Abstract
Two classes of glutamate-activated channels mediate excitation at central synapses: N-methyl-d-aspartic acid (NMDA) receptors and non-NMDA receptors. Despite substantial structural homology, each class generates signals with characteristic kinetics and mediates distinct synaptic functions. In non-NMDA receptors, the strength of inter-subunit contacts within agonist-binding domains is inversely correlated with functional desensitization. Here we test how the strength of these contacts affects NMDA receptor activation by combining mutagenesis and single-channel current analyses. We show that receptors with covalently linked dimers had dramatically lower activity due to high barriers to opening and unstable open states but had intact desensitization. Based on these observations, we suggest that in NMDA receptors rearrangements at the heterodimer interface represent an early and integral step of the opening sequence but are not required for desensitization. These results demonstrate distinct functional roles in the activation of NMDA and non-NMDA glutamate-gated channels for largely conserved inter-subunit contacts.
INTRODUCTION
For all glutamate-gated channels, agonist-binding in the cleft formed by two dynamic lobes, D1 and D2, of ligand-binding domains (LBDs) is the event that energizes resting receptors to either open or desensitize1,2. The present model for glutamate receptor gating postulates that the mechanical tension produced by agonist-induced cleft-closure can be relieved either by pulling on connected pore-lining helices, which results in channel opening, or by disrupting the D1-D1 intersubunit interface within each LBD dimer, which results in channel desensitization3–5. Consistent with this model, in AMPA- and kainate- sensitive glutamate receptors, perturbations of D1-D1contacts that strengthen the dimer interface prevent desensitization whereas those that weaken it facilitate desensitization6–10. By analogy, dimer separation is assumed to represent the physical substrate of NMDA receptor desensitization11; however, direct evidence is lacking.
To test the functional role of a strong heterodimer interface in NMDA receptor gating we set out to delineate how perturbing this interface affects the activation mechanism of NMDA receptors. Furukawa and colleagues showed that in the isolated GluN1/GluN2A LBD heterodimer, N521 and L777 of GluN1 are in close proximity with L780 and E516 of GluN2A, respectively, and substituting pairs of cysteine residues at these four positions in full length GluN1/GluN2A receptors (Mt1/Mt1), results in cell surface receptors that are spontaneously cross-linked but functional, and whose activity is augmented by DTT treatment12. A separate study showed that cross-linked Mt1/Mt1 receptors are resistant to allosteric inhibition and become more sensitive than wild-type receptors upon DTT-treatment, which suggested the possibility that cross-linked receptors cannot desensitize11. To more clearly define the role of intersubunit interactions within NMDA receptor LBDs, we examined the activation mechanism of Mt1/Mt1 receptors in oxidizing and reducing conditions.
Here we find that stabilizing the LBD dimer with disulfide bridges deeply impairs the channel opening reaction but has no effect on desensitization. Conversely, restoring flexibility across the heterodimer interface by reducing the engineered inter-subunit bonds allows Mt1/Mt1 receptors to reach open states that have native-like stabilities. Based on these results, we propose that the NMDA receptor activation sequence but not its desensitization involves the relative repositioning of subunits within ligand-binding domains and we suggest that the intra molecular interfaces of NMDA and non-NMDA receptors may have distinct functional roles.
RESULTS
Cross-linked receptors produce desensitizing currents
Consistent with previous studies11,12, we found that Mt1/Mt1 receptors expressed in HEK293 cells were functional, their activity was strongly potentiated by DTT treatment, and DTT treatment effectively induced dimer dissociation (Figure 1a-c and Supplementary Figure S1). However, contrary to the presumption that cross-linking prevents desensitization, whole-cell currents desensitized with kinetics comparable to those of wild-type (Wt/Wt) receptors, and reducing agents (DTT 10 mM, 2 min) strongly potentiated current amplitudes and reduced macroscopic desensitization (Figure 1b). These effects were fully reversible along several reducing/oxidizing cycles (Figure 1d), indicating that the majority of receptors reaching the membrane were in oxidized form and that the DTT-induced changes in current amplitude and kinetics resulted from increased flexibility at the dimer interface.
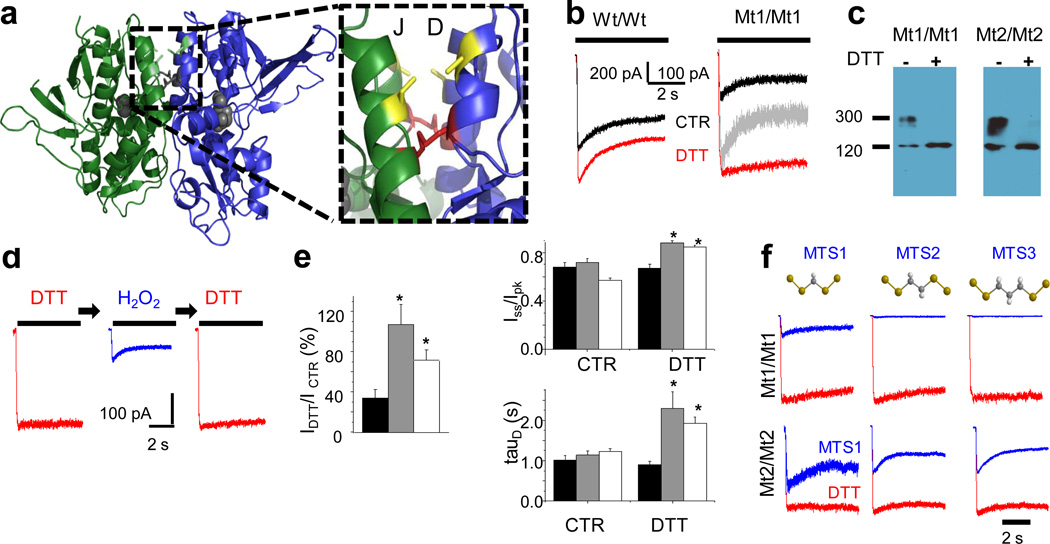
Figure 1. Macroscopic properties of Mt1/Mt1 receptors.
a, Crystal structure of the GluN1/GluN2A LBD heterodimer (PDB: 2A5T) illustrates the residues changed to cysteines in this study. Mt1/Mt1 receptors have cysteines (red) at N521 and L777 of GluN1, and E516 and L780 of GluN2A. Mt2/Mt2 has cysteines (yellow) at Q525 and L774 of GluN1 and E520 and L777 of GluN2A. b, Whole-cell currents recorded from HEK 293 cells expressing Wt/Wt or Mt1/Mt1 receptors in control (CTR, black) and reducing (DTT, red) conditions. CTR normalized to peak of DTT response is in grey. c, Western blot of proteins solubilized from HEK cells expressing Mt1/Mt1 or Mt2/Mt2 receptors and probed with an anti-GluN1 antibody illustrate a DTT-sensitive, high molecular weight band indicative of cross-linked heterodimer formation. d, Kinetics of Mt1/Mt1 receptors are reversibly modulated by reducing (DTT, red)/oxidizing (H2O2, blue) treatment cycles. e, Summary of whole-cell current properties (mean ± s.e.m.) for Wt/Wt (black, n=6), Mt1/Mt1 (grey, n=12), and Mt2/Mt2 (white, n=19), asterisk indicates P<0.05 relative to CTR (Student’s t-test). f, Traces recorded from DTT-treated Mt1/Mt1 or Mt2/Mt2 (red) and after exposure to bifunctional cross-linking reagents (blue).
To determine whether these effects were specific to the positions used to covalently tether the GluN1 and GluN2A subunits, we tested a second mutant whose dimer interface was reinforced with disulfide bridges between Q525 and L774 of GluN1, and E520 and L777 of GluN2A, respectively (Mt2/Mt2, Figure 1a). These residues are located one helix-turn higher along the D and J helices and face each other directly across the heterodimer interface12. In these mutants, the formation of disulfide bridges across subunits would require protomers to move closer together than illustrated by the structural model of the GluN1/GluN2A dimer LBD fragment, but would preserve the relative orientation of protomers within the dimer. Similar to Mt1/Mt1 receptors, Mt2/Mt2 receptors produced normally desensitizing currents and DTT strongly potentiated peak current amplitude (Ipk) and reduced desensitization (Iss/Ipk and τD, Figure 1e). These results indicate that the observed loss of activity originated from a loss of flexibility across the dimer-interface and was not residue-specific. This loss of flexibility did not visibly affect macroscopic desensitization as determined by comparing mutant with wild-type whole-cell current traces. Contrary to expectations, restoring flexibility at the dimer interface by treating either mutant with DTT caused currents to desensitize less and with slower kinetics.
Next, we used bifunctional cross-linkers of several lengths to test whether receptor activation required intersubunit proximity or just interface flexibility. A disulfonate reagent (MTS1), which when bound would allow a maximal separation of ~4.7 Å between the engineered cysteines, decreased whole-cell currents recorded from DTT-treated Mt1/Mt1 receptors to the same extent as direct cross-linking (disulfide bond, ~2 Å). MTS reagents with longer linkers (5.7 Å for MTS2 and 7.1 Å for MTS3) prevented activation even more severely (Figure 1f). Consistently, MTS tethering of subunits produced faster desensitizing macroscopic currents. These results are in contrast with the observation that MTS-tethering of subunits within dimers potentiate current amplitudes and reduce desensitization in AMPA receptors7. Together they indicate that the dimer interface may play a distinct role in NMDA receptor activation, one that requires both protomer proximity and a flexible interface. To investigate this novel hypothesis, we examined the reaction mechanism of Mt1/Mt1 receptors at the single-molecule level.
Cross-linked Mt1/Mt1 receptors open infrequently and briefly
We recorded on-cell single-channel currents from Mt1/Mt1 receptors with high agonist concentrations in the recording pipette (1mM glutamate and 0.1 mM glycine), condition which causes the receptors to be essentially fully liganded (n = 6). In all records, Mt1/Mt1 channels displayed bursting behavior, a direct indication that channels entered desensitized states despite covalent tethering of the dimer interface. Additionally, we observed that overall channel activity was drastically reduced (Figure 2a). The measured open probability (Po) of Mt1/Mt1 receptors was ~ 200-fold lower than that of Wt/Wt receptors. Mean closed time (MCT) was increased >100-fold and mean open time (MOT) was reduced 4-fold (Table 1). Notably, the large increase in MCT reflected entirely longer closures within bursts, while the duration of closures between bursts, which correspond to dwells in desensitized states13, were intact (τD, 3.0 ± 0.4 s for Mt1/Mt1 vs. 2.7 ± 0.3 for Wt/Wt, P>0.05, two-tailed Student's t test) (Figure 2b and Table 1). This result represents strong evidence that cross-linked receptors had severely reduced activity due to longer intra-burst closures and shorter openings but had intact microscopic desensitization.
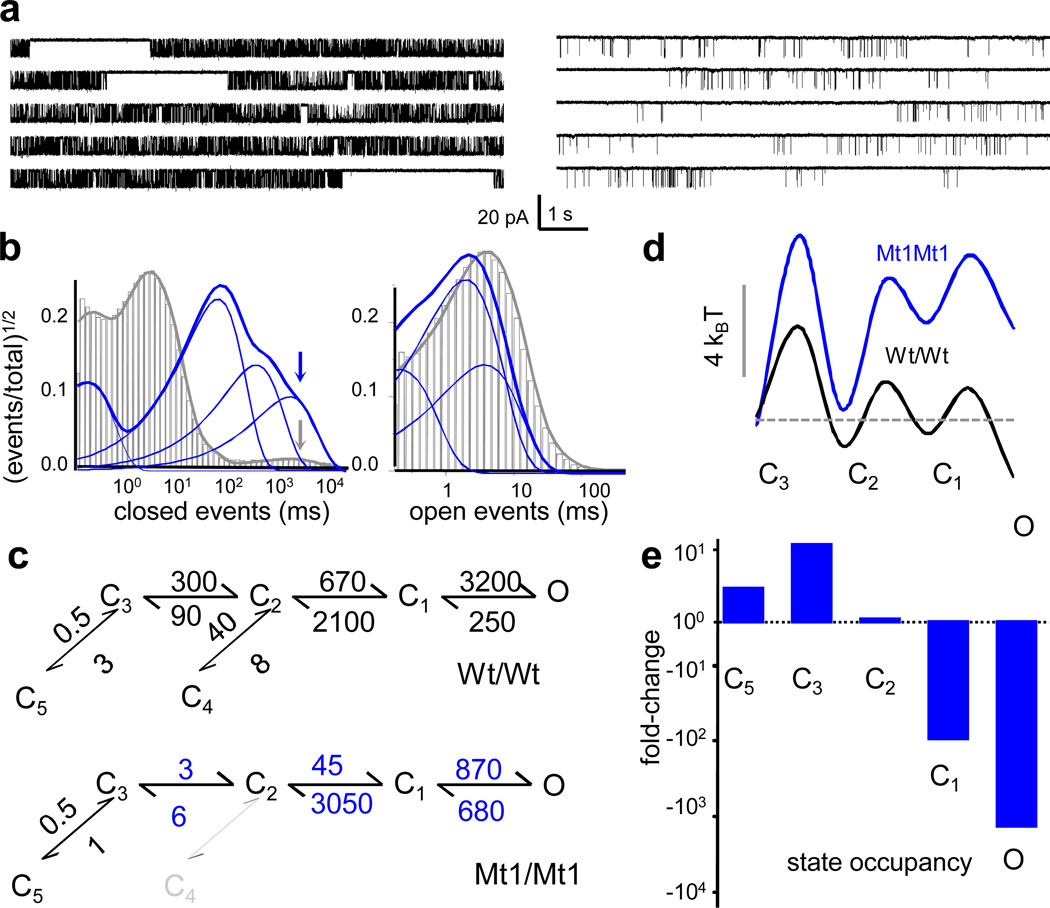
Figure 2. Single molecule activity of NMDA receptors with cross linked dimers.
a, Continuous traces recorded from on-cell patches containing one Wt/Wt (left) or one Mt1/Mt1 receptor (right). b, Overlaid event histograms calculated from the records illustrated in a for Wt/Wt (grey) and Mt1/Mt1 (blue) receptors. Arrows point to the longest closed component, which is of similar duration for both receptors. c, Reaction mechanisms show best-fitting schemes to stationary single-channel activity for Wt/Wt (n=18) and Mt1/Mt1 (n=6) receptors. Transitions into state C4 (grey) were not detected for cross-linked receptors. Rate constants (s−1) are given as rounded means of the values estimated from each data set; blue indicates P<0.05 relative to Wt/Wt (Student's t-test). d, Relative free-energy profiles calculated from the models in b aligned to the first liganded state. e, Change in fractional state occupancies calculated from the models in b.
Table 1.
Single Channel Parameters of NMDA Receptors with Mutations at the Dimer Interface
| GluN1/GluN2A | Amplitude (pA) |
Po | MOT (ms) |
MCT (ms) |
τD (s) |
n | Duration (min) |
Events analyzed |
|---|---|---|---|---|---|---|---|---|
| Wt/Wt | 9.3 ± 0.3 | 0.50 ± 0.03 | 7.1 ± 0.6 | 6.9 ± 0.7 | 2.7 ± 0.3 | 18 | 756 | 6.3×106 |
| Mt1/Mt1 | 9.1 ± 0.6 | 0.003 ± 0.001#* | 1.8 ± 0.2#* | 792 ±213#* | 3.0 ± 0.4 | 6 | 340 | 8.1×104 |
| Wt/Wt +DTT | 9.9 ± 0.4 | 0.58 ± 0.05 | 11.3 ± 1.7* | 9.3 ± 2.9 | 2.1 ± 0.3 | 6 | 257 | 1.6×106 |
| Mt1/Mt1+ DTT | 10.9 ± 0.7 | 0.16 ± 0.02#* | 12.5 ± 1.5* | 73 ± 12#* | 3.3 ± 0.4 | 7 | 178 | 2.5×105 |
| Mt1/Wt | 8.8 ± 0.5 | 0.25 ± 0.03#* | 10.6 ± 1.5* | 34.2 ± 6.3#* | 2.8 ± 0.6 | 6 | 145 | 4.1×105 |
| Wt/Mt1 | 9.7 ± 0.9 | 0.16 ± 0.03#* | 7.5 ± 0.3# | 45.0 ± 7.6#* | 2.8 ± 0.5 | 6 | 204 | 4.5×105 |
| N521Y/Wt | 9.9 ± 0.4 | 0.48 ± 0.07 | 7.6 ± 1.0 | 8.6 ± 1.7 | 2.5 ± 0.2 | 5 | 359 | 2.7×106 |
| Wt/E516Y | 9.9 ± 0.5 | 0.52 ± 0.07 | 8.3 ± 0.9 | 8.4 ± 1.8 | 2.1 ± 0.2 | 8 | 599 | 4.4×106 |
| N521Y/E516Y | 9.9 ± 0.3 | 0.24 ± 0.03* | 4.6 ± 0.4* | 16.2 ± 1.8* | 2.7 ± 0.2 | 8 | 546 | 3.7×106 |
Superscripts denote values that are significantly different from Wt/Wt (*) or Wt/Wt + DTT (#) in a Student’s t-test, ± s.e.m.
We next used statistical analyses of event distributions in combination with kinetic modeling to evaluate the reaction mechanism of cross-linked receptors. previous work has demonstrated that NMDA receptors activate along a complex pathway consisting of three functional steps: agonist binding, receptor activation, and receptor desensitization14,15. Activation can be further decomposed into three sequential rearrangements: C3→C2→C1→O, where C3-1 denote three kinetically distinct pre-open states and O represents several linked open states16–19. Desensitization is often depicted as two separate transitions that diverge from the main activation pathway: C3-C5 and C2-C4, where C5 represents the main, most stable desensitized state and C4 represents a minor, less prominent desensitized state20,21 (Figure 2c). We found that Mt1/Mt1 receptors followed a reaction mechanism similar to wild-type NMDA receptors; the single difference was the absence of the minor desensitized state C4, perhaps due to decreased kinetic resolution in this low-activity mutant. Macroscopic currents simulated with this state model resembled closely the time course of experimentally recorded whole-cell and excised-patch responses from a similar preparation, further reinforcing this model’s validity (Figure 2c and Supplementary Figures S2 and S3).
The model attributes the low Po of cross-linked receptors to substantially slower activation and faster deactivation transitions while clearly demonstrating normal microscopic desensitization (Figure 2c). Based on the relative free-energy profile calculated from this model, we propose that NMDA receptors with cross-linked heterodimer interfaces cannot open properly due to higher energy barriers to activation and substantially destabilized open states (Figure 2d). The implication is that when the D1-D1 interfaces of NMDA receptors are frozen in the arrangement described by Furukawa et al (2005)12 receptors are most likely closed, residing primarily in pre-open C3 and desensitized C5 conformations (Figure 2e).
A flexible dimer interface is required for normal openings
Further, we examined the activation mechanism of three NMDA receptor mutants that had free cysteine residues facing the dimer interface: DTT-treated Mt1/Mt1, Mt1/Wt and Wt/Mt1 receptors. Based on the available structural data, we reasoned that these receptors would have overall weaker dimer interfaces relative to Wt/Wt receptors due to a two-bond deficit in each LBD dimer: one hydrogen bond and one Van der Waals interaction. Based on the marked potentiation of whole-cell Mt1/Mt1 currents by DTT (Figure 1b), we expected that reduced Mt1/Mt1 would generate higher single-channel activity than cross-linked Mt1/Mt1, but we could not anticipate how these activities would compare to Wt/Wt. Indeed, we found that all three receptors investigated, DTT-treated Mt1/Mt1, Mt1/Wt and Wt/Mt1, had substantially higher Po values (>50-fold) than cross-linked Mt1/Mt1; in addition, we were able to determine that their Po values remained substantially lower (<4-fold) than Wt/Wt receptors (Figure 3a, Table 1). Remarkably, the absence of a restraining disulfide bond in these mutant receptors resulted in wild-type-like openings, a clue that restoring flexibility at the dimer interface may have restored open state stabilities (Figure 3b, Table 1).
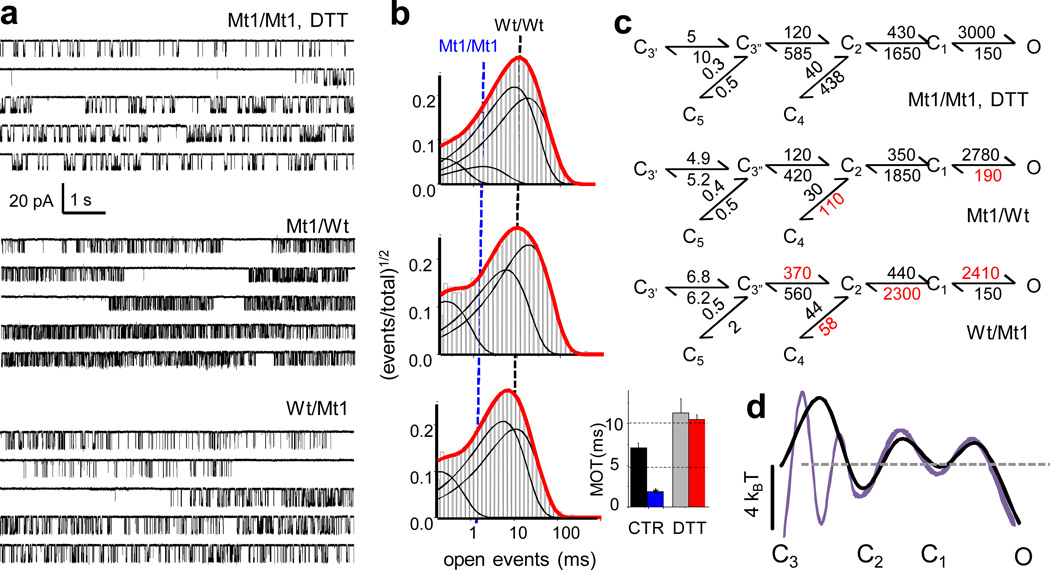
Figure 3. Single-molecule activity of NMDAR receptors with destabilized LBD interfaces.
a, Portions of continuous one-channel activity recorded from receptors that have cysteine residues substituted within the hydrophobic cores of ligand binding domain dimers. b, Open event distributions calculated from the records illustrated in a. Dotted lines show relative position of MOTs calculated for Wt/Wt and Mt1/Mt1 receptors. Bar graph shows summary of MOTs (means ± s.e.m.) from Wt/Wt (black, grey) and Mt1/Mt1 (blue, red) receptors in control (CTR) and reducing (DTT) conditions. c, Reaction mechanisms show best fitting schemes to single channel and macroscopic data in each condition. Rate constants are given in s−1 as the rounded mean of fits to six one-channel records; in red are rates that are different (P<0.05, Student's t-test) relative to reduced Mt1/Mt1 receptors. d, Relative free energy landscapes of the main activation pathway for reduced Mt1/Mt1 (purple) and reduced Wt/Wt (black) receptors were aligned to O state levels, based on equal MOTs.
In contrast to Wt/Wt receptors, event distributions of receptors with cysteine residues facing the dimer interface, revealed six rather than five closed components, an indication that these receptors dwell longer in a closed state that is only briefly populated by Wt/Wt receptors. The scheme illustrated in Figure 3c best described both single channel data and macroscopic responses recorded from these mutants (Supplementary Figures S2b and S3). This scheme postulates that after becoming fully liganded, receptors with reduced Mt1 subunits dwell longer in a pre-open state (C3’) before accessing a state (C3”) from which they can either desensitize or continue on the activation pathway. A simple interpretation of this result is that the cysteine residues engineered at the dimer interface divided the collection of equienergetic conformers that make up the aggregate state C3 of Wt/Wt receptors into two kinetically resolvable states, C3’ and C3” (Figure 3d).
Consistent with this interpretation, substitutions that preserved or even enhanced the hydrophobic nature of the buried interface produced close to wild-type NMDA receptor activity. In the structure represented in Figure 1a, we substituted only one of the two residues in each interacting pair for a tyrosine residue: GluN1(N521Y) for the N521~L780 pair, or/and GluN2A(E516Y) for the symmetrically related L777~E516 pair. Tyrosine substituted subunits produced wild-type activity when paired with a Wt partner and only mildly (2-fold) decreased Po when paired together (GluN1(N521Y) with GluN2A(E516Y) (Table 1 and Supplementary Figure S4)). The lack of kinetic phenotype for these mutants is remarkable because the homologous mutation in AMPA receptors, GluA2 (L483Y), produces largely non-desensitizing receptors7,22. We conclude that introducing an aromatic (tyrosine) but not a polar residue (cysteine) allows rearrangements at the heterodimer interface without delay or interruption, and that this structural plasticity is required for proper receptor activation. However, in contrast to non-NMDA receptors, neither substitution affects NMDA receptor desensitization.
DISCUSSION
In AMPA- and kainate-type glutamate receptors, the physical separation of subunits within LBDs leads to functional desensitization and this can be prevented by bracing the dimer interface with covalent links across subunits5–7,10. Based on the substantial structural homology demonstrated for glutamate receptor classes the current assumption is that the dimer interface serves a similar function in all glutamate-gated channels, including NMDA receptors. To test this hypothesis we investigated how perturbing contacts between GluN1 and GluN2A subunits in the ligand-binding domains of NMDA receptors influenced the receptor’s activation mechanism. We chose to examine substitutions that were previously demonstrated to have dramatic effects on the macroscopic behaviors of AMPA, kainate and NMDA receptors. Further, we used one-channel current recordings, kinetic analyses and state modeling to examine NMDA receptors whose heterodimers were reinforced with disulfide bridges across the D1-D1 interface or had free cysteine or tyrosine residues facing the intersubunit boundary.
Disulfide bridges between LBD protomers prevent AMPA7,10 and kainate5,10 receptor desensitization and render NMDA receptors insensitive to allosteric inhibition11. We found that NMDA receptors whose dimer interfaces were locked with disulfide cross-links in the arrangement described by Furukawa et al.12 produced normally desensitizing currents, whereas reducing the disulfide bonds increased the macroscopic current and reduced its desensitization (Figure 1). These results reveal that the strength of contacts at dimer interfaces within LBDs have opposite effects on the shape of the macroscopic currents recorded from NMDA and non-NMDA receptors.
At central excitatory synapses the shape of the post-synaptic current results from the combined action of NMDA and non-NMDA classes of glutamate receptors. Each class generates currents with distinct temporal profiles: non-NMDA receptors open and desensitize within milliseconds, whereas NMDA receptors do so much slower, within tens and hundreds of milliseconds, respectively1. The characteristic time course of the macroscopic response is rooted in unique reaction mechanisms. Importantly, non-NMDA receptors can open one subunit at a time, indicative of a largely subunit-independent gating mechanism, whereas NMDA receptor subunits are highly coupled during activation. Despite these fundamental differences in kinetics and reaction mechanisms, glutamate receptors share substantial sequence homology and are similar in overall structure1,23,24.
At present, extensive structural data support a conserved mechanism for agonist-induced opening of glutamate-gated channels: upon binding, glutamate changes the shape of the LBD in each subunit and this movement is transmitted to adjoining pore-forming helices as long as the LBD dimer remains intact1,2,4. Similarly, strong evidence exists in support of a common desensitization mechanism for AMPA and kainate receptors: the tension resulting from glutamate-induced change in the shape of the LBD in each subunit can cause subunits within the LBD dimers to separate and allows the pore to close while glutamate is still bound6–8,10. In this context, our results, which clearly show that NMDA receptors with covalently locked dimers desensitize normally, strongly support the assertion that NMDA and non-NMDA receptors have distinct desensitization mechanisms. This is consistent with the marked difference in intersubunit coupling during gating for these receptor types and points to a key role for intersubunit contacts in the divergent reaction mechanisms of structurally similar glutamate-gated channels.
The reaction mechanism of NMDA receptor is known in sufficient detail to allow accurate attribution of macroscopic current features such as amplitude and decay time course to microscopic kinetic transitions inferred from the single-channel record, a task not yet possible for non-NMDA receptors. We examined the reaction mechanism of cross-linked NMDA receptors and concluded that indeed receptors with covalently joined subunits had intact microscopic desensitization but their activation was deeply impaired: they had 200-fold lower open probabilities due to increased barriers to activation and unstable open states (Figure 2). Reducing the disulfide bonds restored open state stabilities but not the height of the activation barriers, which retained receptors in early pre-open conformations (Figure 3). These results led us to hypothesize that in contrast to non-NMDA receptors, a frangible intersubunit interface within the LBDs of NMDA receptors is necessary for activation rather than desensitization.
However, simply increasing dimer separation by tethering subunits at fixed lengths with MTS reagents failed to reproduce the activity seen in reducing conditions (Figure 1c), an indication that flexibility at the dimer interface as well as inter-subunit proximity are required for effective channel opening. Based on these results, we conclude that NMDA receptor activation but not desensitization requires rearrangements at the dimer interface described by Furukawa el al.12, which likely represents the ligand-binding unit of a closed receptor, an atomic arrangement adopted early in the activation sequence. Taken together, our results demonstrate that changes in the relative positions of residues facing the dimer interface are integral to the activation sequence of NMDA receptors and demonstrate fundamental differences in the activation mechanism of glutamate receptor classes.
Methods
Molecular Biology
Plasmids expressing GluN1-1a, GluN2A and GFP were transfected into HEK 293 cells as described in detail previously20. Cysteine or tyrosine substitutions were obtained with standard molecular biology procedures and were verified by full-insert sequencing.
Macroscopic current recordings and analyses
Macroscopic NMDA receptor currents were recorded with the excised-patch or the whole-cell patch clamp techniques where intracellular solutions contained (in mM): 135 CsF, 33 CsOH, 2 MgCl2, 1 CaCl2, 10 HEPES, 11 EGTA, adjusted to pH 7.4 (CsOH) and the holding potential was -70 mV. All extracellular solutions contained (mM): 150 NaCl, 2.5 KCl, 0.5 CaCl2, 0.01 EDTA, and 10 HEPBS adjusted to pH 8.0 (NaOH) plus 0.1 mM Gly. Currents were elicited by switching the cell or excised patch into solutions containing Glu (1 mM)20,25. When specified, DTT (10 mM) or H2O2 (0.5%) was also included. Bifunctional cross-linkers (Toronto Research Chemicals) were prepared from dimethylformamide stocks to 0.5 mM final concentration. Decay time course (tauD) was evaluated by fits to single-exponential function; steady-state to peak current ratio (Iss/Ipk) was calculated in pClamp 10.2 (Molecular Devices).
Single-channel current recordings and analyses
Activity from individual NMDA receptors was recorded using the cell-attached patch-clamp technique with extracellular solutions that contained (in mM): 150 NaCl, 2.5 KCl, 10 HEPBS, 1 EDTA, 1 Glu, 0.1 Gly adjusted to pH 8 (NaOH), and applying +100 mV through the recording electrode. Currents were analog filtered at 10 kHz and digitally sampled at 40 kHz directly into digital files using QUB software (www.qub.buffalo.edu). Only records originating from one-channel patches were kept and used for analyses20,26. Idealization, modeling and simulations were done in QUB with a 0.15 ms dead time27.
Simulations
Macroscopic responses were calculated as time-dependent occupancies of open states from the models obtained in this study after appending glutamate-binding steps, as previously described19,25,28,29. All channels were started in the resting state and glutamate pulses were simulated as instantaneous steps into 1-mM Glu. For each condition, the simulated traces were analyzed to extract desensitization time constants (tauD) and Iss/Ipk ratios as for experimentally recorded traces.
Supplementary Material
Acknowledgements
Funding was from NINDS (F31 to WFB and RO1 to GKP).
Footnotes
Author contributions
W.F.B performed whole-cell and cell-attached electrophysiological and simulation experiments; S.E.M recorded fast responses from outside-out patches; E.M.K. generated all mutations and the Western blot data; W.F.B and G.K.P designed the experiments, interpreted the results and wrote the paper.
References
- 1.Traynelis SF, et al. Glutamate Receptor Ion Channels:Structure, Regulation, and Function. Pharmacological Reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer ML. Structure and mechanism of glutamate receptor ion channel assembly, activation and modulation. Current Opinion in Neurobiology. 2011;21:283–290. doi: 10.1016/j.conb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouaux E. Structure and function of AMPA receptors. J Physiol. 2004;554:249–253. doi: 10.1113/jphysiol.2003.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 5.Priel A, Selak S, Lerma J, Stern-Bach Y. Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron. 2006;52:1037–1046. doi: 10.1016/j.neuron.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 8.Horning MS, Mayer ML. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379–388. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer ML. Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization. Embo J. 2009;28:1518–1530. doi: 10.1038/emboj.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- 11.Gielen M et al. Structural Rearrangements of NR1/NR2A NMDA Receptors during Allosteric Inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 13.Sakmann B, Patlak J, Neher E. Single AChR channels show burst-kinetics in presence of desensitizing concentrations of agonist. Nature. 1980;286:71–73. doi: 10.1038/286071a0. [DOI] [PubMed] [Google Scholar]
- 14.Howe JR, Colquhoun D, Cull-Candy SG. On the kinetics of large-conductance glutamate-receptor ion channels in rat cerebellar granule neurons. Proc R Soc Lond B Biol Sci. 1988;233:407–422. doi: 10.1098/rspb.1988.0030. [DOI] [PubMed] [Google Scholar]
- 15.Lester RA, Jahr CE. J Neurosci. 1992;Vol. 12:635–643. doi: 10.1523/JNEUROSCI.12-02-00635.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nat Neurosci. 2003;6:144–152. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- 17.Popescu G, Auerbach A. Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci. 2003;6:476–483. doi: 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- 18.Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- 20.Kussius CL, Kaur N, Popescu GK. Pregnanolone Sulfate Promotes Desensitization of Activated NMDA Receptors. J. Neurosci. 2009;29:6819–6827. doi: 10.1523/JNEUROSCI.0281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dravid SM, Prakash A, Traynelis SF. Activation of recombinant NR1/NR2C NMDA receptors. J Physiol. 2008;586:4425–4439. doi: 10.1113/jphysiol.2008.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stern-Bach Y, Russo S, Neuman M, Rosenmund C. A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron. 1998;21:907–918. doi: 10.1016/s0896-6273(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 23.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of Subunits in Functional NMDA Receptors. J Neurosci. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Howe JR, Popescu GK. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat Neurosci. 2008;11:1373–1375. doi: 10.1038/nn.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colquhoun D, Hawkes AG. Stochastic properties of ion channel openings and bursts in a membrane patch that contains two channels: evidence concerning the number of channels present when a record containing only single openings is observed. Proc R Soc Lond B Biol Sci. 1990;240:453–477. doi: 10.1098/rspb.1990.0048. [DOI] [PubMed] [Google Scholar]
- 27.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. J Gen Physiol. 1997;109:181–189. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amico-Ruvio S, Popescu G. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophysical Journal. 2010;98:1160–1169. doi: 10.1016/j.bpj.2009.12.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kussius CL, Popescu GK. NMDA receptors with locked glutamate-binding clefts open with high efficacy. J Neurosci. 2010;30:12474–12479. doi: 10.1523/JNEUROSCI.3337-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.