Abstract
The innate-like T cells expressing Vγ1.1 and Vδ6.3 represent a unique T cell lineage sharing features with both the γδ T and the invariant NKT cells. The population size of Vγ1.1+Vδ6.3+ T cells is tightly controlled and usually contributes to a very small proportion of thymic output, but the underlying mechanism remains enigmatic. Deletion of Id3, an inhibitor of E-protein transcription factors, can induce an expansion of the Vγ1.1+Vδ6.3+ T cell population. This phenotype is much stronger on the C57Bl/6 background than on the 129/sv background. Using quantitative trait linkage analysis, we identified Id2, a homologue of Id3, to be the major modifier of Id3 in limiting Vγ1.1+Vδ6.3+ T cell expansion. The Vγ1.1+Vδ6.3+ phenotype is attributed to an intrinsic weakness of Id2 transcription from Id2 C57Bl/6 allele, leading to an overall reduced dosage of Id proteins. However, complete removal of both Id2 and Id3 genes in developing T cells suppressed the expansion of Vγ1.1+Vδ6.3+ T cells due to decreased proliferation and increased cell death. We showed that conditional knockout of Id2 alone is sufficient to promote a moderate expansion of γδ T cells. These regulatory effects of Id2 and Id3 on Vγ1.1+Vδ6.3+ T cells are mediated by titration of E protein activity, since removing one or more copies of E protein genes can restore Vγ1.1+Vδ6.3+ T cell expansion in Id2 and Id3 double conditional knockout mice. Our data indicated that Id2 and Id3 collaboratively control survival and expansion of the γδ lineage through modulating a proper threshold of E-proteins.
Keywords: Id3, Id2, γδ T cells, E-proteins, PLZF, modifier mapping
Introduction
γδ T cells are a subset of T lymphocytes generated in the thymus that function between the innate and adaptive immune system. They have features of the adaptive immune system, such as the expression of variable rearranged γδ T cell receptors, but they also have features of the innate immune system, such as the ability to respond to stimulation rapidly (1). They can directly lyse infected or stressed cells as well as interact with αβ T cells, B cells and dendritic cells and regulate their functions (2). As a result, γδ T cells are involved in a broad range of immune processes, such as infection, inflammation, autoimmunity, tumor surveillance and tissue maintenance (1, 3). These cells are produced in large numbers in the fetal and neonatal stages in mammals, disseminating and forming stable populations in internal organs, mucosal and body surfaces, but their thymic production is gradually replaced by αβ T cells when the animal matures (4). The mechanism that controls the developmental switch from γδ to αβ T cell production in the thymus is not fully understood.
Among γδ T cells, the cells that express the Vγ1.1 and Vδ6.3 segments of the γδ T cell receptor belong to a unique subset. In mice, these cells are produced in large numbers in the neonatal thymus (5) and are capable of rapidly producing multiple cytokines, including IFNγ and IL-4, upon stimulation (6). They express the transcription factor PLZF that is also found in NKT cells (7). Like NKT cells, they also have a significant presence in the liver (8). The semi-invariant nature of their T cell receptor and their response pattern lead to the classification of these cells as “innate-like γδ T cells.” Although their function is not clearly understood, several studies pointed out that these cells may play an important role in attenuating excessive inflammation during infection and autoimmune processes due to their unique ability among γδ T cells to produce Th2-like cytokines (as reviewed by Carding SR. and Egan PJ. (3)). The population size of Vγ1.1+Vδ6.3+ γδ T cells varies between mouse strains; they are particularly abundant in the DBA strain (in which usually a Vδ6.4 segment is expressed) but relatively rare in the B6 strain (6). However, in the absence of a helix-loop-helix transcription regulator, Id3, it has been shown that Vγ1.1+Vδ6.3+ γδ T cells can also expand dramatically in mice with B6 genetic background (9).
Id3 has been implicated to play both positive and negative roles in the developmental control of γδ T cells. It has been shown that in developing DN thymocytes, if a cell successfully rearranges the γδ T cell receptor genes, the surface expression of γδ T cell receptor can send a strong signal into the cell and up-regulate Id3, promoting the cell to adopt a γδ T cell fate (10). However, Id3 also plays a distinct inhibitory role controlling the development of Vγ1.1+Vδ6.3+ γδ T cells because this population is dramatically expanded in Id3 deficient mice. More interestingly, this expansion is limited to the neonatal window and cannot be recapitulated by transferring Id3-deficient bone marrow cells into adult wild type B6 animals (11). The expansion also requires a pure B6 genetic background; in a B6/129 mix background, the expansion is variable and often greatly diminished (9). The latter phenomenon suggests that additional gene(s) specific to the B6 background is also critical in the development of Vγ1.1+Vδ6.3+ γδ T cells in the absence of Id3.
While the importance of Id3 in regulating the development and population size of γδ T cells has been firmly established, the underlying mechanism is still poorly defined. This strain- and genotype-specific expansion of Vγ1.1+Vδ6.3+ γδ T cells represents a unique opportunity to identify novel players in the developmental control of γδ T cells. We designed a backcross experiment between B6 and 129 Id3-deficient mice with a goal to identify the background genes in regulating the Vγ1.1+Vδ6.3+ γδ T cells. We found that another member of the Id protein family, Id2, was the major modifier of Id3 involved in the control of γδ T cell population size. Id2 129 allele is expressed more in γδ T cells than Id2 B6 allele; it is highly expressed in Vγ1.1+Vδ6.3+ γδ T cells and mature γδ T cells in general. Conditional knockout of Id2 leads to expansion of γδ T cells not limited to the Vγ1.1+Vδ6.3+ subset. Paradoxically, if both Id2 and Id3 are completely deleted, the Vγ1.1+Vδ6.3+ γδ T cells actually fail to accumulate, possibly due to attenuated proliferation and increased cell death induced by unrestricted E protein activity. We further showed that these phenomena may occur after γδ T cell lineage commitment, thus separating them from the role Id3 plays in the initial TCR signaling and lineage choice processes. These results clearly demonstrated the interweaving roles of Id proteins and E proteins in the control of γδ T cell development.
Materials and Methods
Mice
The Id3−/− (12), Id2GFP (13), Id2f/f (14), Id3f/f (15), E2Af/f (16), HEBf/f (17) and LckCre transgenic (18) mice have been described previously and all maintained on pure B6 background. C57BL/6J, 129X1/SvJ mice were purchased from The Jackson Laboratory. CD4Cre transgenic mice on B6 background were purchased from Taconic. Animals were bred and maintained in the SPF facility managed by Duke University Division of Laboratory Animal Research. All animal procedures were approved by the Duke University Institutional Animal Care and Use Committee.
Flow cytometry
The antibodies used in the flow cytometry analyses were as follows: anti-mouse CD4 (GK1.5), anti-mouse CD8a (53-6.7), anti-mouse B220 (RA2-6B2), anti-mouse/human CD44 (IM7), anti-mouse CD25 (3C7), anti-mouse NK-1.1(PK136), anti-mouse Ly-6G/Ly-6C(Gr-1) (RB6-8C5), anti-mouse CD11b(M1/70), anti-mouse TCRγ/δ(GL3), anti-mouse TCR Vγ1.1 (2.11), anti-mouse CD24 (M1/69) and anti-mouse TCRβ (H57-597) were purchased from Biolegend. The PE anti-mouse Vδ 6.3/2 (8F4H7B7) antibody, annexin V and the APC BrdU Flow Kit were purchased from BD Biosciences. 7-Aminoactinomycin D (7-AAD) was purchased from Life Technologies.
Single-cell suspensions were prepared from thymus, spleen and peripheral lymph nodes, and suspended in cold FACS buffer (1×PBS supplemented with 5% bovine calf serum). 1×106 cells were stained with antibodies in the dark at 4°C for 30 min. After washing with cold FACS buffer, cell suspensions were analyzed on a FACSCanto II flow cytometer (BD Biosciences). FlowJo software (Tree Star) was used for data analysis. Cell sorting was performed with a FACS DiVa sorter (BD Biosciences).
Quantitative trait linkage analysis
Id3−/− mice on B6 background were crossed with 129X1/SvJ mice to generate Id3+/− F1 mice. F1 mice were backcrossed with Id3−/− mice on B6 background to generate Id3−/− F2 mice. The genomic DNA was extracted from toes of Id3−/− F2 mice and sent to Genomic Analysis Facility at Duke University for single nucleotide polymorphism (SNP) analysis using a 377 genome-wide mouse SNP panel (Illumina). Genome-wide scans were plotted using J/QTL mapping program (version 1.3) (http://churchill.jax.org/software/jqtl.shtml), and genomic regions with significant linkage to the expansion of Vγ1.1+ Vδ6.3+ γδ T cells (>1% of total thymocyte) was determined using methods previously described (19). Additional DNA primers were designed to PCR-amplify regions on chromosome 12 near the centromere end, and the PCR products were sequenced to determine the status of additional SNP markers around this region.
Real-time PCR analysis
TCRγδ+ Vδ 6.3+ cells were sorted from mouse thymus, and total RNA was extracted with RNAqueous micro kit (Life Technologies). Reverse transcription was performed with M-MLV reverse transcriptase (Life Technologies). SYBR-based real-time PCR was performed to quantitatively compare gene expression, normalized to β-actin. QPCR primer sequences are available upon requests.
Restriction fragment length polymorphism analysis
A 481bp fragment was PCR amplified from Id2 cDNA made from thymic TCRγδ+ Vδ 6.3+ cells. The PCR product was digested with EcoRI. The 481bp PCR product from B6 Id2 allele does not contain any EcoRI restriction site. The product from 129 Id2 allele contains one EcoRI site, and digestion with the enzyme will generate one 393bp fragment and one 88bp fragment.
In vitro stimulation of γδ T cells
Thymic GFP-negative TCRγδ+ cells from Id2GFP/+ mice were sorted and cultured in OP9-DL1 cell covered wells with MEM-α medium, supplemented with 10% fetal bovine serum and 5 ng/mL IL-7, with or without 1 μg/mL anti-TCRγδ (clone UC7-13D5, Biolegend). Cells were harvested after 5 days for FACS analysis.
In vivo BrdU incorporation assays
1 mg of BrdU was i.p. injected to each mouse 15 hours (for Id2f/f CD4Cre+ vs. Id2f/f CD4Cre− mice experiments) or 4 hours (for Id2 and Id3 double conditional knockout experiments) before sacrificing the mice. Cells were harvested and processed with the BrdU Flow Kit (BD Biosciences) according to manufacturer protocol.
Cell death analysis
For Id2f/f CD4Cre+ vs. Id2f/f CD4Cre− mice experiments, thymocytes were harvested and directly analyzed with 7AAD/annexin V for cell death according to manufacturer protocol (BD Biosciences). For Id2f/f Id3f/f CD4Cre+ vs. Id2f/B Id3f/f CD4Cre+ mice experiments, TCRγδ+ Vδ 6.3+ cells were first sorted from mouse thymus and cultured in RPMI1640 medium supplemented with 10% fetal bovine serum and 55μM of 2-mercaptoenthanol for 24 hours prior to 7AAD/annexin V analysis.
Statistical analysis
Sample data was compared using Student's t test, and p value less than 0.05 was considered significant.
Results
Backcross mapping identifies a single locus that modulates numbers of Vδ6.3+ cells in Id3 knockout mice
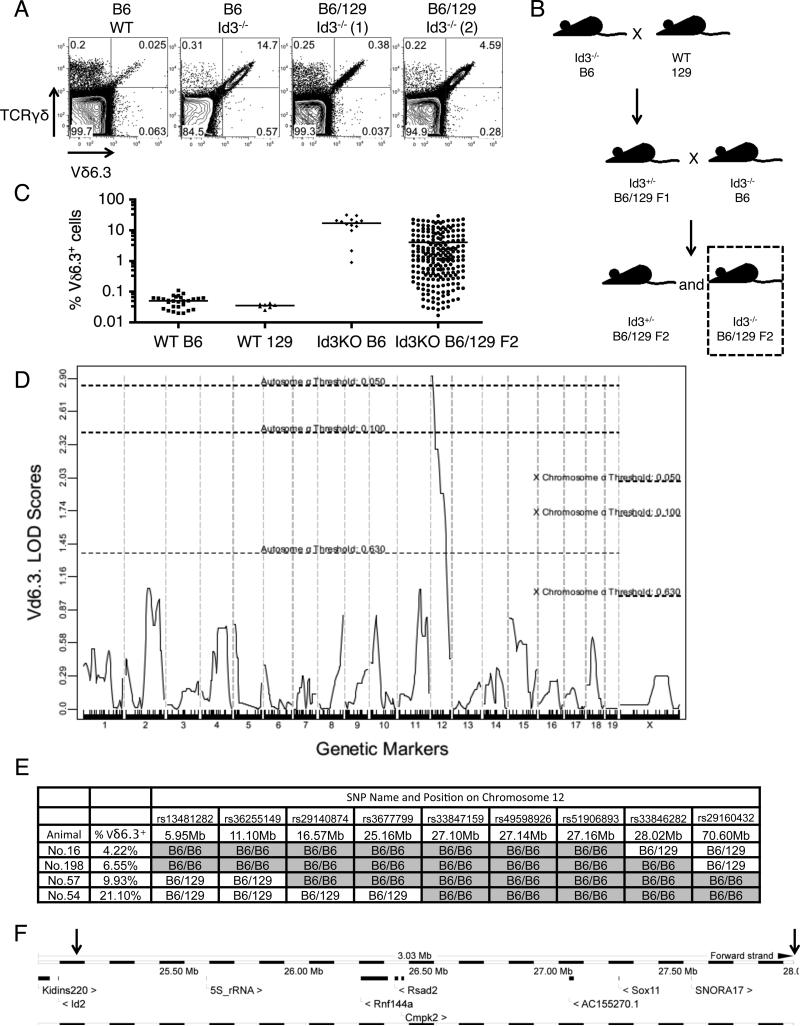
We analyzed the thymus of Id3 deficient mice on B6 or B6/129 mix background and showed that the population of Vγ1.1+Vδ6.3+ γδ T cells is consistently large in mice with B6 background but variable in mice with B6/129 mix background (Fig 1A, S1A). Because Vδ6.3 usage is uniquely associated with the Vγ1.1+Vδ6.3+ population found in Id3 deficient mice(11), Vδ6.3 was used as a lineage marker in subsequent genetic analysis. We hypothesized that an Id3-modifying gene(s) is responsible for the phenotypic difference observed between these two strains. We designed a backcrossing strategy to further verify our hypothesis (Fig 1B). First, Id3−/− mice on B6 background were crossed with wild type 129X1/SvJ mice. The resulting F1 mice (Id3+/− with mixed background) were further backcrossed with Id3−/− mice on B6 background. Half of the F2 progeny from the backcross were expected to be Id3−/−, which were analyzed for their Vδ6.3+ γδ T cell percentage in the thymus. Among all the 226 Id3−/− F2 mice analyzed, we found a wide range of distribution in Vδ6.3+ γδ T cell percentage (Fig 1C). As a comparison, most Id3−/− mice on B6 background have >1% Vδ6.3+ γδ T cell among thymocytes (Fig 1C). This result suggests the existence of one or more possible Id3 modifier gene, with the 129 allele being dominant (inhibit Vδ6.3+ γδ T cell expansion), and the B6 allele being recessive (permit Vδ6.3+ γδ T cell expansion). In order to identify the potential gene(s), we performed genome-wide SNP analysis in 25 Id3−/− F2 mice. We found that B6 homozygosity of a single 30 Mb region on chromosome 12 near the centromere was strongly linked with the presence of high numbers (>1%) of Vδ6.3+ γδ T cells (LOD score >2) (Fig 1D). Focusing on one SNP on chromosome 12 location 30Mb, we analyzed additional 138 Id3−/− F2 mice and showed that B6/B6 homozygosity of this location was significantly correlated with the accumulation of Vδ6.3+ γδ T cells (Fig. S1B). We further analyzed these 138 Id3−/− F2 mice, determining their genotype of two SNP markers on chromosome 12 (location around 3Mb and 30Mb) flanking the region identified in the genome-wide SNP analysis. We looked for mice showing discordant genotype at these two loci (one being B6/B6, the other being B6/129) and with more than 1% Vδ6.3+ γδ T cells in their thymi. We then performed detailed study of additional SNP markers inside this region and narrowed down the critical interval to a 3 Mb region (chromosome 12 location 25.16Mb-28.02Mb) (Fig 1E). There are only four known protein coding genes in this interval, including Sox11, Cmpk2, Rsad2, and Rnf144a (Fig 1F). The other features (SNORA17, AC155270.1, 5s_rRNA) are either RNA coding genes or putative genes. Id2, a homolog of Id3, locates immediately outside of the boundary (25.09Mb), leaving the possibility that its expression may still be influenced by regulatory elements inside this interval.
Figure 1.
A single locus on mouse chromosome 12 strongly influences the development of Vδ6.3+ γδ T cells in Id3-deficient mice. (A) Deficiency of Id3 induces the accumulation of a large population of Vδ6.3+ γδ T cells in the thymus of mice with pure B6 background. However, this phenomenon persists in some but is absent in other mice with a B6/129 mix background, as two representative mice are showing here. (B) A breeding scheme was established to dissect potential modifying gene(s) in the B6 and 129 genetic backgrounds. Only F2 B6/129 hybrid Id3−/− mice were used in the linkage analysis. (C) Percentages of Vδ6.3+ γδ T cells among total thymocytes were scored for individual mice of indicated genotype group. Each dot represents one mouse. Horizontal line indicates mean of the genotype group. (D) SNP analysis of the B6/129 hybrid Id3−/− mice showed that one location on chromosome 12 is strongly correlated with the presence of more than 1% of Vδ6.3+ γδ T cells in the thymus. (E) Detailed SNP analysis of four mice with more than 1% of Vδ6.3+ γδ T cells in the thymus showed a linkage to B6 homozygocity within a 3 Mb region on chromosome 12. (F) A map of known features around the critical region on chromosome 12; arrows indicate the border of the region (25.16Mb-28.02Mb) as determined in E. Note that Id2 and Kidins220 are immediately outside of the border of this region.
Id2 is a major modifier of Id3 in regulating the population size of Vδ6.3+ cells
To determine which gene in this region on chromosome 12 is truly responsible for the accumulation of Vδ6.3+ γδ T cells in Id3−/− mice with B6 background, we first examined allelic variations between 129 and B6 mice in terms of their protein-coding sequences. We did not find non-synonymous changes or splice site SNPs for Sox11, Cmpk2, Rnf144a and Id2 genes. Several non-synonymous changes were identified in the Rsad2 gene. However, transgenic rescue tests showed that the 129 allele of Rsad2 cannot prevent Vδ6.3+ γδ T cells from accumulating when introduced into conditional Id3 knockout mice on B6 background (Fig S2 A, B).
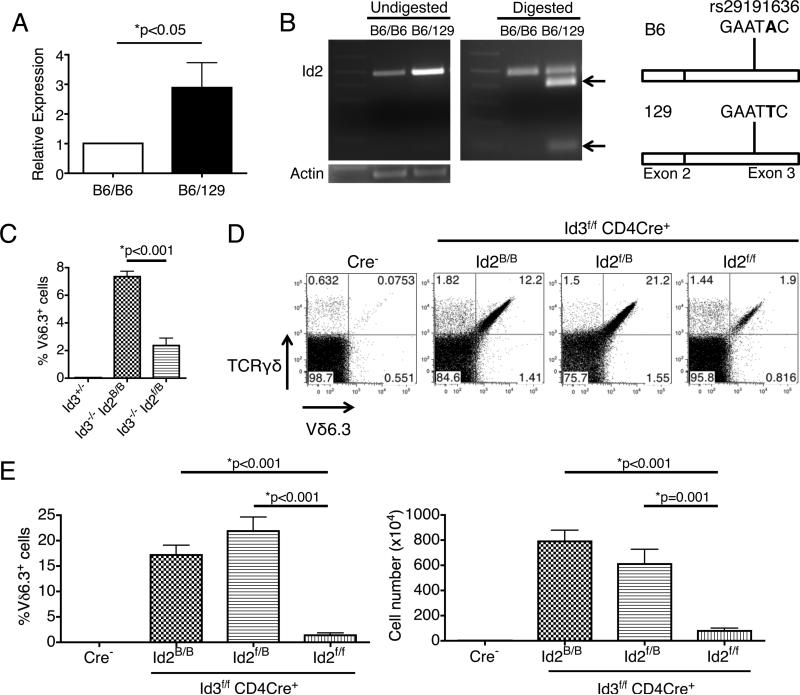
We considered the alternative possibility that variation in non-protein coding sequences between 129 and B6 mice may affect the expression level of one of these candidate genes. We compared the mRNA expression level of all the aforementioned candidate genes in thymic Vδ6.3+ γδ T cells from Id3−/− mice with either pure B6 or B6/129 F2 mix background on chromosome 12 position 25.16Mb-28.02Mb. We found that only Id2 is expressed differently between the groups, being more highly expressed in mice with B6/129 F2 mixed background (Fig 2A and Fig S2C). In order to verify that the increased expression comes from the 129 Id2 allele, we performed restriction fragment length polymorphism analysis of the Id2 cDNA made from the γδ T cells. The 129 Id2 allele contains an EcoRI restriction site in the 3’ UTR which is absent in the B6 allele. We amplified the Id2 cDNA region surrounding the EcoRI site by PCR and digested the product with EcoRI, and we observed abundant restriction fragments generated from the 129 allele (Fig 2B). These results indicate that a potential regulatory element on chromosome 12, between position 25.16Mb-28.02Mb, acts in cis to control Id2 expression. The 129 version of the element may induce a higher level of Id2 expression than the B6 version, and this higher Id2 level may inhibit the expansion of Vδ6.3+ γδ T cells in a dominant manner.
Figure 2.
Id2 is a major modifier of Vδ6.3+ γδ T cell development in Id3-deficient mice. (A) QPCR analysis showed that Id2 mRNA expression in Vδ6.3+ γδ T cells from Id3−/− mice with B6 background on the chromosome 12 region encompassing Id2 is lower than those with B6/129 mix background. n=3 for independent sorting of each genotype group. (B) Restriction enzyme analysis of the SNP marker rs29191636 within the exon 3 of the Id2 gene. EcoRI digestion of Id2 cDNA made from Vδ6.3+ γδ T cells with B6/129 mix background generated abundant product specific to the 129 allele (marked by two arrows), indicating that the higher Id2 expression in these cells came from that allele. Data representative of 3 experiments. (C) Replacement of one copy of the Id2 B6 allele with the Id2f allele of 129 origin is sufficient to reduce the population size of Vδ6.3+ γδ T cells. Id2B indicates the wild type Id2 allele in B6 background. n≥3 for each group. (D) Analysis of Vδ6.3+ γδ T cells with various combinations of Id2B and Id2f alleles on Id3f/f CD4Cre+ background. Data representative of 3 mice in each group. (E) Percentage and number of Vδ6.3+ γδ T cell in the thymus of genotype each group shown in D. N≥4 in each group. All error bars indicate SD.
In order to further examine this hypothesis, we took advantage of the Id2f allele (as compared to Id2B, indicating the wild type allele in B6 background). This floxed allele was generated with mouse embryonic stem cells from the 129 strain (14). Although this allele has been backcrossed to the B6 genetic background for more than 10 generations, the region on chromosome 12 around Id2, including location 25.16Mb-28.02Mb, remained of 129 origin. In the absence of Cre, this allele behaves similarly to the wild type allele from the 129 strain. By introducing this allele into the Id3−/− mouse on B6 background, we can specifically investigate the role of this region on the development of Vδ6.3+ γδ T cells. We found that introducing one copy of the Id2f allele was sufficient to significantly suppress the accumulation of Vδ6.3+ γδ T cells (Fig 2C). In order to determine whether it is Id2 itself that is limiting the development of Vδ6.3+ γδ T cells, we knocked out Id2 and Id3 in T cells specifically with CD4Cre. The Id2B/B Id3f/f CD4Cre+ mice behaved similarly to Id3−/− mice, showing significant accumulation of Vδ6.3+ γδ T cells (Fig 2D). However, unlike the Id2f/B Id3−/− mice, the 129 genetic material in the Id2f/B Id3f/f CD4Cre+ mice could not suppress the accumulation of Vδ6.3+ γδ T cells, demonstrating that it is indeed Id2 that is playing the inhibitory role (Fig 2D). These results indicate that genetic material on chromosome 12 location 25.16Mb-28.02Mb from the 129 background may influence the expression of Id2, thus inhibiting the accumulation of Vδ6.3+ γδ T cells together with Id3.
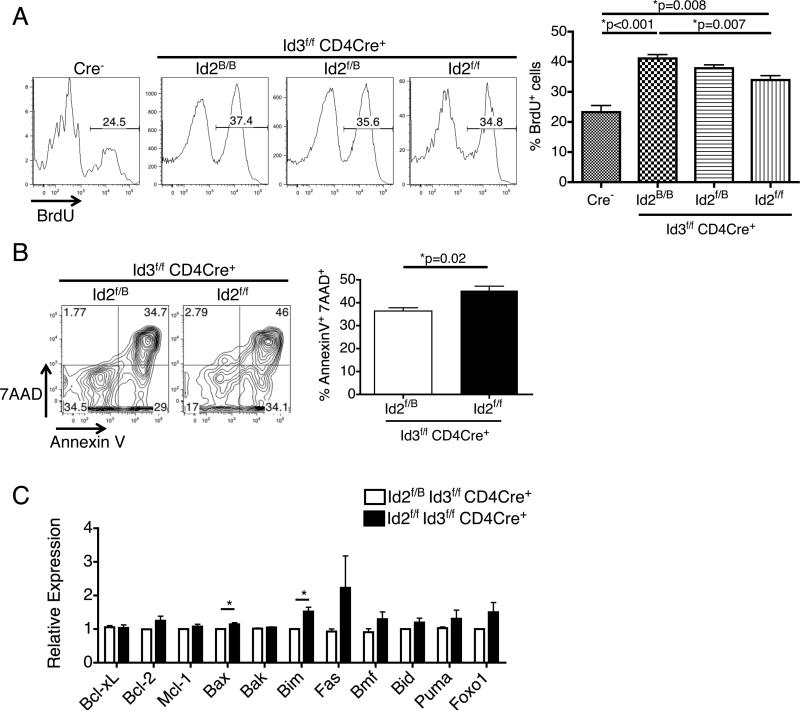
We hypothesized that double deletion of Id2 and Id3 with CD4Cre might lead to further increase of the Vδ6.3+ γδ T cells. To our surprise, Id2f/f Id3f/f CD4Cre+ mice actually have very few of these cells (Fig 2D, E). Because the Vδ6.3+ γδ T cells proliferate vigorously during the neonatal stage, we examined thymocytes from 7 days old mice and found that, regardless of the Id2 genotype, Vδ6.3+ γδ T cells from Id3f/f CD4Cre+ mice were more highly proliferative than those from the Cre− controls, but the proliferation is slightly attenuated in Id2f/f Id3f/f CD4Cre+ mice (Fig 3A). When we cultured the cells in vitro for 24 hours, we found that cells from Id2f/f Id3f/f CD4Cre+ mice showed more rapid cell death than those from Id2f/B Id3f/f CD4Cre+ mice (Fig 3B). We performed QPCR for a panel of cell-death-related genes and found that two proapoptotic genes, Bax and Bim, were significantly up-regulated in Id2f/f Id3f/f CD4Cre+ cells (Fig 3C). These results indicate that complete removal of both Id2 and Id3 proteins can actually be inhibitory for Vδ6.3+ γδ T cell survival and proliferation, implying that the maximal output of Vδ6.3+ γδ T cells requires an optimal level of Id proteins.
Figure 3.
Conditional knockout of both Id2 and Id3 impairs the proliferation and survival of Vδ6.3+ γδ T cells. (A) In the neonatal thymus, Vδ6.3+ γδ T cells from Id3f/f CD4Cre+ mice are more highly proliferative than the Cre− controls as shown by BrdU incorporation assay, regardless of their Id2 genotype. However, cells from the Id2f/f Id3f/f CD4Cre+ mice show a small but significant decrease in BrdU+ cell percentage compared to those from Id2B/BId3f/fCD4Cre+ mice. n=3 for each group. (B) Vδ6.3+ γδ T cells were sorted from the thymus of neonatal mice and cultured for 24 hours. Id2f/f Id3f/f CD4Cre+ cells showed increased cell death by 7AAD and Annexin V staining compared to Id2f/B Id3f/f CD4Cre+ cells. n=3 in each group. (C) QPCR analysis of a panel of cell death-related genes showed that Id2f/f Id3f/f CD4Cre+ Vδ6.3+ γδ T cells express more mRNA of pro-apoptotic genes Bim and Bax. n=3 in each group. *p<0.05. All error bars indicate SD.
Id2 functions as an inhibitor of γδ T cell development
Since complete knockout of both Id2 and Id3 is detrimental to the accumulation of Vδ6.3+ γδ T cells, one possible explanation for the expansion of these cells in Id2B/B Id3−/− mice on B6 background is that the lower expression level from the B6 version of Id2 allele results in a higher, but not too high, activity of E proteins to drive Vδ6.3+ γδ T cell expansion. If so, removing Id2 alone may also result in an expansion of γδ T cells -- at least in some subsets -- even in the presence of Id3.
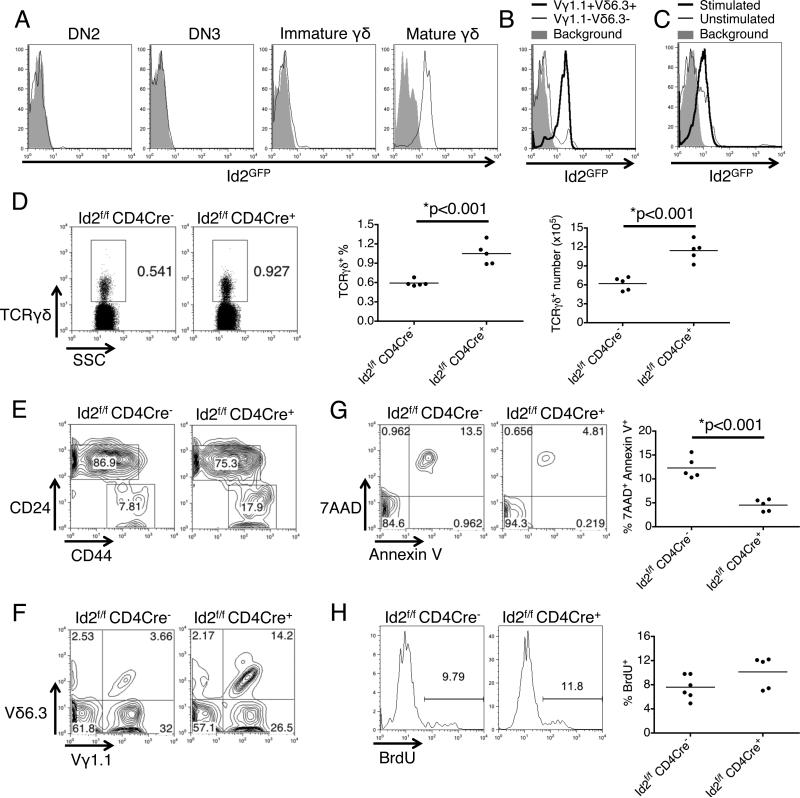
To further characterize the expression of Id2 in γδ T cells, we utilized the Id2GFP reporter mouse. The mouse was also generated with 129 ES cells (13) and subsequently backcrossed to the B6 background for more than 10 generations; however, region around the Id2 locus still retain genetic material from the 129 background, possibly also including the putative regulatory region we identified here. In thymus, we found that Id2 is not expressed in developing γδ T cells, but is highly expressed in their mature stage (TCRγδ+CD24−CD44high) (20) (Fig 4A). We also found that Id2 expression is higher in the Vγ1.1+Vδ6.3+ γδ T cells compared to Vγ1.1−Vδ6.3− γδ T cells (Fig 4B). When GFP-negative γδ T cells were sorted and cultured with OP9-DL1 cells and IL-7, stimulation with anti-TCRγδ antibody further up-regulated the expression of GFP, suggesting that the expression of Id2 may be controlled by TCR signaling (Fig 4C).
Figure 4.
Conditional knockout of Id2 alone results in expansion of γδ T cells. (A) Examination of Id2 expression in developing γδ T cells in the thymus with an Id2GFP reporter showed that Id2 is up-regulated at the mature stage. DN2: Lin−CD25+CD44+. DN3: Lin−CD25+CD44−. Immature: TCRγδ+CD24+CD44low. Mature: TCRγδ+CD24−CD44high. (B) Id2 expression is higher in the Vγ1.1+Vδ6.3+ cells than in other γδ T cells. (C) In vitro culturing of sorted Id2GFP negative γδ T cells from the thymus for 5 days with IL-7 and anti-TCR γδ stimulation resulted in more significant up-regulation of Id2 compared to culturing with IL-7 alone. For A-C, data representative of 3 mice in each group. (D) Id2f/f CD4Cre+ mice have more γδ T cells in the spleen compared to Id2f/f CD4Cre− mice. Bar graphs show the percentage and number of γδ T cells in the spleen of mice in each group. Each dot represents one mouse. (E) Id2f/f CD4Cre+ mice have a higher percentage of CD24−CD44high mature γδ T cells in the thymus. Pre-gated on TCRγδ+ cells. (F) Id2f/f CD4Cre+ mice have a higher percentage of Vγ1.1+Vδ6.3+ γδ T cells in the thymus. Pre-gated on TCRγδ+ cells. (G) The mature thymic γδ T cells from Id2f/f CD4Cre+ mice show decreased cell death by 7AAD and Annexin V staining. (H) The mature thymic γδ T cells from Id2f/f CD4Cre+ mice show similar proliferation rate to Id2f/f CD4Cre− mice in BrdU incorporation assay. Data representative of 3 mice in each group in E and F.
We next examined the effect of Id2 deficiency alone on the development of γδ T cells using the Id2f/f CD4Cre single conditional knockout model. We found that Id2f/f CD4Cre+ mice indeed have more γδ T cells in the spleen compared to Id2f/f CD4Cre− mice (Fig 4D). In the thymus, although the percentage and number of total γδ T cells are similar between the groups, Id2f/f CD4Cre+ mice have more mature γδ T cell (TCRγδ+CD24−CD44high) compared to Id2f/f CD4Cre− mice (Fig 4E). They also have more Vγ1.1+Vδ6.3+ γδ T cells, although unlike in Id3−/− mice, these Vγ1.1+Vδ6.3+ γδ T cells still contribute to only a minority of γδ T cells (Fig 4F). These findings supported the hypothesis that Id2 functions as an inhibitor of γδ T cell development, although its effect is not limited to Vγ1.1+Vδ6.3+ γδ T cells. This inhibition effect of Id2 is attributed to increased cell death, as mature γδ T cells from Id2f/f CD4Cre+ mice showed decreased cell death in 7AAD/annexin V analysis but showed no difference in BrdU incorporation assays (Fig 4G,H).
Id proteins control γδ T cell development through inhibition of E proteins in a developmental stage-specific manner
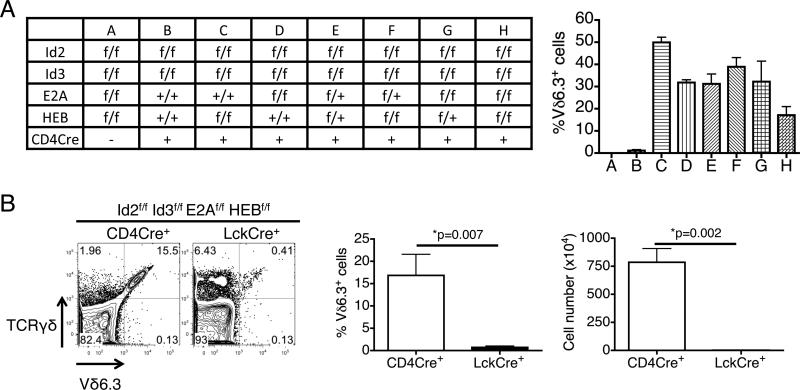
All evidence thus far pointed to the conclusion that Id2 and Id3 collaboratively act as “dual safety” in limiting the expansion of Vδ6.3+ γδ T cells, and a higher, but not too high, activity of E proteins is required to permit the expansion of this population. To demonstrate that Id2 and Id3 indeed function in γδ T cells through inhibiting E proteins, namely E2A and HEB (21), we sought to combine different floxed alleles of E proteins and Id proteins with CD4Cre and determine whether reduction of E protein dosage can counter the effect of the loss of Id proteins (Fig 5A). We found that removing any two to four of the E2A and HEB alleles can result in expansion of Vδ6.3+ γδ T cells in Id2f/f Id3f/f CD4Cre+ mice, although not all combinations result in the same degree of expansion. Nevertheless, this finding indicates that when both Id2 and Id3 are deleted in Vδ6.3+ γδ T cells, it is the excessive activity of E proteins that limits the size of this population.
Figure 5.
The effect of Id proteins on Vδ6.3+ γδ T cells is mediated by E proteins and is developmental stage-specific. (A) Although conditional knockout of both Id2 and Id3 limits the accumulation of Vδ6.3+ γδ T cells in the thymus, further deletion of HEB and/or E2A can restore the accumulation of those cells. N≥3 for each group. (B) Deletion of all alleles of HEB, E2A, Id2 and Id3 by CD4Cre can induce accumulation of Vδ6.3+ γδ T cells, but deletion by LckCre fails to induce a similar phenotype. N≥3 for each group. All error bars indicate SD.
We next sought to investigate whether this regulation of γδ T cells by Id proteins and E proteins occurs before or after γδ lineage specification. Although E2Af/f HEBf/f Id2f/f Id3f/f CD4Cre+ mice can accumulate a significant number of Vδ6.3+ γδ T cells in their thymus, deletion of these four genes by LckCre, which becomes active earlier in the DN3 stage, blocked the development of Vδ6.3+ γδ T cells (Fig 5B). The results indicate that E protein and Id protein play different roles before and after γδ lineage specification.
Discussion
In this study, we found that the dramatic expansion of Vγ1.1+Vδ6.3+ γδ T cells observed in Id3−/− mice is contingent on B6 homozygocity in a small region on chromosome 12, which possibly contains a regulatory element that leads to lower expression of the nearby Id2 gene. Using the Id2f allele that is of 129 origin, we showed that this region alone is capable of suppressing Vγ1.1+Vδ6.3+ γδ T cell accumulation, and this suppression is dependent on the Id2 gene itself. However, complete loss of Id2 and Id3 actually reduces the Vγ1.1+Vδ6.3+ γδ T cell population size, indicating that unrestrained E protein activity is also detrimental to these cells. We further showed that conditional knock-out of Id2 alone was sufficient to induce a moderate expansion of γδ T cells. All of these phenomena occurred when CD4Cre was used to delete the Id and E protein genes; when the genes were deleted earlier in T cell development with LckCre, even removal of E proteins cannot restore the Vγ1.1+Vδ6.3+ γδ T cells in Id2 and Id3 conditional knockout mice, emphasizing the stage-specific nature of these genetic regulations.
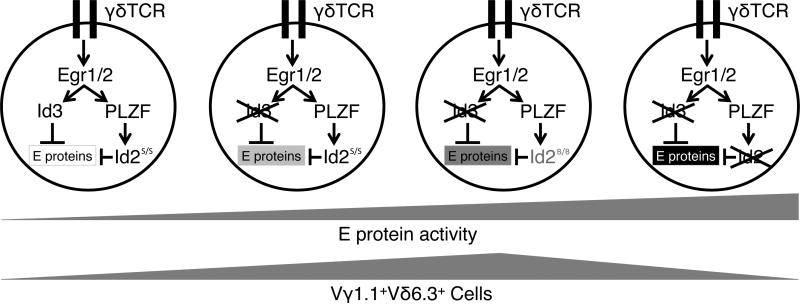
We propose a dual safety model to summarize the above findings (Fig 6). In this model, Id3 and Id2 are differentially regulated by the TCR signals. Egr is the major transcription factor acting between the TCR signal and Id3 in T cell development (10). PLZF is a unique transcription factor involved in the development of innate-like lymphocytes such as iNKT and Vγ1.1+Vδ6.3+ γδ T cells (22, 23). Id2 has been shown to be activated by PLZF, which is a direct target of Egr2 in iNKT cell development (24). When both Id2 and Id3 are present, they respond to the TCR signal and keep E protein activity low, and consequently prevent the expansion of Vγ1.1+Vδ6.3+ γδ T cells. When Id3 is deleted, Id2 will assume a safety role to control E-protein activity. However, this safety role of Id2 is compromised by the hypomorphic allele of Id2 in the B6 background, allowing an increase in E protein activities to the optimal level for driving Vγ1.1+Vδ6.3+ γδ T cell expansion. When both Id2 and Id3 are completely deleted, E protein activity becomes too high and again limits the Vγ1.1+Vδ6.3+ γδ T cell population.
Figure 6.
A schematic diagram of Vγ1.1+Vδ6.3+ γδ T cell developmental control by Id2 and Id3. In the developing thymus, γδ T cells that express the Vγ1.1 and Vδ6.3 TCR segments receive strong TCR signaling, up-regulating Id2 and Id3 through Egr1/2 and PLZF. The Id proteins inhibit activity of E proteins, affecting the survival and proliferation of Vγ1.1+Vδ6.3+ γδ T cells. When Id3 is present, and Id2 is expressed from a more active allele, such as the one from the 129 genetic background (Id2s, “strong”), E protein activity is very low and Vγ1.1+Vδ6.3+ γδ T cell population size is small. If Id3 is absent, and Id2 is expressed from a less active allele, such as the one from the B6 background (Id2B, “B6”), E protein activity becomes higher and the Vγ1.1+Vδ6.3+ γδ T cells expand dramatically. However, if both Id2 and Id3 are completely absent, E protein activity becomes too high and again impairs the survival and proliferation of Vγ1.1+Vδ6.3+ γδ T cells, limiting its population size.
Our study indicated that the level of E protein activity, regulated by Id2 and Id3 expression levels, is crucial for γδ T cell development, especially during the “maturation” stage. Both very high and very low E protein activity can limit the accumulation of γδ T cells, especially the Vγ1.1+Vδ6.3+ γδ T cells. Since Id proteins are up-regulated by TCR signaling, a developmental restrain imposed by high Id protein level and low E activity can be interpreted as a mechanism the body uses to limit the number of γδ T cells that can recognize self antigen in the thymus, reiterating the idea that Id2 and Id3 are “dual safety” involved in the negative selection of γδ T cells (10). However, this “negative selection” seems to be affected by age of the animal and TCR V segment usage. The Vγ1.1+Vδ6.3+ γδ T cells, but not other autoreactive γδ T cells, dramatically expand during the neonatal period in Id3 deficient mice on B6 background. Why is this specific population particularly sensitive to Id protein regulation? One possibility is the presence of its cognate antigen. Vγ1.1+Vδ6.3+ γδ T cells have been shown to recognize HSP60 of both mouse and Mycobacteria origin (25). Expression level of this antigen or other possible ligands of the Vγ1.1+Vδ6.3+ γδ TCR may change in the thymus during development, thus making these γδ T cells specifically prone to expand during the neonatal window, unless the Id proteins prevent them from doing so. Alternatively, the expression of Id proteins in response to TCR signaling may be different between cells generated in the neonatal period versus those generated in the adult stage, and different in cells utilizing other TCR V segments; mechanisms other than Id and E proteins may be more important in restraining autoreactive γδ T cells in those conditions, so they are less affected by Id protein deletion. Nevertheless, in the Id2 single conditional knockout mouse, γδ T cells other than those expressing Vγ1.1 and Vδ6.3 also expanded, indicating that Id2 is broadly involved in the suppression of γδ T cell expansion.
However, our study also showed that total loss of Id2 and Id3 can impair γδ T cell proliferation and survival. Unrestricted E protein activity can lead to death of T cells, especially effector and memory T cells, which is well documented in the studies of peripheral CD4 and CD8 αβ T cells (26, 27). γδ T cells are considered innate-like cells, and many of them have an effector phenotype even in the thymus (1). Therefore, it is not surprising that they share the same requirement of Id protein activity with effector αβ T cells.
What is the physiological consequence of having a larger pool of autoreactive γδ T cells? The Id3 deficient mice spontaneously develop an autoimmune disease similar to human Sjogren's syndrome (28). The large population of Vγ1.1+Vδ6.3+ γδ T cells in these mice is potentially involved in the pathogenesis. However, previous report also showed that these cells can play a role in suppressing tissue inflammation (3). More tests are required to further clarify the impact of the expanded γδ T cell population in mice with Id protein deficiency.
Supplementary Material
Acknowledgements
We thank Drs. Lasorella and Iavarone for sharing the Id2f strain, Drs. Sehoon Keum and Douglas Marchuk for help in performing SNP analysis, the Flow Cytometry Facility of Duke Comprehensive Cancer Center for cell sorting, and the Transgenic Mouse Facility of Duke Comprehensive Cancer Center for generating the GFP-floxedSTOP-Rsad2 transgenic mice.
This work has been supported by the National Institute of Health grants R01GM-059638 and R21RR-032742 to YZ and Duke University Medical Center Bridge Fund.
Footnotes
Conflict of Interest
Authors declare that they have no conflict of interests.
References
- 1.Bonneville M, O'Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nature reviews. Immunology. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 2.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nature reviews. Immunology. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nature reviews. Immunology. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 4.Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunological reviews. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 5.Grigoriadou K, Boucontet L, Pereira P. Most IL-4-producing gamma delta thymocytes of adult mice originate from fetal precursors. J Immunol. 2003;171:2413–2420. doi: 10.4049/jimmunol.171.5.2413. [DOI] [PubMed] [Google Scholar]
- 6.Azuara V, Grigoriadou K, Lembezat MP, Nagler-Anderson C, Pereira P. Strain-specific TCR repertoire selection of IL-4-producing Thy-1 dull gamma delta thymocytes. European journal of immunology. 2001;31:205–214. doi: 10.1002/1521-4141(200101)31:1<205::AID-IMMU205>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, Sant'Angelo DB. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber DJ, Azuara V, Levraud JP, Huang SY, Lembezat MP, Pereira P. IL-4-producing gamma delta T cells that express a very restricted TCR repertoire are preferentially localized in liver and spleen. J Immunol. 1999;163:3076–3082. [PubMed] [Google Scholar]
- 9.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zuniga-Pflucker JC, Wiest DL. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent “innate” gammadelta T cells. PLoS One. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Molecular and cellular biology. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M, Nam HS, Zhuang Y, Benezra R, Di Bernardo D, Iavarone A, Lasorella A. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nature cell biology. 2012;14:477–487. doi: 10.1038/ncb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Z, Li H, Han M, Xu T, Wu X, Zhuang Y. Modeling Sjogren's syndrome with Id3 conditional knockout mice. Immunology letters. 2011;135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- 17.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keum S, Marchuk DA. A locus mapping to mouse chromosome 7 determines infarct volume in a mouse model of ischemic stroke. Circulation. Cardiovascular genetics. 2009;2:591–598. doi: 10.1161/CIRCGENETICS.109.883231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nature immunology. 2006;7:995–1003. doi: 10.1038/ni1371. [DOI] [PubMed] [Google Scholar]
- 21.Kee BL. E and ID proteins branch out. Nature reviews. Immunology. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 22.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta M, Kraus ZJ, Gomez-Rodriguez J, Hwang SH, Cannons JL, Cheng J, Lee SY, Wiest DL, Wakeland EK, Schwartzberg PL. A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes. J Immunol. 2013;190:2121–2128. doi: 10.4049/jimmunol.1202145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleimer M, von Boehmer H, Kreslavsky T. PLZF Controls the Expression of a Limited Number of Genes Essential for NKT Cell Function. Frontiers in immunology. 2012;3:374. doi: 10.3389/fimmu.2012.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien RL, Fu YX, Cranfill R, Dallas A, Ellis C, Reardon C, Lang J, Carding SR, Kubo R, Born W. Heat shock protein Hsp60-reactive gamma delta cells: a large, diversified T-lymphocyte subset with highly focused specificity. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4348–4352. doi: 10.1073/pnas.89.10.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin YY, Jones-Mason ME, Inoue M, Lasorella A, Iavarone A, Li QJ, Shinohara ML, Zhuang Y. Transcriptional regulator Id2 is required for the CD4 T cell immune response in the development of experimental autoimmune encephalomyelitis. J Immunol. 2012;189:1400–1405. doi: 10.4049/jimmunol.1200491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nature immunology. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Dai M, Zhuang Y. A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity. 2004;21:551–560. doi: 10.1016/j.immuni.2004.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.