (NES=2.08; p<0.001; Figure 5A), demonstrating that the HDAC inhibitor reverses the EWS/FLI-mediated direct transcriptional repressive signature in Ewing sarcoma cells. Interestingly, LOX was most correlated with derepression by vorinostat. These data strongly suggest that HDACs are involved in direct transcriptional repression of LOX and TGFBR2 (and other genes) by EWS/FLI in Ewing sarcoma.
Figure 5. Transcriptional repression by EWS/FLI is mediated by HDACs.
(A) Gene set enrichment analysis (GSEA) using vorinostat-regulated genes in A673 Ewing sarcoma cells as the rank-ordered dataset and the 100 EWS/FLI direct downregulated targets as the geneset. The positions of the 100 genes are indicated as black vertical lines in the center portion of the panel. The normalized enrichment score (NES) and p-value are shown.
(B) qRT-PCR analysis of LOX and TGFBR2 in A673 cells treated with increasing concentrations of the HDAC-inhibitor vorinostat. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control vehicle treated condition, with the data in each condition normalized to an internal housekeeping control gene GAPDH. Error bars indicate standard deviations.
(C, D) qRT-PCR analysis of LOX and TGFBR2 in A673 cells expressing a control RNAi or EWS/FLI RNAi construct (EF-2-RNAi) treated with increasing concentrations of the HDAC inhibitor vorinostat. Error bars indicate standard deviations.
(E, F) Co-immunoprecipitation of EWS/FLI and HDACs. Endogenous EWS/FLI was knocked-down in A673 cells (with the EF-2-RNAi) and replaced with 3X-FLAG-tagged versions of the indicated cDNAs that were resistant to the RNAi construct. Luc-RNAi is a negative control. Relative band intensities were quantified using ImageQuant.
Quantitative RT-PCR analysis demonstrated a dose-dependent increase in the expression of LOX and TGFBR2 following vorinostat treatment (Figure 5B). This effect was absolutely dependent on the expression of EWS/FLI, as the effect was lost following knockdown of EWS/FLI (Figures 5C, 5D), and neither LOX nor TGFBR2 was upregulated by vorinostat in HEK 293 cells (Human embryonic kidney cells that do not express EWS/FLI; Figure S5B). Taken together, these data demonstrate that full transcriptional repression by EWS/FLI requires HDAC activity.
These data also suggested a model in which EWS/FLI interacts with one or more HDACs (either directly, or through binding to an HDAC-containing complex) to mediate transcriptional repression. To test this model, we performed co-immunoprecipitation experiments in A673 cells expressing a 3X-FLAG tagged version of EWS/FLI in place of the wild-type fusion. We found that EWS/FLI co-immunoprecipitates with HDAC2 and HDAC3, but not HDAC1, in Ewing sarcoma cells (Figure 5E). These data were consistent with siRNA experiments that showed that knock-down of HDAC2 or HDAC3, but not HDAC1, derepressed both LOX and TGFBR2 (Figures S5C, S5D).
If HDAC2 and/or HDAC3 were truly involved in EWS/FLI-mediated transcriptional repression, we would predict that these proteins would interact with EWS/FLI mutants that mediate transcriptional repression, but not with inactive mutants. To test this, we knocked-down endogenous EWS/FLI in Ewing sarcoma cells and rescued with RNAi-resistant cDNAs encoding wild-type EWS/FLI, the Δ22 mutant (that does not mediate transcriptional repression), or mutant 9 (that retains transcriptional repression activity). We found that wild-type EWS/FLI and mutant 9 both bind HDAC2 and HDAC3, while the Δ22 mutant does not (Figure 5F). This correlation between transcriptional repression and HDAC binding suggests that repression may be mediated via EWS/FLI-bound HDACs at specific genomic loci. Of interest, we previously found that vorinostat-mediated HDAC blockade also disrupts the growth of Ewing sarcoma cells in tissue culture, and the ability of those cells to form colonies in soft-agar assays (9).
NuRD interacts with EWS/FLI to mediate transcriptional repression
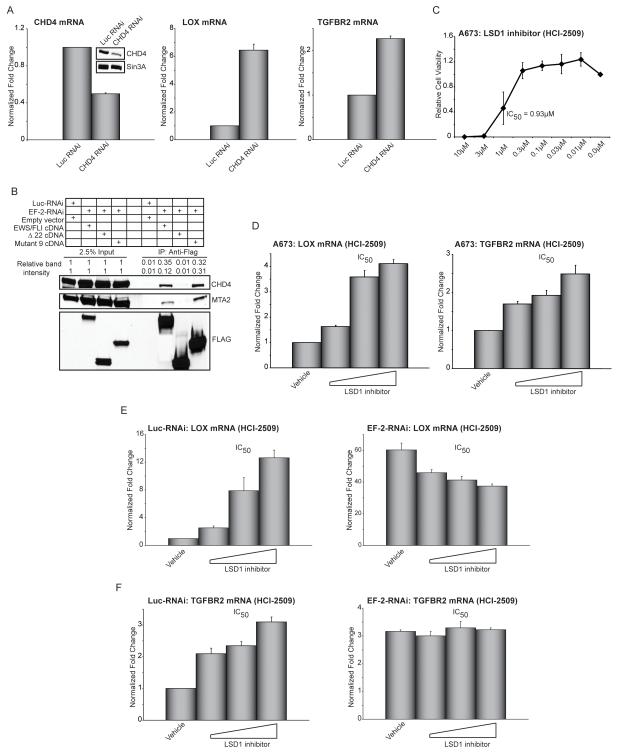
HDAC2 and HDAC3 are class I HDACs. Class I HDACs serve as catalytic subunits for various multiprotein transcriptional repressor complexes, including Sin3A, NuRD, NCoR/SMRT and CoREST (see Figure 7 for complex composition) (22). To test if any of these complexes are involved in EWS/FLI-mediated transcriptional repression, we asked whether depletion of any of these complexes (using RNAi) resulted in derepression of LOX and/or TGFBR2. Although we achieved good reduction of both RNA and protein levels for Sin3A, REST, and NCoR/SMRT in A673 cells, there were no changes in LOX or TGFBR2 expression with any of these manipulations (Figures S6A-S6C). In contrast, ~50% knockdown of the NuRD complex component Chromodomain Helicase DNA-Binding Protein 4 (CHD4) resulted in significant upregulation of both LOX and TGFBR2 (Figure 6A). These data suggest that the NuRD complex is involved in EWS/FLI-mediated transcriptional repression.
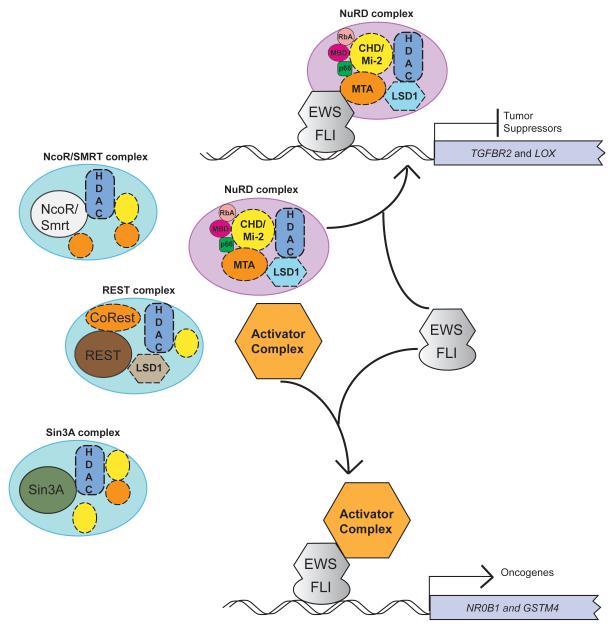
Figure 7. Binary switch model for EWS/FLI mediated transcriptional regulation.
At directly repressed genes, such as LOX and TGFBR2, EWS/FLI may preferentially recruit transcriptional repressor complexes, such as the NuRD complex with its associated HDACs and LSD1, to transcriptionally inhibit gene expression. At other repressed loci, other repressor complexes could be recruited. In contrast, at directly activated genes, such as NR0B1 and GSTM4, EWS/FLI may preferentially recruit (yet to be determined) activator complexes to transcriptionally upregulate gene expression. The mechanism by which preferential recruitment occurs is not yet known.
Figure 6. EWS/FLI interacts with members of the NuRD co-repressor complex.
(A) qRT-PCR analysis of CHD4, LOX, and TGFBR2 in A673 cells following knock-down of the NuRD component CHD4. Western blot analysis to demonstrate efficiency of CHD4 knockdown, Sin3A was used as the loading control. Error bars indicate standard deviations. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control Luc-RNAi condition, with the data in each condition normalized to an internal housekeeping control gene GAPDH.
(B) Co-immunoprecipitation of 3X-FLAG EWS/FLI, or the indicated mutants, and NuRD complex components CHD4 and MTA2. Relative band intensities were quantified using ImageQuant.
(C) Relative cell viability of A673 cells treated with increasing concentrations of the LSD1 inhibitor (HCI-2509). The IC50 is indicated. Error bars indicate standard deviations.
(D) qRT-PCR analysis of LOX and TGFBR2 following 72 hours of treatment with increasing concentrations of the LSD1 inhibitor (HCI-2509). The dose corresponding to the IC50 is indicated. Error bars indicate standard deviations.
(E, F) qRT-PCR analysis of LOX and TGFBR2 in A673 cells expressing Luc-RNAi (negative control) or EWS/FLI RNAi (EF-2-RNAi) following 72 hours of treatment with increasing concentrations of the LSD1 inhibitor HCI-2509. The dose corresponding to the IC50 is indicated. Error bars indicate standard deviations.
The simplest model to explain these findings is that EWS/FLI binds the NuRD complex directly. We found that full-length EWS/FLI and the repressive mutant 9 allele bind both CHD4 and MTA2 NuRD complex components in co-immunoprecipitation experiments, while the non-repressive Δ22 protein does not (Figure 6B). These data support the hypothesis that EWS/FLI interacts (directly or indirectly) with the NuRD complex to mediate transcriptional repression at loci such as LOX and TGFBR2.
The NuRD complex also contains the LSD1 histone demethylase (23). To determine if LSD1 activity is required for transcriptional repression mediated by EWS/FLI, we used two recently-described small molecule reversible inhibitors of LSD1 (HCI-2509 and HCI-2528) (Venkataswamy et al., submitted). Both of these inhibitors decreased the viability of A673 cells (IC50 of 0.93 μM and 0.47 μM respectively) and upregulated both LOX and TGFBR2 (Figures 6C, 6D; Figures S6D, S6E). Similar results were observed in three other Ewing sarcoma cell lines (Figures S6F-S6H). De-repression of LOX and TGFBR2 was absolutely dependent on the expression of EWS/FLI, as the effect was lost following knockdown of the fusion protein (Figures 6E, 6F), and was not observed in the EWS/FLI-deficient cell line HEK 293 (Figure S6I). Dose-dependent increases in expression of both LOX and TGFBR2 with siRNA knock-down of LSD1 in A673 cells confirmed the specificity of the effect observed with the small-molecule inhibitors (Figure S6J). Taken together, these data highlight a central role for the NuRD complex in the development of Ewing sarcoma and indicate a possible therapeutic strategy for targeting a critical transcriptional function of EWS/FLI via NuRD/LSD1/HDAC blockade.
DISCUSSION
Early studies using heterologous systems suggested that EWS/FLI functions as a transcriptional activator to mediate oncogenic transformation. Later data from Ewing sarcoma patient-derived cell lines suggested that EWS/FLI might also function as a transcriptional repressor. However, the mechanism of transcriptional repression by EWS/FLI and the role of repression in oncogenesis have been uncertain. We have now demonstrated that EWS/FLI functions as both a transcriptional activator, and as a transcriptional repressor, in Ewing sarcoma. Importantly, we demonstrated that the repressive function of EWS/FLI is absolutely required for the oncogenic function of the fusion. Finally, we showed that transcriptional repression by EWS/FLI is mediated through direct binding of the NuRD complex, and that the NuRD-associated histone deacetylase and LSD1 functions are key components of this activity.
These data have important implications for our understanding of Ewing sarcoma development. First, these data once again indicate that the NIH3T3 model does not recapitulate critical features of Ewing sarcoma oncogenesis, because in that model, transcriptional activation by EWS/FLI was sufficient for oncogenic transformation (2, 6). A recent study indicated that some ETS family members mimic the RAS/MAPK pathway by activating genes that are regulated by this oncogenic pathway (24). It is possible that EWS/FLI upregulates a similar set of genes in NIH3T3 cells, but a distinct set in Ewing sarcoma. Indeed, transcriptional profiling studies support the notion that EWS/FLI regulates different genes in each cell type (18, 20).
The second important implication is that in addition to key upregulated genes, such as NR0B1, NKX2.2, CAV1, GSTM4, etc., the downregulated transcriptional signature is also of critical importance to the development of Ewing sarcoma. In support of this argument, we validated previous data suggesting an important role for inhibition of TGFBR2 in Ewing sarcoma development, and extended these data by demonstrating a critical role for the inhibition of LOX expression as well. Interestingly, LOX has been suggested to function as a tumor suppressor, or as an oncogene, depending on the cellular context (25). In Ewing sarcoma, our data indicate its tumor suppresive function dominates. The ability of EWS/FLI to inhibit the expression of tumor suppressors in Ewing sarcoma suggests that mutation of these genes may be unnecessary for the development of this tumor. Indeed, relatively few genes have been identified that are mutationally inactivated in Ewing sarcoma (26). We speculate that direct transcriptional repression of tumor suppressors by EWS/FLI diminishes the need for mutational inactivation.
Third, binding of NuRD to EWS/FLI demonstrates that the transcriptional repressive function of EWS/FLI is both direct and active. We previously demonstrated that some EWS/FLI upregulated targets, such as NKX2.2 and NR0B1, function as transcriptional repressors in Ewing sarcoma (9, 10). Transcriptional repression of some genetic loci is therefore “indirect” (i.e., EWS/FLI upregulates transcriptional repressors, and these repressors mediate inhibition at other loci). However, part of the transcriptional repressive signature of EWS/FLI is directly mediated by the fusion protein itself. Further work will be required to determine the relative contributions of direct versus indirect transcriptional repression.
Additionally, a direct contribution by NuRD and its associated HDACs and LSD1 indicate an “active” transcriptional repressive mechanism mediated by EWS/FLI. Such a mechanism stands in contrast to “passive” mechanisms of transcriptional repression, such as non-productive competition for ETS-family DNA binding sites by EWS/FLI, or “squelching” mechanisms (where EWS/FLI could compete for limiting transcriptional co-activators, thereby preventing these proteins and complexes from being recruited to some target loci). Furthermore, the finding that de-repression of LOX and TGFBR2 with an HDAC inhibitor or the LSD1 inhibitor in Ewing sarcoma cells was completely dependent on EWS/FLI expression further supports the ‘direct” repression model at these promoters. Identification of an “active” transcriptional repressive mechanism for EWS/FLI does not negate the possibility for passive repression at some loci. Further work would be required to determine whether such passive mechanisms are operative, and if so, what their relative contributions are to EWS/FLI-mediated oncogenesis in Ewing sarcoma. It is also worth noting that even though the NuRD complex typically contains both HDAC1 and HDAC2, we instead found HDAC2 and HDAC3 associated with EWS/FLI and required for repression of LOX and TGFBR2. This suggests that there may be EWS/FLI-associated sub-complexes of the NuRD co-repressor in Ewing sarcoma cells that contains HDAC3 instead of HDAC1.
One question that arises from this work is how the choice between EWS/FLI-mediated transcriptional activation versus transcriptional repression is made at specific genetic loci. Our finding that the activation and repressive functions of the fusion protein are currently inseparable suggests that a binary-switch model might be operative (Figure 7). For example, at some loci EWS/FLI would bind transcriptional activators (such as p300/CBP), while at other loci the fusion will bind NuRD (or even other repressors at other loci). This decision could be dictated by the specific EWS/FLI response element present, nearby bound transcriptional regulators, and/or the local chromatin environment. Interestingly, we previously demonstrated that EWS/FLI binds GGAA-microsatellite elements near genes that are transcriptionally activated by the oncoprotein, and that GGAA-microsatellites are enriched near EWS/FLI-upregulated genes, but are depleted near downregulated genes (12). Thus, the GGAA-microsatellite appears to serve as an EWS/FLI-activating response element. Similarly, using an unbiased motif identification tool (MEME) we were unable to identify any significant enrichment of the consensus high-affinity ETS site at the EWS/FLI direct-repressed gene promoters (data not shown). Most members of the ETS family bind to DNA at sequences containing either a GGAA or GGAT core sequence (3). At the EWS/FLI direct repressed gene promoters analyzed there were several such potential ETS core motifs with flanking sequences that resembled low-affinity binding sites for other transcription factors (composite sites). It is tempting to speculate that such composite sites function as in vivo EWS/FLI binding elements at direct repressed genes. In support of this concept are genome-wide localization studies that have identified two classes of in vivo ETS-binding sites, (1) high-affinity binding sites and (2) lower-affinity binding sites found in close proximity to low affinity binding sites for other transcription factors, allowing for co-operative binding and regulation (27). Our current data do not address whether a specific EWS/FLI-repressive response element exists in the promoter or enhancer regions of downregulated target genes. Additional studies will be required to characterize the EWS/FLI binding sites at repressed genes.
Finally, the importance of EWS/FLI-mediated transcriptional repression to the oncogenic phenotype of Ewing sarcoma suggests that inhibition of the repressive function might be a therapeutic strategy for this disease. Indeed, the contribution of HDACs and LSD1 to the repressive function suggests an immediate avenue to exploit this strategy via the administration of HDAC and/or LSD1 inhibitors to patients with Ewing sarcoma. HDAC inhibition has already been shown to block Ewing sarcoma cell growth, transformation, and survival in ex vivo settings (9). In vivo, HDAC inhibitors have shown efficacy in some preclinical xenograft models (28) but not in others (29). Our data show that LSD1 specific inhibitors also block growth and survival of multiple Ewing sarcoma patient-derived cell lines. Of note, a recent report documented LSD1 expression in Ewing sarcoma tumors, and demonstrated that inhibition of LSD1 activity (with a monoamine oxidase inhibitor) blocked Ewing sarcoma cell growth in tissue culture (30). LSD1 inhibition in xenograft models has yet to be reported. It is possible that combination approaches would be useful, such as a combination of HDAC and LSD1 inhibitors, or combinations of these inhibitors with other established therapeutic approaches (such as chemotherapy). Additional preclinical studies are warranted to test this hypothesis.
In summary, we have shown that EWS/FLI functions as a transcriptional repressor, and that this function is also critical for oncogenic transformation mediated by this fusion oncoprotein. This provides a mechanistic explanation for recent transcriptional profiling data and the occasional report of putative EWS/FLI direct-downregulated genes. These data also provide a biochemical rationale for the evaluation of NuRD inhibitors (such as HDAC and LSD1 inhibitors) as a new therapeutic approach for Ewing sarcoma.
MATERIALS AND METHODS
Constructs and Retroviruses
The Luc-RNAi, EF-2-RNAi, EWS/FLI, Δ22, R2L2, mutants 2, 3 and 9 have been described previously (6, 7, 15). The mutants were 3X-FLAG tagged, R2L2 was 1X-FLAG tagged and sub-cloned into the MSCV retroviral system (Clontech). 3X-FLAG mutants 10 and 11 were generated by PCR and sub-cloned into the MSCV vector. The 2x-VP16/FLI construct in the SRα retroviral vector was previously described (6). 3X-FLAG cDNAs of LOX and TGFBR2 were generated and cloned into the MSCV retroviral vector. REST shRNA was designed and cloned into the pMKO.1 retroviral vector. SMARTpool siRNAs targeting NcoR/SMRT, HDAC1, HDAC2, HDAC3 and LSD1 were purchased from Dharmacon.
Cell culture
HEK 293EBNA cells and Ewing sarcoma cell lines (A673, TC71, TC32 and SK-N-MC) were infected with retrovirus, and polyclonal populations were grown in the appropriate selection media, as previously described (31, 32). Growth assays (3T5) were performed as previously described (31).
Soft agar and Methylcellulose assays
Soft agar assays were performed as described previously (31). Methylcellulose assays were performed by seeding 1 × 105 cells per 6 cm plate in the absence or presence of appropriate antibiotic selection media in 1% final concentration of methylcellulose.
Xenograft assay
TC32 cells infected and selected with an empty vector, LOX or TGFBR2 cDNA were injected into the flanks of nude mice at 1 × 106 cells per flank. A673 cells infected with a control or EWS/FLI RNAi and re-expressing an empty vector, RNAi-resistant EWS/FLI, Δ22 or 2x VP16/FLI cDNA constructs were selected and injected into the flanks of nude mice at 5 × 105 cells per flank. Five mice were used per condition. Tumors were measured using digital calipers and 3-dimensional tumor volumes were calculated using the equation (Length × Width × Depth)/2. Survival curve were plotted using GraphPad Prism. Animal experiments were performed following approval from the University of Utah Institutional Animal Care and Use Committee.
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
Total RNA from cells was amplified and detected using SYBR green fluorescence for quantitative analysis (31). Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control condition (either Luc-RNAi or empty vector), with the data in each condition normalized to an internal housekeeping control gene GAPDH. Primer sequences used for qRT-PCR analysis for all target genes are included in the supplementary information (Table S2).
Antibodies
The following antibodies were used for immunodetection: M2-anti-FLAG (HRP; Sigma A8592), anti-FLI-1 (Santa-Cruz sc-356X), anti-α-Tubulin (Calbiochem CP06), anti-HDAC1 (Santa-Cruz sc-7872), anti-HDAC2 (Santa-Cruz sc-7899), anti-HDAC3 (Santa-Cruz sc-11417), anti-CHD4/Mi2 beta (BETHYL Laboratories Inc. A301-081A), anti-MTA-2 (Abcam A8106), anti-LOX (Santa-Cruz sc-66947), anti-EWS (Santa-Cruz sc-48404), anti-REST (Santa-Cruz sc-374611), anti-NcoR (Abcam ab80856), anti-LSD1 (Cell Signaling C69G12), anti-NKX2.2 (Santa-Cruz sc-15015) and anti-NR0B1 (Abcam ab24552).
Directed ChIP
Directed ChIPs were performed as previously described (27) using anti-FLI-1 and anti-ELK-1 antibodies (sc-356X and sc-355 respectively; Santa Cruz Biotechnology, Inc.). Quantitative RT-PCR was performed with LOX, TGFBR2 primers and with ALB and BCL2L1 as normalization controls (12). See Table S2 for primer sequences.
Electrophoretic mobility shit assay (EMSA)
Nuclear extracts were prepared from 293EBNA cells transfected with 3X-FLAG wild-type EWS/FLI, 3X-FLAG deletion mutants of EWS/FLI, or empty vector control expression plasmids. Twenty milligrams of nuclear extract protein, 5 nM [32P]-labeled probe called DNA duplex (I), containing a high-affinity EWS/FLI-binding site, called “ETS2 probe” (2), one hundred fold excess (500nM) of specific unlabeled competitor DNA duplex (I) and 1× Gel Shift Binding Buffer (Promega Corporation) were used in each reaction to determine specific binding. For details refer to (12).
Co-immunoprecipitation assays
A673 cells infected with the Luc-RNAi or EF-2-RNAi construct were transduced with an empty vector or 3X-FLAG EWS/FLI cDNA. Nuclear extracts were prepared. Co-immunoprecipitation experiments were conducted as previously described (33) using anti-FLAG-M2-Magnetic Beads (Sigma M8823).
Cell Viability Assay
ATPlite from PerkinElmer was used to determine cell viability. Ewing sarcoma cell lines were seeded in 384-well plates (1,000 cells per well) and treated with different concentrations of the inhibitors (0.1% final DMSO concentration). After 96-hours of incubation, an equal volume of ATPlite was added directly to the culture well. Luminescence was read 5 minutes later on an Envision plate reader. Viability was calculated relative to untreated cells, and IC50 values were calculated using GraphPad Prism.
Microarray and ChIP-Chip analysis
Overlaps between the different gene sets were performed using the Venn Master program (http://www.informatik.uni-ulm.de/ni/mitarbeiter/HKestler/vennm/doc.html). Statistical significance of the overlaps was determined using Chi square analysis. Gene set enrichment analysis (GSEA) was performed using GSEA2.07 program (http://www.broad.mit.edu/gsea/) (34). Analysis of the Schaefer sarcoma dataset was done using ONCOMINE (https://www.oncomine.com/resource/login.html).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Cairns, Engel and Bhaskara for discussions and critical reading of this manuscript and members of the Lessnick laboratory, and Drs. Denny and Ayer, for discussions and reagents. SS acknowledges support from the HHMI Med into Grad program at the University of Utah (U2M2G). This work was supported by NIH/NCI grants R01 CA140394 (to SLL) and P30 CA042014 (to Huntsman Cancer Institute) and funding from Imaging Diagnostics and Therapeutics Program at Huntsman Cancer Institute (to Sunil Sharma).
Footnotes
CONFLICT OF INTEREST
Sorna Venkataswamy, Hariprasad Vankayalapati, Steven L. Warner, Sunil Sharma, and David J. Bearss filed United States patent application 61/523,801 on 08/15/2011: Substituted (E)-N’-(1-Phenylethylidene)Benzohydrazide Analogs as Histone Demethylase Inhibitors.
Supplementary information is available at Oncogene’s website.
REFERENCES
- 1.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Petermann R, Mossier BM, Aryee DN, Khazak V, Golemis EA, Kovar H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 5.Ramakrishnan R, Fujimura Y, Zou JP, Liu F, Lee L, Rao VN, et al. Role of protein-protein interactions in the antiapoptotic function of EWS-Fli-1. Oncogene. 2004;23:7087–7094. doi: 10.1038/sj.onc.1207927. [DOI] [PubMed] [Google Scholar]
- 6.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 7.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS ONE. 2008;3:e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinsey M, Smith R, Iyer AK, McCabe ER, Lessnick SL. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing’s sarcoma. Cancer Res. 2009;69:9047–9055. doi: 10.1158/0008-5472.CAN-09-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG, et al. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet. 1999;23:222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- 12.Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer KL, Eisenacher M, Braun Y, Brachwitz K, Wai DH, Dirksen U, et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. European journal of cancer. 2008;44:699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Kauer M, Ban J, Kofler R, Walker B, Davis S, Meltzer P, et al. A molecular function map of Ewing’s sarcoma. PLoS ONE. 2009;4:e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailly RA, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, et al. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arvand A, Welford SM, Teitell MA, Denny CT. The COOH-terminal domain of FLI-1 is necessary for full tumorigenesis and transcriptional modulation by EWS/FLI-1. Cancer Res. 2001;61:5311–5317. [PubMed] [Google Scholar]
- 17.May WA, Gishizky ML, Lessnick SL, Lunsford LB, Lewis BC, Delattre O, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle. 2006;5:2753–2759. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- 19.Gangwal K, Lessnick SL. Microsatellites are EWS/FLI response elements: genomic “junk” is EWS/FLI’s treasure. Cell Cycle. 2008;7:3127–3132. doi: 10.4161/cc.7.20.6892. [DOI] [PubMed] [Google Scholar]
- 20.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7:250–256. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 21.Owen LA. Lessnick SL. Identification of Target Genes in Their Native Cellular Context: An Analysis of EWS/FLI in Ewing’s Sarcoma. Cell Cycle. 2006;5:2049–2053. doi: 10.4161/cc.5.18.3213. [DOI] [PubMed] [Google Scholar]
- 22.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nature reviews Genetics. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 24.Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, Graves BJ. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes & development. 2011;25:2147–2157. doi: 10.1101/gad.17546311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nature reviews Cancer. 2012;12:540–552. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- 26.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaboin J, Wild J, Hamidi H, Khanna C, Kim CJ, Robey R, et al. MS-27-275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002;62:6108–6115. [PubMed] [Google Scholar]
- 29.Keshelava N, Houghton PJ, Morton CL, Lock RB, Carol H, Keir ST, et al. Initial testing (stage 1) of vorinostat (SAHA) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2009;53:505–508. doi: 10.1002/pbc.21988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennani-Baiti IM, Machado I, Llombart-Bosch A, Kovar H. Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing’s sarcoma, osteosarcoma, and rhabdomyosarcoma. Human pathology. 2012;43:1300–1307. doi: 10.1016/j.humpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 32.Kinsey M, Smith R, Lessnick SL. NR0B1 Is Required for the Oncogenic Phenotype Mediated by EWS/FLI in Ewing’s Sarcoma. Mol Cancer Res. 2006;4:851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nature protocols. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.