Abstract
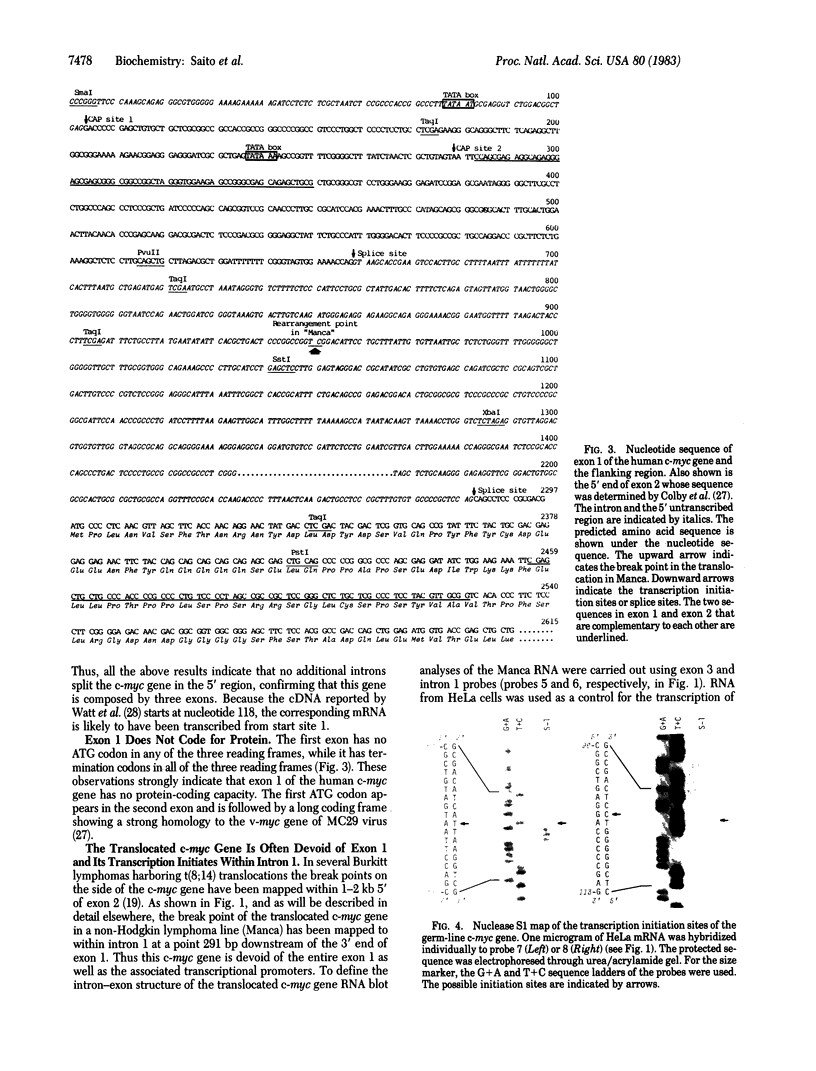
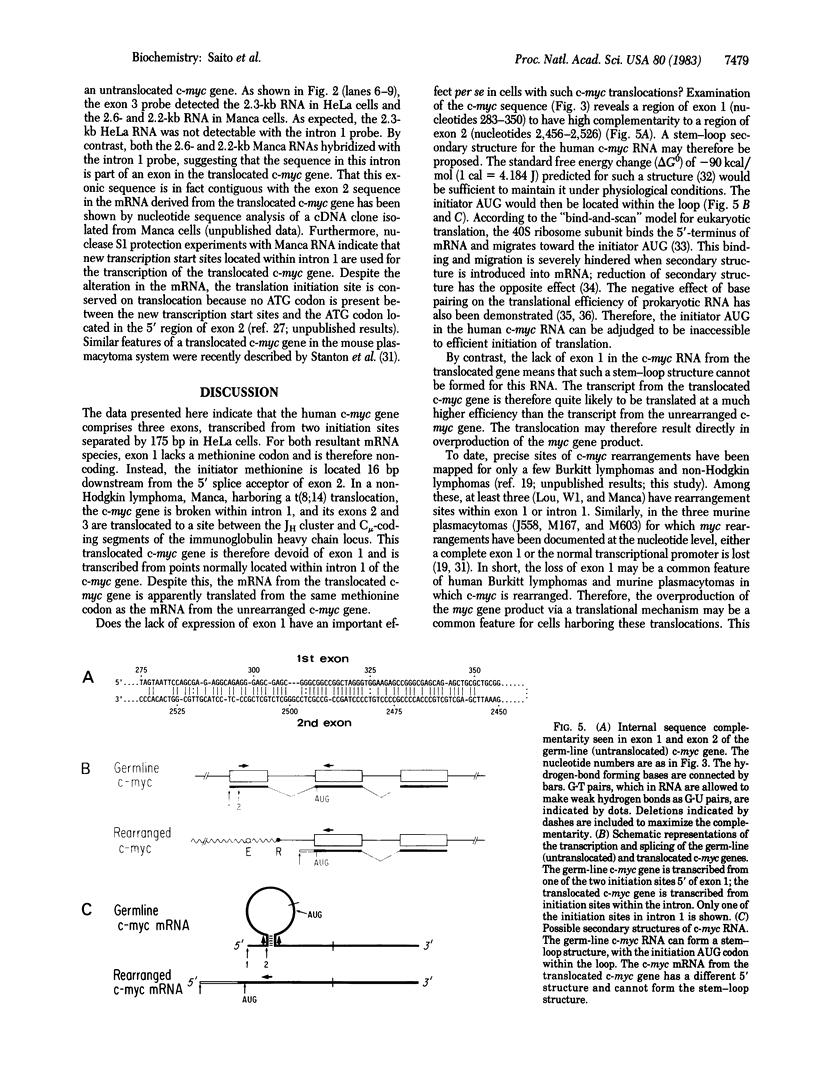
We have shown that the human cellular oncogene c-myc is composed of three exons and is transcribed from two initiation sites separated by 175-base-pair DNA in HeLa cells. For both resulting mRNA species, exon 1 composes the 5' untranslated region and the initiator methionine is located 16 base pairs down-stream from the 5' splice acceptor of exon 2. In a non-Hodgkin lymphoma, Manca, harboring a t(8; 14) translocation, c-myc gene is broken within intron 1, and its exons 2 and 3 are translocated to a site between the heavy chain joining region cluster and C mu-coding DNA segment of the immunoglobulin heavy chain locus. The translocated c-myc gene is transcribed from points within intron 1 but is apparently still translated from the same methionine codon as the mRNA from the unrearranged c-myc gene. The nucleotide sequence of the c-myc gene shows that a region of exon 1 is highly complementary to a region of exon 2. Thus the mRNA from the untranslocated c-myc gene, as opposed to that of the translocated c-myc gene, could form a stable stem-loop structure (delta Go = -90 kcal/mol; 1 cal = 4.184 J) where the initiator AUG would be located within the loop. In view of the bind-and-scan model for the initiation of eukaryotic translation, we propose that such a secondary structure will severely hinder the translation. We further propose that the c-myc gene is often activated by translocation through the escape from such a translational suppression.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Gerondakis S., Webb E., Corcoran L. M., Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. M., Gerondakis S., Webb E., Mitchell J., Bernard O., Cory S. Transcriptionally active DNA region that rearranges frequently in murine lymphoid tumors. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6966–6970. doi: 10.1073/pnas.79.22.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Bishop J. M., Smith D. H., Chen E. Y., Colby W. W., Levinson A. D. Nucleotide sequence to the v-myc oncogene of avian retrovirus MC29. Proc Natl Acad Sci U S A. 1983 Jan;80(1):100–104. doi: 10.1073/pnas.80.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Schwab M., Lin C. C., Varmus H. E., Bishop J. M. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1707–1711. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bister K., Hayman M. J., Vogt P. K. Defectiveness of avian myelocytomatosis virus MC29: isolation of long-term nonproducer cultures and analysis of virus-specific polypeptide synthesis. Virology. 1977 Oct 15;82(2):431–448. doi: 10.1016/0042-6822(77)90017-4. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calame K., Kim S., Lalley P., Hill R., Davis M., Hood L. Molecular cloning of translocations involving chromosome 15 and the immunoglobulin C alpha gene from chromosome 12 in two murine plasmacytomas. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6994–6998. doi: 10.1073/pnas.79.22.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby W. W., Chen E. Y., Smith D. H., Levinson A. D. Identification and nucleotide sequence of a human locus homologous to the v-myc oncogene of avian myelocytomatosis virus MC29. Nature. 1983 Feb 24;301(5902):722–725. doi: 10.1038/301722a0. [DOI] [PubMed] [Google Scholar]
- Collins S., Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982 Aug 12;298(5875):679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Bregni M., Erikson J., Patterson D., Gallo R. C., Croce C. M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R., Gelmann E. P., Martinotti S., Franchini G., Papas T. S., Gallo R. C., Wong-Staal F. Cloning and characterization of different human sequences related to the onc gene (v-myc) of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1982 Nov;79(21):6497–6501. doi: 10.1073/pnas.79.21.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R., Wong-Staal F., Gallo R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982 Sep 2;299(5878):61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- Fung Y. K., Crittenden L. B., Kung H. J. Orientation and position of avian leukosis virus DNA relative to the cellular oncogene c-myc in B-lymphoma tumors of highly susceptible 15I5 X 7(2) chickens. J Virol. 1982 Nov;44(2):742–746. doi: 10.1128/jvi.44.2.742-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Klein G. The role of gene dosage and genetic transpositions in carcinogenesis. Nature. 1981 Nov 26;294(5839):313–318. doi: 10.1038/294313a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980 Jan;19(1):79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- Kozak M., Nathans D. Translation of the genome of a ribonucleic acid bacteriophage. Bacteriol Rev. 1972 Mar;36(1):109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire R. T., Robins T. S., Thorgeirsson S. S., Heilman C. A. Expression of cellular myc and mos genes in undifferentiated B cell lymphomas of Burkitt and non-Burkitt types. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1947–1950. doi: 10.1073/pnas.80.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Harris L. J., Stanton L. W., Erikson J., Watt R., Croce C. M. Transcriptionally active c-myc oncogene is contained within NIARD, a DNA sequence associated with chromosome translocations in B-cell neoplasia. Proc Natl Acad Sci U S A. 1983 Jan;80(2):519–523. doi: 10.1073/pnas.80.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushinski J. F., Bauer S. R., Potter M., Reddy E. P. Increased expression of myc-related oncogene mRNA characterizes most BALB/c plasmacytomas induced by pristane or Abelson murine leukemia virus. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1073–1077. doi: 10.1073/pnas.80.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel B. G., Gasic G. P., Rogler C. E., Skalka A. M., Ju G., Hishinuma F., Papas T., Astrin S. M., Hayward W. S. Molecular analysis of the c-myc locus in normal tissue and in avian leukosis virus-induced lymphomas. J Virol. 1982 Oct;44(1):158–166. doi: 10.1128/jvi.44.1.158-166.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel B. G., Jhanwar S. C., Chaganti R. S., Hayward W. S. Two human c-onc genes are located on the long arm of chromosome 8. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7842–7846. doi: 10.1073/pnas.79.24.7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Robins T., Bister K., Garon C., Papas T., Duesberg P. Structural relationship between a normal chicken DNA locus and the transforming gene of the avian acute leukemia virus MC29. J Virol. 1982 Feb;41(2):635–642. doi: 10.1128/jvi.41.2.635-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J. D. Identification of the constant chromosome regions involved in human hematologic malignant disease. Science. 1982 May 14;216(4547):749–751. doi: 10.1126/science.7079737. [DOI] [PubMed] [Google Scholar]
- Saito H., Richardson C. C. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981 Dec;27(3 Pt 2):533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- Shen-Ong G. L., Keath E. J., Piccoli S. P., Cole M. D. Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell. 1982 Dec;31(2 Pt 1):443–452. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Taub R., Kirsch I., Morton C., Lenoir G., Swan D., Tronick S., Aaronson S., Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vennstrom B., Sheiness D., Zabielski J., Bishop J. M. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol. 1982 Jun;42(3):773–779. doi: 10.1128/jvi.42.3.773-779.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]