Abstract
Zebrafish transgenesis is increasingly popular owing to the optical transparency and external development of embryos, which provide a scalable vertebrate model for in vivo experimentation. The ability to express transgenes in a tightly controlled spatio-temporal pattern is an important prerequisite for exploitation of zebrafish in a wide range of biomedical applications. However, conventional transgenesis methods are plagued by position effects: the regulatory environment of genomic integration sites leads to variation of expression patterns of transgenes driven by engineered cis-regulatory modules. This limitation represents a bottleneck when studying the precise function of cis-regulatory modules and their subtle variants or when various effector proteins are to be expressed for labelling and manipulation of defined sets of cells. Here, we provide evidence for the efficient elimination of variability of position effects by developing a PhiC31 integrase-based targeting method. To detect targeted integration events, a simple phenotype scoring of colour change in the lens of larvae is used. We compared PhiC31-based integration and Tol2 transgenesis in the analysis of the activity of a novel conserved enhancer from the developmentally regulated neural-specific esrrga gene. Reporter expression was highly variable among independent lines generated with Tol2, whereas all lines generated with PhiC31 into a single integration site displayed nearly identical, enhancer-specific reporter expression in brain nuclei. Moreover, we demonstrate that a modified integrase system can also be used for the detection of enhancer activity in transient transgenesis. These results demonstrate the power of the PhiC31-based transgene integration for the annotation and fine analysis of transcriptional regulatory elements and it promises to be a generally desirable tool for a range of applications, which rely on highly reproducible patterns of transgene activity in zebrafish.
Keywords: Zebrafish, Integrase, Transgenesis, Tol2, Enhancer, Position effects
INTRODUCTION
Transgenesis is one of the fastest growing technologies in the zebrafish model system. ZFIN, the Zebrafish Model Organism Database (Bradford et al., 2011), lists 10,867 transgenic zebrafish lines. Transgenic zebrafish are produced for a variety of reasons, from in vivo labelling of cells and tissues to tissue-specific cell ablation, analysis of gene function, protein dynamics and localisation, generation of disease models through mis-expression of disease-associated genes, or manipulation of gene activities for developmental and physiological analysis (e.g. Gilmour et al., 2002; Langenau et al., 2005; Curado et al., 2008; Wyart et al., 2009; Hans et al., 2011).
With the publication of the ENCODE project and the discovery of the pervasiveness of predicted non-coding cis-regulatory modules (CRMs) in vertebrate genomes, emphasis is put on the functional validation of these computationally and biochemically predicted elements. Small laboratory fish models, such as zebrafish and medaka, with external development of optically transparent embryos and relatively easy transgenesis stand to make a major contribution to this area (Rada-Iglesias et al., 2011; Bernstein et al., 2012) and can meet the high-throughput capabilities demanded by the vast genomics data generated by next generation sequencing technologies (e.g. Gehrig et al., 2009). For functional analysis, predicted CRMs are often placed in front of a minimal promoter driving fluorescent protein expression. The construct is injected into zebrafish embryos and expression of the fluorescent reporter is used as a readout of CRM activity (reviewed by Ishibashi et al., 2013). In most cases, the CRM-fluorescent reporter cassette is placed into a Tol2 transposon vector to facilitate genomic integration of the transgene (Kawakami, 2004; Fisher et al., 2006; Kwan et al., 2007). However, the regulatory environment of the locus of the transgene integration may influence the expression of the transgenic reporter, leading to substantial variability of reporter expression through ectopic enhancement or silencing. These observations are collectively termed position effects (Jaenisch et al., 1981; Wilson et al., 1990; Rossant et al., 2011). Throughout this manuscript we use the term ‘variability of position effects’, to describe expression variation among transgenic lines with different integration sites to distinguish from ‘position effect variegation’ (PEV), a phenomenon characterised by variable changes (silencing) of a transgene among cells of an organism (Schotta et al., 2003). Variability of position effects are utilised in enhancer trapping, or enhancer detection, in which the regulatory landscape of the genome is probed by a weak expression cassette (Wilson et al., 1990; Balciunas et al., 2004; Parinov et al., 2004; Ellingsen et al., 2005; Davison et al., 2007; Scott et al., 2007; Asakawa et al., 2008). In most, if not all, other applications, where predesigned and controlled expression of transgenes is intended, random transgene integration and position effects are major obstacles. Such negative observations are often quoted in scientific meetings and discussions but are seldom published. We have found that 29 out of 30 lines generated with eight constructs using Tol2 transgenesis contained substantial variation of gene expression among lines with the same construct, probably caused by position effects (Y.H. and F.M., unpublished). Furthermore, flanking an expression cassette by insulator elements does not seem to reduce position effects (Grajevskaja et al., 2013). Taken together, the consequence of variable position effects is that often a large number of transgenic lines have to be generated with a single DNA construct to be able to obtain information on tissue-specific activity or to conclude correctly on the function of a tested cis-regulatory element.
Site-specific transgene integration using PhiC31 integrase has the potential to limit variability of position effects, leading to much faster and more precise analysis of putative CRMs in the zebrafish model system. Thus, site-specific integration would enable more efficient and high-throughput analysis of putative CRMs with increased reproducibility. Importantly, a robust targeted integration system would aid in the exploitation of the transgenic zebrafish model in the functional analysis of single nucleotide polymorphism (SNP) variants in CRMs identified in large numbers by recent genome-wide disease association studies (Ragvin et al., 2010; Maurano et al., 2012), particularly in those cases in which the SNP was expected to cause only subtle, quantitative changes in gene expression (e.g. Gaulton et al., 2010).
Several methods for site-specific integration of transgenes may be applied in zebrafish. Cre and Flp recombinases have been used for site-specific integration of transgenes in animal models (reviewed by Branda and Dymecki, 2004), but transgene integration potential of these recombinases has not yet been tested in zebrafish. A transcription activator-like effector nuclease (TALEN)-based method for site-specific transgene integration has been described but suffers from very low efficiency of germline transmission (Zu et al., 2013).
Based on successful application in the mouse and especially the fruit fly (Olivares et al., 2002; Groth et al., 2004; Venken and Bellen, 2012), we decided to develop a PhiC31-based method for analysis of enhancer activity in zebrafish. The natural function of PhiC31 integrase is to facilitate the integration of the phage genome into the bacterial host genome (Kuhstoss and Rao, 1991). PhiC31 integrase binds to two recognition sites, attP and attB, and exchanges DNA strands between them (Smith et al., 2004). Lister (Lister, 2010) demonstrated that PhiC31 integrase can facilitate intramolecular excision of a DNA sequence flanked by attP/attB sites in zebrafish (Lister, 2010; Lister, 2011). More recently, targeted integration of a transgene by PhiC31-mediated cassette exchange and insertion of circular plasmid were demonstrated in the genome of zebrafish (Hu et al., 2011; Mosimann et al., 2013).
We have developed a simple phenotype-based strategy using eye colour change to monitor site-specific integration of a transgene into the zebrafish genome. We have reproducibly achieved an ∼10% germline transmission rate with site-specific transgenesis using PhiC31. Compared with Tol2-mediated random integration, PhiC31-mediated site-specific integration resulted in reduced variability and aided in the detection of the tissue-specific activity of a novel neural enhancer from the locus of the esrrga gene. A modified phenotyping strategy was also developed to detect enhancer function by the analysis of targeted, mosaic transgene activity in microinjected founder embryos.
RESULTS
In vivo detection system for targeted integration of reporter constructs in zebrafish
We have adapted a site-specific integration system with PhiC31 integrase consisting of two components: ‘recipient transgenic lines’ containing docking attP site(s) in the genome, and targeting plasmids containing attB site(s) (Thorpe and Smith, 1998). In mouse and human cells, integrations were observed to also occur into ‘pseudo’ attP sites, i.e. endogenous sequences with similarity to the attP site (reviewed by Keravala and Calos, 2008). To facilitate screening for desired site-specific integrations, we designed a phenotypic selection based on fluorescent reporter colour change in the lens (Fig. 1A). Our recipient transgenic lines [Tg(Xla.crygc:attP-GFP)] contain a lens-specific crygc:GFP cassette (Davidson et al., 2003) with an attP site placed between the lens-specific Xenopus laevis gamma-crystalline promoter and GFP (henceforth crygc:attP-GFP for brevity). To monitor site-specific integration, our targeting vectors contain an attB site followed by a red fluorescent reporter (either mRFP or mCherry). Site-specific integration into crygc:attP-GFP docking site is expected to produce a crygc:attR-Red recombinant site, which can be scored by red fluorescence in the lens. Random integration of this attB-Red targeting vector into the genome is extremely unlikely to result in RFP expression.
Fig. 1.
In vivo detection system for targeted integration of reporter constructs in zebrafish. (A) Schematic of PhiC31 targeted integration system. Transgenic embryos containing an attP docking site have a green lens due to the gamma-crystalline promoter driving a GFP reporter. ITR labels recognition sequences of either Tol2 or Sleeping beauty transposases used in generating the recipient transgenic lines with docking site. Injection of a circular plasmid with attB and a red reporter (targeting vector) into recipient line eggs leads to eye colour change upon PhiC31-targeted integration in larvae. (B) Detection of eye colour change upon PhiC31-mediated integration in the transgenic recipient Tg(Xla.crygc:attP-GFP)uobL6 line. Top row shows a transgenic larva with PhiC31-mediated recombination of the attB-mCherry cassette into the attP-GFP docking site. Bottom row shows transgenic sibling from the recipient line without targeted integration. Side views on cranial end of 5 dpf larvae with anterior to the left. Scale bar: 100 μm. Arrows indicate reporter expression in the lens. (C) Sequence of the tpl102 recipient docking site with germline integration of targeting construct tnnT2:attB-mRFP. Sequence shows the attR site in the crygc:attR-Red recombination product. The three lower case nucleotides denote the recombination site.
Two different targeting site vectors were constructed: a Tol2-based vector pTol2/Xla.crygc:attP-GFP (pDB896) containing miniTol2 arms and a Sleeping Beauty-based vector pT2/Xla.crygc:attP-GFP (pKW1) (Fig. 1A). Docking site vectors were co-injected into zebrafish embryos with in vitro-transcribed Tol2 (Balciunas et al., 2006) or SB100X (Mátés et al., 2009) mRNA. Injected embryos positive for lens GFP fluorescence at 3 dpf were raised to adulthood. Founder adults were crossed and their embryos were screened for lens GFP fluorescence at 3-5 days post-fertilisation (dpf). GFP-positive embryos were raised to establish the F1 generation. Five different founders produced GFP-positive progeny in Tol2-mediated transgenesis and three different founders produced GFP-positive embryos for SB-mediated transgenesis (supplementary material Table S1). We molecularly characterised the five genomic loci harbouring the docking site by inverse PCR and Southern blotting (supplementary material Fig. S1 and Table S1) and confirmed linkage of the identified integration event with GFP fluorescence (tpl102, tpl103, tpl104, uobL6 and uobL12). The docking loci represent a variety of genomic landscapes: 700-kb intergenic region 100 kb away from nearest gene (uobL6), middle of a 100-kb intergenic region (uobL12), 5-kb intergenic region with two divergently transcribed genes (tpl102), and within a gene (tpl103 and tpl104) (supplementary material Table S1 and Fig. S2). Third generation homozygous individuals of recipient lines uobL6 and uobL12 have been outcrossed and resulting offspring were raised to adulthood without notable reduction in fecundity and viability, whereas incrosses of the lines tpl102, tpl103 and tpl104 did not lead to developmental abnormalities in their offspring up to 6 dpf, arguing against mutagenicity of the docking site transgenes (data not shown).
High frequency of PhiC31-mediated targeting detected by lens reporter activity
Next, we investigated whether we can visualise PhiC31 integrase-mediated insertion of a targeting plasmid containing an attB site into an attP site in zebrafish using an in vivo detection system of reporter activity in the lens. In vitro-transcribed PhiC31 mRNA was injected along with attB-Red constructs into one-cell zebrafish embryos from the Tol2-based transgenic recipient line Tg(Xla.crygc:attP-GFP). When transgenic embryos containing the docking site were co-injected with PhiC31 mRNA and the targeting construct pJET-attB-mCherry, up to 66.7% of normal developing larvae were RFP-positive in one or both lenses (n=258, Fig. 1B). By contrast, 0% (n=302) of recipient transgenic embryos injected with targeting vector DNA without PhiC31 mRNA were RFP-positive in the lens, indicating that the eye colour change was due to integrase-mediated recombination events at docking sites in the recipient transgenic locus. We confirmed integrase-dependent transgene integration by PCR amplification and sequencing of the attL and attR sites, which arise by recombination of attP and attB sites (Fig. 1A,C).
In accordance with previously published observations (Hu et al., 2011), we observed that PhiC31 RNA in vitro transcribed from a pT3TS (Hyatt and Ekker, 1999) or pCS2+ based vectors was highly toxic to zebrafish embryos (supplementary material Table S2). A significant percentage of abnormal/dead embryos was observed using as little as 15 pg of integrase RNA (supplementary material Table S2). It was recently demonstrated that addition of nanos1 3′ UTR to PhiC31 mRNA leads to improved embryo survival and germline transmission of recombination events in zebrafish (Hu et al., 2011). Indeed, we observed improved survival upon injection of PhiC31-nos1-3′UTR compared with PhiC31 without nanos1 3′ UTR (supplementary material Table S2). Injection of PhiC31-nos1-3′UTR mRNA did not substantially reduce the viability of primordial germ cells (supplementary material Table S2). However, co-injection of PhiC31-nos1-3′UTR into SB-based recipient lines resulted in varying percentage of lens RFP-positive embryos. We observed 48% (33/69), 14% (6/43) and 36% (9/25) RFP-positive embryos upon injection into tpl102, tpl103 and tpl104, respectively (data not shown). RFP-positive embryos were raised to adulthood, outcrossed and resulting embryos were screened for lens RFP. Rates of site-specific germline transgenesis are ∼10%, and they slightly varied between experiments using different docking loci and different types of integrase RNA (supplementary material Table S3).
Identification of enhancer-specific expression features obtained in various genomic loci
To test if site-specific integration would help in reducing the variability of position effects in experiments aiming to characterise enhancer-driven tissue-specific reporter gene activity, we analysed reporter activity driven by a putative highly conserved enhancer after integration into the genome either by Tol2 or by PhiC31 integrase. In order to select candidate enhancers, we have identified highly conserved syntenic sequences with >80% sequence identity between the zebrafish and human genome (see Materials and Methods). They do not overlap any known exon of annotated coding or noncoding genes. We choose EL161 for this study, which resides in the last intron of the esrrga gene in zebrafish (supplementary material Fig. S3). Based on the conserved synteny and conservation around the esrrga gene, we postulated that EL161 may regulate esrrga expression (Bardet et al., 2004; Thisse et al., 2004). In a preliminary study, EL161, when linked to the krt4 gene minimal promoter with minimal transcriptional activity (Gehrig et al., 2009) was able to activate reporter transgenes in the brain; however, the expression was highly variable due to position effects (our unpublished data).
The Tol2/EL161-krt4:YFP cassette containing the EL161 element was then used with both Tol2- and PhiC31-mediated transgenesis. Tol2-mediated transgenesis is expected to integrate only the Tol2/EL161-krt4:YFP cassette into the genome (Fig. 2A, top), whereas PhiC31-mediated transgenesis integrates the whole vector containing Tol2/EL161-krt4:YFP and attB:mCherry (Fig. 2B, top). We screened F1 embryos of 20 founders injected with Tol2 by fluorescence microscopy, and recovered ten different YFP expression patterns, suggesting multiple integration sites and that Tol2/EL161-krt4:YFP is highly susceptible to position effects (Fig. 2A). We screened the offspring of all 29 adult founders from uobL6 and uobL12 injected with PhiC31 integrase and showing eye colour change during larval development. Four of these founders gave progeny with red lenses together with YFP expression pattern, indicating site-specific integration events (Fig. 2B; supplementary material Tables S3, S4). Notably, the three founders with targeted integration into the uobL6 line showed remarkable similarity in their expression patterns, in contrast to Tol2-mediated transgene integrations, which were characterised by widely varying patterns (Fig. 2; supplementary material Table S5).
Fig. 2.
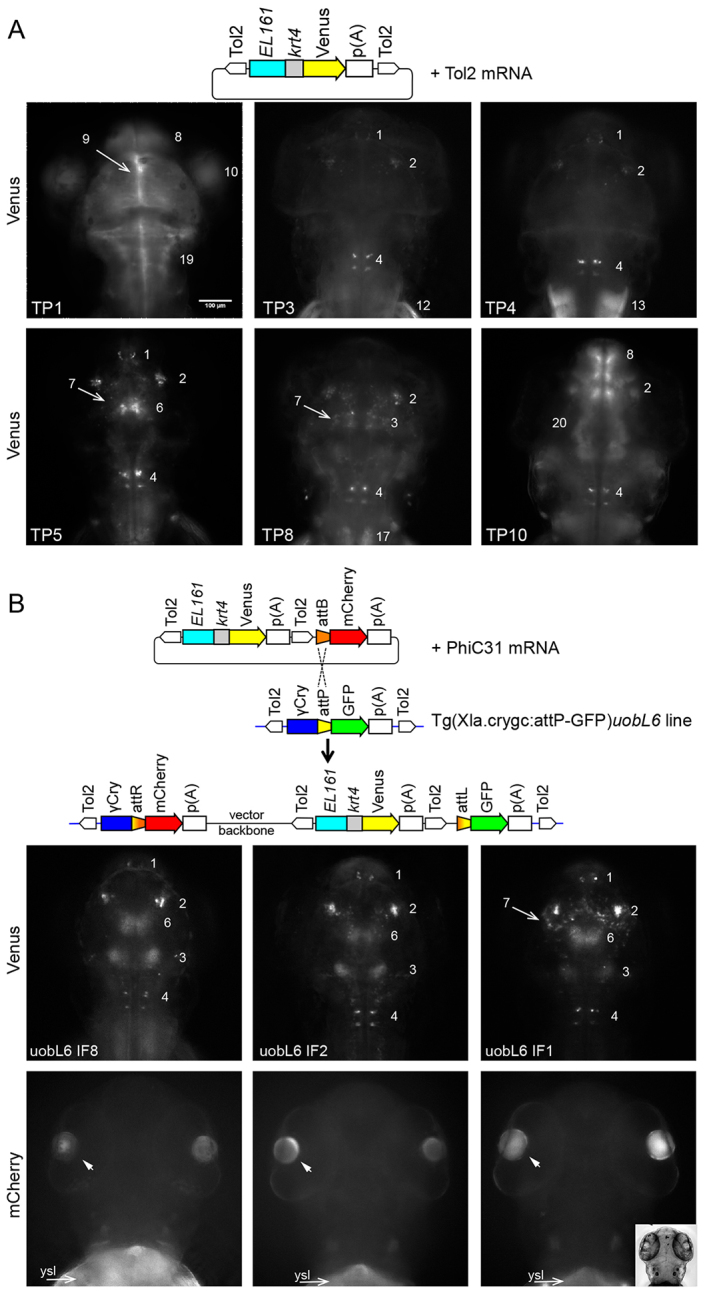
Variability of position effects on reporter activity is sharply reduced among PhiC31 integrase-mediated transgenic lines compared with Tol2 transgenesis. (A) Schematic of the pTol2/EL161-krt4:Venus construct injected with Tol2 transposase mRNA. Below are examples of expression patterns of transgenic F1 larvae (3 dpf). TP indicates Tol2-mediated expression patterns. Numbers refer to domains of Venus expression activity (listed in Fig. 3B). (B) Schematic indicates the recombination of the targeting plasmid pattB-mCherry,Tol2/EL161-krt4:Venus into the attP docking site of the transgenic recipient line Tg(Xla.crygc:attP-GFP)uobL6. Larvae from three different founders (IF1, IF2, IF8) targeted with PhiC31 integrase are shown. Venus expression domains are labelled as in Fig. 3B. Lens activity driven by mCherry (arrowheads in bottom row) demonstrates PhiC31-mediated integration. Insert in bottom right shows brightfield image of the head region. Arrows labelled ‘ysl’ in red channel indicate auto-fluorescence of the yolk syncytial layer. Dorsal views of larvae head are shown. Scale bar: 100 μm.
To evaluate the specific reporter activity driven by EL161 element and to dissect enhancer-driven activity from position effects, we have identified distinct expression domains and analysed the frequency of their occurrence in targeted and random transgene integration loci. We have identified a total of 20 domains of activity labelled 1-20 (Figs 2, 3; supplementary material Figs S4, S5). The PhiC31 integrase-mediated expression activity was present in seven distinguishable expression domains in the neural tube (Fig. 3). Six out of seven expression domains were shared between the three lines with targeting events in the same L6 docking site and only one additional domain (Domain 7) registered in one line indicating mild variation (Fig. 3B). By contrast, Tol2 lines showed a total of 21 expression domains with a variation between seven and 12 domains per pattern, which included a variety of tissues such as somatic muscle, pectoral fins and heart besides neural tube activity (Fig. 3B). This result indicates that integrase-mediated targeting of a single locus (in uobL6) led to reduced variability of expression patterns induced by a neural enhancer among transgenic lines.
Fig. 3.
Reproducible Venus expression patterns upon targeted integration of EL161 enhancer construct demonstrate cis-regulatory function in the brain. (A) Brain-specific enhancer effect of a transgene inserted in different integration sites. Domains of Venus activity are specified in B. Dorsal (top) and lateral (bottom) views of the head of 3 dpf F1 transgenic larvae. Scale bar: 100 μm. (B) Domains of Venus reporter expression and their distribution among targeted (PhiC31 integrase) and randomly (Tol2 transposase) transgenic lines. IF, integrase-injected founders; IP, integrase-mediated expression patterns; TF, Tol2-injected founders; TP, Tol2-mediated expression patterns. Blue depicts expression domains overlapping with esrrga activity, whereas red colour indicates additional domains. Light blue indicates weak expression.
The expression domains 1-6 were shared among uobL6 and uobL12 integration loci and were present in eight out of ten Tol2-mediated transgene loci, which together suggest autonomous enhancer activity in these neural domains. This result demonstrates that the shared expression patterns are unlikely to be due to position effects at the uobL6 locus and are autonomous property of the targeting transgene (Fig. 3A,B). Notably, these six shared expression domains, including epiphysis, diencephalic and hindbrain nuclei and tegmentum, overlap with expression domains of the esrrga gene (supplementary material Fig. S6) (also see www.zfin.org) (Bardet et al., 2004), suggesting that EL161 may contribute to the activity of esrrga in these neural subdomains. Low level of variability was still observed in these transgenic lines, at least in part explained by variation in focal planes of imaging as well as by potential differences in developmental stage of the individuals imaged. For stage-dependent variation of transgene activity, see supplementary material Fig. S7. The variability of expression patterns among PhiC31-mediated targeted integrants was significantly lower than those found among Tol2-mediated transgene integrants (likelihood ratio=15.0, DF=1, P=0.0001). Taken together, the low variability of transgene activity observed in PhiC31-targeted integration events demonstrate superior reproducibility and robustness of enhancer-driven expression patterns compared with that obtained by transposase-mediated integration.
Transient transgenesis strategy for rapid evaluation of enhancer activity upon PhiC31-mediated targeting
Transient transgenic zebrafish with mosaic reporter activity have often been used for the analysis of enhancer function because they allows high-throughput analysis of cis-regulatory elements (Gehrig et al., 2009; Rada-Iglesias et al., 2011). Because transient transgenesis is likely to suffer from variability, we tested whether the PhiC31-based integration strategy could also be applied for analysis of enhancer activity in mosaic transgenic embryos. To ensure that only targeted integration events are detected by reporter activity, we have designed a modified targeting strategy (Fig. 4), whereby an attB site is cloned adjacent to an enhancer-promoter combination, and can only lead to GFP activity driven by these cis-regulatory elements when PhiC31 integrase targets the donor construct upstream to the GFP in the recipient line (Fig. 4A). A set of well-characterised embryonic midline-specific enhancers, ar-A, B and C in introns 1 and 2 and ar-D in the upstream sequence of the promoter from the sonic hedgehog (shh) gene (Müller et al., 1999; Hadzhiev et al., 2007), were joined to the attB with or without mCherry reporter. These donor vectors were injected with PhiC31o-nos1-3′UTR integrase mRNA into embryos of incross of uobL12 recipient line, in which the multicopy docking site maximised the probability of targeting by PhiC31 integrase. Expression of gfp in tissues [floor plate, ventral brain, retina (Shkumatava et al., 2004)] in which shh enhancers are active was detected by whole-mount in situ hybridisation in 52.0% (n=25) and 46.2% (n=91) of donor construct-injected embryos with or without mCherry, respectively at high-pec stage with green lens. No embryos without lens reporter (n=5 and n=23, respectively) showed any gfp expression (Fig. 4B,C; supplementary material Table S6). These results indicate that gfp expression driven by shh regulatory elements was due to landing at the docking site. Expression was highly specific to the shh pattern, only 15.4% (n=13) and 6.3% (n=16) of analysed embryos with shh pattern showing ectopic gfp activity in a small number of cells (data not shown) and no embryos showing ectopic activity without shh pattern, respectively. The activity of the GFP reporter protein was also detected by fluorescence albeit at lower frequency, 3.8% (n=53) and 4.4% (n=91) of lens GFP-positive embryos [3.8% (n=53) and 4.4%, (n=91), respectively], compared with 0% (n=23) of lens GFP-negative embryos (Fig. 4C). These results together demonstrate a proof of principle application for enhancer testing in mosaic transgenic zebrafish embryos using PhiC31 integrase-mediated transgenesis.
Fig. 4.
Transient transgenesis strategy for rapid evaluation of enhancer activity upon PhiC31-mediated targeting. (A) Schematic of PhiC31-targeted integration system, designed for cis-regulatory element analyses in transient transgenic embryos. ‘-2.4 shh’, 2.4 kb upstream region of shh gene; ‘shhABC’, shh complete introns 1 and 2, with ar-A, ar-B, ar-C enhancers (Müller et al., 1999; Ertzer et al., 2007). (B) Detection of reporter activity in embryos injected with construct containing the shh cis-regulatory elements by in situ hybridisation (B) and fluorescence microscopy (C). Arrows indicate activity in shh expression domains, resulting from successful integration of the shh cis-regulatory cassette in to the attP site of uobL12, whereas arrowheads indicate the activity of the crygc promoter in uobL12 recipient line. Scale bars: 150 μm.
DISCUSSION
We tested the ability of PhiC31 integrase to site-specifically integrate transgenes into the zebrafish genome. We devised a simple phenotypic screening strategy based on change of lens fluorescence, and demonstrated germline transmission of site-specific integration rates in >10% of injected founders using different targeting transgenes, different recipient lines and two different versions of in vitro transcribed PhiC31 transposase. The simple phenotyping tool based on lens colour change provides an easily detectable measure of targeted integration events, yet is restricted to the lens with little interference with imaging anatomical features commonly studied by reporter expression-based assays. We chose the crystalline promoter for screening integrants because it is representative of a specialised class of promoters characteristic of highly active tissue-specific genes with no interaction with long-range enhancers (reviewed by Lenhard et al., 2012). As expected, no interference was detected from the crystallin promoter on the tissue-specific enhancer construct inserted in the vicinity.
We have demonstrated a reproducible expression pattern driven by a predicted transcriptional enhancer using targeted integration of an expression cassette. The observed expression domains appeared to match broadly with expected activity of esrrga, the predicted target gene for the enhancer. Given that enhancers can act at distances as far as 1 Mb (reviewed by Krivega and Dean, 2012), unambiguous identification of the target gene(s) remains challenging. Nevertheless, we were able to identify several esrrga expression domains with reproducible activity, which were attributable to the enhancer in independent genomic integration sites. Tol2 transgenics also showed several of the enhancer-specific patterns but were mostly coupled to a variety of additional ectopic patterns. It is unlikely that the additional patterns are due to Tol2 sequences as the PhiC31 donor vector also included them.
uobL6 appeared to be an excellent landing site for the analysis of the EL161 enhancer; however, this may not be the case for other CRMs. It is expected that genomic context and enhancer promoter interaction specificity (Gehrig et al., 2009) will affect different CRMs to varying degrees. To identify optimal sites for a broad range of CRMs, more landing sites will need to be generated and evaluated in the future. Towards this long-term goal we have established and molecularly characterised five different recipient lines. The five ‘docking’ loci cover a variety of locations with respect to potential position effects, including intragenic as well as large (0.7 and 0.1 Mb) intergenic regions, and provide a variety of genomic contexts for future analyses of position effects. Many pseudo-att sites may exist in the genome (Hu et al., 2013). Integration of the donor plasmid into a pseudo-att site is expected to yield YFP fluorescence in the absence of RFP fluorescence. We observed 100% concordance between red lens colour and YFP expression pattern, which indicates that such non-specific events are extremely rare, consistent with a recent publication by Mosimann and colleagues (Mosimann et al., 2013).
We chose insertion of full donor constructs as opposed to the previously published strategy of cassette exchange (Hu et al., 2011). Cassette replacement has the advantage that the vector backbone is not integrated, eliminating the potential impact of bacterial vector backbone on transgene expression. However, the advantage of the insertion strategy applied here lies in the simplicity of the transgene design and the fact that there appears to be no size limit for the sequence to be integrated (Venken et al., 2009), making it potentially applicable for analysis of large vectors such as bacterial artificial chromosomes to test regulatory function of super-enhancers, CRMs acting at large distance and multiple CRMs acting synergistically (Kikuta et al., 2007; White et al., 2008).
In accordance with previously published work, we find that wild-type PhiC31 integrase RNA is toxic to zebrafish embryos (Hu et al., 2011). Application of PhiC31-nos1-3′UTR resulted in substantially increased embryo survival and somewhat reduced activity in somatic tissues, similarly to previously published work (Hu et al., 2011). Interestingly, application of PhiC31-nos1-3′UTR did not result in a significant increase in germline transmission of targeting events. It is also reasonable to expect that different recipient loci may have different accessibility for PhiC31-mediated integration. In this light, the fact that our site-specific transgenesis rate was reproducible using different recipient lines, different targeting vectors and two different integrase expression vectors is very encouraging for future reproducibility of this method.
We have demonstrated in a proof of principle experiment that a PhiC31 integrase-mediated targeting strategy can also aid in generating transient transgenic embryos with reproducible patterns of mosaic expression of transgene reporter driven by tissue-specific cis-regulatory elements. Transient transgenics are immensely useful for fast and high-throughput analysis of a large number of potential cis-regulatory modules (Woolfe et al., 2005; Gehrig et al., 2009; Bernstein et al., 2012). The targeted integration system described herein will improve the reliability and thus the utility of transient transgenesis-based enhancer validation data. The targeting strategy presented here has the advantage of unambiguous identification of integration events by tissue-specific activity of the GFP reporter. However, it is suboptimal for fluorescence microscopy potentially owing to reduced translational efficiency of the reporter when preceded by an attR site. Future experiments will be required to test alternative integration strategies that do not impact on translational efficiency (e.g. a system whereby reconstitution of mRNA splicing occurs upon targeted integration) and/or test potentially more sensitive, enzymatic reporters, such as lacZ.
It should be noted that reproducibility of transgene expression offered by site-specific integration has a much broader range of potential application than functional characterisation of CRMs. It will enable investigators to first integrate a cassette containing a fluorescent reporter into a specific locus and determine precise spatiotemporal expression pattern of the transgene. Subsequently, a cassette in which the fluorescent reporter has been replaced with an effector transgene (e.g. Cre recombinase, calcium channel rhodopsin, calcium sensor, nitroreductase or dominant-negative signalling protein) integrated into the very same locus will provide highly reproducible spatiotemporal expression activity. Furthermore, site-specific integration will simplify comparison of phenotypic effects of several different transgenes expressed in the same spatiotemporal pattern at comparable levels.
During the revision of this manuscript, a similar report has been published showing enhancer testing with PhiC31 integrase-mediated integration in the medaka (Oryzias latipes), signifying the importance of this technology in vertebrate model organisms (Kirchmaier et al., 2013).
MATERIALS AND METHODS
Generation of plasmid vectors for PhiC31-mediated integration system in zebrafish
We generated two recipient vectors with an attP docking site for the PhiC31 integrase. pTol2/Xla.crygc:attP-GFP (pDB896) vector contains miniTol2 arms (Balciunas et al., 2006), ∼390 bp of the X. laevis gamma-crystallin promoter (Davidson et al., 2003), an 83 bp attP integrase recognition site (Thorpe and Smith, 1998), GFP coding sequence and a SV40 pA tail. pT2/Xla.crygc:attP-GFP (pKW1) is identical to the Sleeping Beauty vector pT2/gCry:GFP (pDB387) (Davidson et al., 2003), but has an 83 bp attP site inserted between BamHI and KpnI sites. The targeting vector pJET-attB-mCherry was constructed by joining an 84 bp attB sequence (Thorpe and Smith, 1998) to the mCherry coding sequence with polyA tail by PCR. This PCR product was then cloned into pJET1.2 using a blunt-ended cloning kit (Thermo Scientific). The targeting vector tnnT2(3.2)-attB:mRFP contains a heart-specific 3.2 kb tnnT2 promoter followed by 83 nucleotide attB site and monomeric RFP (mRFP) (Campbell et al., 2002) for potential phenotypic screening of heart RFP activity indicating random or illegitimate integration events). The pCS2+PhiC31 plasmid was generated by amplifying PhiC31 coding sequence from plasmid pPhiC31o (Addgene) using the following primers: F: 5′-AAAAGGATCCATGGATACCTACGCCGGAG-3′; R: 5′-AAATTCTCGAGTCACACTTTCCGCTTTTTCTTAG-3′. This sequence was then cloned into pCS2+ vector using BamHI and XhoI sites. pCS2+PhiC31 plasmid was linearised with NotI (New England Biolabs) and this product was used to synthesise PhiC31 mRNA using the in vitro transcription kit mMESSAGE mMACHINE (Ambion), according to the manufacturer’s instructions. The pT3TS-PhiC31 (pRG3) plasmid contains an integrase open reading frame from pCS2+PhiC31 cloned between BglII and SpeI sites of in vitro transcription vector pT3TS (Hyatt and Ekker, 1999). The plasmid was linearised using XbaI and in vitro transcribed using mMESSAGE mMACHINE (Ambion) in vitro transcription kit and T3 RNA polymerase. After RNA synthesis and DNase digestion, RNA was purified using RNeasy MiniElute RNA purification kit (Qiagen). pCS2+PhiC31o-nos1-3′UTR plasmid was a kind gift from Dr Shannon Fisher (University of Pennsylvania, PA, USA). The mRNA for microinjection was made as described by Hu et al. (Hu et al., 2011), except that after synthesis and DNase digestion, RNA was purified using RNeasy MiniElute RNA purification kit (Qiagen). Constructs described in this article will be made available through Addgene.
Identification conservation analysis and cloning of esrrga enhancer
The zebrafish (Zv9) and human (GRCh37) repeated masked version of the genomes were downloaded from the Ensembl FTP site (Flicek et al., 2013). The Ensembl gene annotations (coordinates of the genes and the exons and homology relationships) were downloaded from BioMart using R and the Bioconductor package BiomaRt (ensembl mart version 60) (Durinck et al., 2005; Durinck et al., 2009). The zebrafish genome was aligned against the human genome using blastn (Camacho et al., 2009) from the blast+ suite (version 2.2.24) with a word_size parameter of 50 bp. The resulting conserved regions were mapped according to the overlapping/flanking genes. Sixteen conserved elements showing the following features were considered: (1) minimum 500 bp length, (2) minimum 80% identity, (3) overlapping and/or directly flanking at least one pair of homologous genes between zebrafish and human and (4) presence of the element in the same genomic context (intronic or intergenic) in the two species. The 794 bp conserved element EL161 from the last intron of the esrrga gene was selected from the list and amplified by PCR from zebrafish genomic DNA using the oligos 5′-CATCCTAAATTTGGTGTCCTCTCTG-3′ (forward) and 5′-TGCGTCAACAAGATGAGATGAAC-3′ (reverse) and cloned upstream of the minimal promoter from the keratin 4 (krt4) gene and a Venus reporter gene (Gehrig et al., 2009) using Gateway Multisite System (Urasaki et al., 2006; Roure et al., 2007) resulting in the vector pTol2/EL161-krt4:Venus. For PhiC31 integrase-mediated experiments, the attB-mCherry cassette from pJET-attB-mCherry vector was cloned into the NotI site of pTol2/EL161-krt4:Venus construct using PCR In-Fusion System (Clontech), according to the manufacturer’s instructions. The primers used for amplifying attB-mCherry cassette were: F: 5′-AGTTCTAGAGCGGCCGCATTGACGGTCTCGAAGCCG-3′ and R: 5′-ACCGCGGTGGCGGCCGCAAAAAACCTCCCACACCTCCC-3′. The resulting vector was named pattB-mCherry,Tol2/EL161-krt4:Venus.
Production and microinjection of zebrafish fertilised eggs
Fertilised zebrafish eggs were generated by natural crossings. The zygote stage embryos were injected with either with 3 nl of water solution containing 8.3 ng/μl DNA and 8.3 ng/μl of PhiC31 integrase transcribed from pT3TS or with 15 ng/μl of DNA and 15 ng/μl of PhiC31 integrase transcribed from pCS2+ vector. After injections, embryos were incubated in embryo water containing 0.1 g/l of Instant Ocean salts or E3 medium at 28-29°C (Westerfield, 1993).
Fluorescence reporter expression analysis
Transgenic embryos and larvae [60-96 hours post-fertilisation (hpf)] were imaged using Olympus ScanR high content screening microscope (Olympus) as described (Gehrig et al., 2009). Anaesthetised embryos were oriented in an agarose template made with a brass mold (Peravali et al., 2011). Stacks containing 100 slices of 4 μm thickness were taken, out of focus slices were manually removed and maximum projections were made using an extended depth of field plug-in for ImageJ software (Forster et al., 2004). Expression domains were identified by comparing to anatomies and gene expression data in ZFIN database (www.zfin.org). Statistical analysis of variation of expression domains was carried out by binomial regression using a generalised linear mixed-effects model.
Whole-mount in situ hybridisation
Whole-mount in situ hybridisation was carried out as previously described (Thisse and Thisse, 2008). Digoxigenin (DIG)-labelled antisense probes were produced by PCR using the following primers: Venus probe: F: 5′-CAAGGGCGAGGAGCTGTT-3′ and R: 5′-AACTCCAGCAGGACCATGT-3′; esrrga probe: F: 5′-CACTAACGGACAGCGTAAATCA-3′ and R: 5′-TCTCCAGCTTCATGCTCTTG-3′; GFP probe: F: 5′-ACACTTGTCACTACTTTCGC-3′ and R: 5′-AACTCAAGAAGGACCATGTG-3′. Embryos were mounted in glycerol and imaged on an AxioScope microscope (Carl Zeiss).
Analysis of shh enhancer activity in transient transgenic embryos
The reporter constructs -2.4shh:attB:mCherry:ABC and -2.4shh:attB:ABC were generated by replacing the GFP:poly(A) cassette from the -2.4shh:gfp:ABC vector (Ertzer et al., 2007) with PCR-amplified attB:mCherry:p(A) or attB cassettes from the pJET-attB-mCherry vector, using XhoI/NotI restriction sites. About 2 nl of injection solution, containing 20 ng/μl reporter plasmid DNA, 15 ng/μl PhiC31nos mRNA and 0.1% Phenol Red, was injected into zygotes of the landing lines. The injected embryos were analysed for reporter expression by fluorescence microscopy and whole-mount in situ hybridisation using GFP DIG-labelled probe at 48 hpf. The reporter-positive embryos identified by the in situ hybridisation were categorised according to number and type of positive cells in relation to the expected pattern of reporter construct activity.
Analysis of primordial germ cell (PGC) survival
PGC numbers were assessed at 24 hpf in embryos injected with PhiC31 and PhiC31o-nos1-3′UTR mRNA (15 and 30 ng/μl, respectively) by anti-Vasa antibody staining (kind gift from Holger Knaut, Skirball Institute, NY, USA) as previously described (Nusslein-Volhard and Dahm, 2002). Antibodies used were anti-Vasa (rabbit, 1:500) (Knaut et al., 2000) and secondary antibody conjugated with HRP (goat, 1:150; A10260, Life Technologies). Detection was carried out with DAB Peroxidase Substrate Kit (Vector Laboratories) following the manufacturer’s instructions.
Molecular analysis of integration events
Genomic DNA was prepared from batches of lens GFP-positive and lens GFP-negative embryos from an F1 or F2 outcross of Tg(pT2/Xla.crygc:attP-GFP)tpl102, Tg(pT2/Xla.crygc:attP-GFP)tpl103, Tg(pT2/Xla.crygc:attP-GFP)tpl104, and from an F3 outcross of Tg(Xla.crygc:attP-GFP)uobL6 and Tg(Xla.crygc:attP-GFP)uobL12 lines. To map pT2/Xla.crygc:attP-GFP integrations in lines tpl102, tpl103 and tpl104 integrations, genomic DNA was digested in two separate reactions with TaqI, and a mix of BamHI/BclI/BglII restriction enzymes. The digested DNA was ligated overnight as described (Hermanson et al., 2004; Clark et al., 2011). Amplification of genomic sequences flanking pT2/Xla.crygc:attP-GFP was achieved by carrying two nested PCR reactions. For TaqI-digested DNA, the 5′ end of the construct was amplified using the primer pairs LP1 (5′-GTGTCATGCACAAAGTAGATGTCC-3′) and GFPF3 (5′-TGAGTAAAGGAGAAGAACTTTTCAC-3′) in the first PCR reaction followed by the second PCR reaction with LP2 (5′-ACTGACTTGCCAAAACTATTGTTTG-3′) and GFPF1a (5′-TTCACTGGAGTTGTCCCAATTCTTG-3′). Similarly, the 3′ end of the construct was amplified using nRP1 (5′-CTAGGATTAAATGTCAGGAATTGTG-3′) and SBR1 (5′-ACACTCAATTAGTATTTGGTAGCAT-3′) primers in the first PCR, and RP2 (5′-GTGAGTTTAAATGTATTTGGCTAAG-3′) and SBR2 (5′-CATTGCCTTTAAATTGTTTAACTTG-3′) in the second PCR reaction. For BamHI/BclI/BglII-digested DNA, the 5′ end of the construct was amplified with LP1 (see above) and SBF1 (5′-AATTCCAGTGGGTCAGAAGTTTACA-3′), followed by second PCR with LP2 (see above) and SBF2 (5′-GTTGACTGTGCCTTTAAACAGCTTG-3′) primers. The 3′ end of the construct was isolated using nRP1 and GFPR1 (5′-CAAGAGTGCCATGCCCGAA-3′) as well as RP2 (see above) and GFPR2 (5′-TTGCGAGATACCCAGATCATA-3′) in first and second PCRs, respectively. To map Tg(Xla.crygc:attP-GFP)uobL6 and uobL12 integrations, genomic DNA was digested in three separate reactions: TaqI, NlaIII, and BamHI/BclI/BglII. After ligation, inverse PCRs were carried out to amplify the 3′ end of the construct only using the same primers for all three differently digested genomic DNA sources: Tol2-R3 (5′-ACTGGGCATCAGCGCAATTCAATTG-3′) and S1/3′ No1 (5′-GTTGTTAACTTGTTTATTGCAGCTT-3′), as well as Tol2-R4 (5′-ATAATACTTAAGTACAGTAATCAAG-3′) and S1/3′ No2 (5′-CAAACTCATCAATGTATCTTATCAT-3′) in first and second PCRs, respectively. In addition to the transposase-mediated integration recovered in Tg(Xla.crygc:attP-GFP)uobL12, this line also has a non-transposase-mediated integration even of pTol2/Xla.crygc:attP-GFP. Transgenic lines with integrase docking sites will be made available from the zebrafish stock centres at the Zebrafish International Resource Center (ZIRC) and the Karlsruhe Institute of Technology (KIT).
Southern blotting
DNA from GFP-positive and GFP-negative embryos from the five recipient lines was digested with PvuII and PstI. Specificity and completeness of digestions was verified by carrying out a parallel digestion with added plasmid DNA. Digested DNA was separated on 1% agarose gel and transferred to Whatman SPC Nytran membrane sheets (ThermoFisher Scientific). The GFP probe was produced by carrying out a PCR using primers GFP-F3 (5′-TGAGTAAAGGAGAAGAACTTTTCAC-3′) and S1/3No2 (5′-CAAACTCATCAATGTATCTTATCAT-3′), then digesting the PCR product with PstI and purifying it with ThermoFisher Fermentas Gel Extraction Kit. The PCR fragment was then labelled using DECAPrimeII DNA labelling kit (Ambion) and alphaP32 dCTP (Perkin Elmer) and purified using Illustra ProbeQuant G-50 Micro Columns (GE Healthcare).
To analyse the copy number of the uobL12 transgenic recipient line, the nylon membrane was exposed overnight, blocked by light for 30 minutes and transferred to a Storage Phosphor Screen (Molecular Dynamics). It was scanned on a Typhoon 9200 Variable Mode Imager (Amersham Biosciences) with Typhoon Scanner Control software (v5.0) and analysed using ImageQuant 5.1.
Supplementary Material
Acknowledgments
We thank Shannon Fisher for the pCS2/PhiC31-nos1-3′UTR plasmid, Vladimir Korzh for original Tol2 transposase vectors, Holger Knaut for the anti-Vasa antibody, Zsuzsanna Izsvak and Zoltan Ivics for SB100X constructs. We thank Allen Nicholson for enabling us to carry out Southern hybridisation, James Bull for assistance with statistical analysis, Nan Li for the esrrga probe and help with WISH, Amy Singer for advice regarding transgene and transgenic line nomenclature, and Jochen Gehrig and Urban Liebel for help with imaging tool development and image processing analysis.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
D.B. and F.M. conceived the study, J.A.R., I.M.-E., Y.H., D.B. and F.M. designed experiments, J.A.R., I.M.-E., K.J.S., K.T.W., Y.H., E.K.M., J.B. and D.B. performed experiments, R.S. and E.S. performed computational analyses. J.A.R., I.M.-E., Y.H., J.B., D.B. and F.M. analysed the data, D.B. and F.M. wrote the manuscript with input from J.A.R., I.M.-E., Y.H. and J.B.
Funding
This work was funded by ‘BOLD’ Marie-Curie Initial Training Network; and ‘ZF-Health’ Integrating project in the Framework 7 programme of the European Commission; University of Birmingham (F.M.); Temple University; and the National Institutes of Health (NIH) [HD061749 to D.B.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.100347/-/DC1
References
- Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., Kawakami K. (2008). Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 105, 1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Davidson A. E., Sivasubbu S., Hermanson S. B., Welle Z., Ekker S. C. (2004). Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5, 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Wangensteen K. J., Wilber A., Bell J., Geurts A., Sivasubbu S., Wang X., Hackett P. B., Largaespada D. A., McIvor R. S., et al. (2006). Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet P. L., Obrecht-Pflumio S., Thisse C., Laudet V., Thisse B., Vanacker J. M. (2004). Cloning and developmental expression of five estrogen-receptor related genes in the zebrafish. Dev. Genes Evol. 214, 240–249 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Birney E., Dunham I., Green E. D., Gunter C., Snyder M.; ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D. G., Knight J., Mani P., Martin R., Moxon S. A., et al. (2011). ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 39, D822–D829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda C. S., Dymecki S. M. (2004). Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. J., Balciunas D., Pogoda H. M., Ding Y., Westcot S. E., Bedell V. M., Greenwood T. M., Urban M. D., Skuster K. J., Petzold A. M., et al. (2011). In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat. Methods 8, 506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S., Stainier D. Y., Anderson R. M. (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. E., Balciunas D., Mohn D., Shaffer J., Hermanson S., Sivasubbu S., Cliff M. P., Hackett P. B., Ekker S. C. (2003). Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 263, 191–202 [DOI] [PubMed] [Google Scholar]
- Davison J. M., Akitake C. M., Goll M. G., Rhee J. M., Gosse N., Baier H., Halpern M. E., Leach S. D., Parsons M. J. (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., Huber W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingsen S., Laplante M. A., König M., Kikuta H., Furmanek T., Hoivik E. A., Becker T. S. (2005). Large-scale enhancer detection in the zebrafish genome. Development 132, 3799–3811 [DOI] [PubMed] [Google Scholar]
- Ertzer R., Müller F., Hadzhiev Y., Rathnam S., Fischer N., Rastegar S., Strähle U. (2007). Cooperation of sonic hedgehog enhancers in midline expression. Dev. Biol. 301, 578–589 [DOI] [PubMed] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L., Urasaki A., Kawakami K., McCallion A. S. (2006). Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 1, 1297–1305 [DOI] [PubMed] [Google Scholar]
- Flicek P., Ahmed I., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., et al. (2013). Ensembl 2013. Nucleic Acids Res. 41, D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster B., Van De Ville D., Berent J., Sage D., Unser M. (2004). Complex wavelets for extended depth-of-field: a new method for the fusion of multichannel microscopy images. Microsc. Res. Tech. 65, 33–42 [DOI] [PubMed] [Google Scholar]
- Gaulton K. J., Nammo T., Pasquali L., Simon J. M., Giresi P. G., Fogarty M. P., Panhuis T. M., Mieczkowski P., Secchi A., Bosco D., et al. (2010). A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig J., Reischl M., Kalmár E., Ferg M., Hadzhiev Y., Zaucker A., Song C., Schindler S., Liebel U., Müller F. (2009). Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat. Methods 6, 911–916 [DOI] [PubMed] [Google Scholar]
- Gilmour D. T., Maischein H. M., Nüsslein-Volhard C. (2002). Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron 34, 577–588 [DOI] [PubMed] [Google Scholar]
- Grajevskaja V., Balciuniene J., Balciunas D. (2013). Chicken β-globin insulators fail to shield the nkx2.5 promoter from integration site effects in zebrafish. Mol. Genet. Genomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadzhiev Y., Lang M., Ertzer R., Meyer A., Strähle U., Müller F. (2007). Functional diversification of sonic hedgehog paralog enhancers identified by phylogenomic reconstruction. Genome Biol. 8, R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S., Freudenreich D., Geffarth M., Kaslin J., Machate A., Brand M. (2011). Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev. Dyn. 240, 108–115 [DOI] [PubMed] [Google Scholar]
- Hermanson S., Davidson A. E., Sivasubbu S., Balciunas D., Ekker S. C. (2004). Sleeping Beauty transposon for efficient gene delivery. Methods Cell Biol. 77, 349–362 [DOI] [PubMed] [Google Scholar]
- Hu G., Goll M. G., Fisher S. (2011). ϕC31 integrase mediates efficient cassette exchange in the zebrafish germline. Dev. Dyn. 240, 2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. P., Chen L. S., Jia C. Y., Zhu H. Z., Wang W., Zhong J. (2013). Screening of potential pseudo att sites of Streptomyces phage ϕC31 integrase in the human genome. Acta Pharmacol. Sin. 34, 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt T. M., Ekker S. C. (1999). Vectors and techniques for ectopic gene expression in zebrafish. Methods Cell Biol. 59, 117–126 [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Mechaly A. S., Becker T. S., Rinkwitz S. (2013). Using zebrafish transgenesis to test human genomic sequences for specific enhancer activity. Methods 62, 216–225 [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Jähner D., Nobis P., Simon I., Löhler J., Harbers K., Grotkopp D. (1981). Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell 24, 519–529 [DOI] [PubMed] [Google Scholar]
- Kawakami K. (2004). Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 77, 201–222 [DOI] [PubMed] [Google Scholar]
- Keravala A., Calos M. P. (2008). Site-specific chromosomal integration mediated by phiC31 integrase. Methods Mol. Biol. 435, 165–173 [DOI] [PubMed] [Google Scholar]
- Kikuta H., Laplante M., Navratilova P., Komisarczuk A. Z., Engström P. G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., et al. (2007). Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 17, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier S., Höckendorf B., Möller E. K., Bornhorst D., Spitz F., Wittbrodt J. (2013). Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase. Development 140, 4287–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H., Pelegri F., Bohmann K., Schwarz H., Nüsslein-Volhard C. (2000). Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 149, 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega I., Dean A. (2012). Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev. 22, 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhstoss S., Rao R. N. (1991). Analysis of the integration function of the streptomycete bacteriophage phi C31. J. Mol. Biol. 222, 897–908 [DOI] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 [DOI] [PubMed] [Google Scholar]
- Langenau D. M., Feng H., Berghmans S., Kanki J. P., Kutok J. L., Look A. T. (2005). Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 102, 6068–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B., Sandelin A., Carninci P. (2012). Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13, 233–245 [DOI] [PubMed] [Google Scholar]
- Lister J. A. (2010). Transgene excision in zebrafish using the phiC31 integrase. Genesis 48, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister J. A. (2011). Use of phage φC31 integrase as a tool for zebrafish genome manipulation. Methods Cell Biol. 104, 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L., Chuah M. K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D. P., Schmitt A., Becker K., Matrai J., et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753–761 [DOI] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Puller A.-C., Lawson K. L., Tschopp P., Amsterdam A., Zon L. I. (2013). Site-directed zebrafish transgenesis into single landing sites with the phiC31 integrase system. Dev. Dyn. 242, 949–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Chang B., Albert S., Fischer N., Tora L., Strähle U. (1999). Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development 126, 2103–2116 [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Dahm R. (2002) Zebrafish: A Practical Approach, Oxford: Oxford University Press; [Google Scholar]
- Olivares E. C., Hollis R. P., Chalberg T. W., Meuse L., Kay M. A., Calos M. P. (2002). Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 20, 1124–1128 [DOI] [PubMed] [Google Scholar]
- Parinov S., Kondrichin I., Korzh V., Emelyanov A. (2004). Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 [DOI] [PubMed] [Google Scholar]
- Peravali R., Gehrig J., Giselbrecht S., Lütjohann D. S., Hadzhiev Y., Müller F., Liebel U. (2011). Automated feature detection and imaging for high-resolution screening of zebrafish embryos. Biotechniques 50, 319–324 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragvin A., Moro E., Fredman D., Navratilova P., Drivenes O., Engström P. G., Alonso M. E., de la Calle Mustienes E., Gómez Skarmeta J. L., Tavares M. J., et al. (2010). Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc. Natl. Acad. Sci. USA 107, 775–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Nutter L. M., Gertsenstein M. (2011). Engineering the embryo. Proc. Natl. Acad. Sci. USA 108, 7659–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure A., Rothbächer U., Robin F., Kalmar E., Ferone G., Lamy C., Missero C., Mueller F., Lemaire P. (2007). A multicassette Gateway vector set for high throughput and comparative analyses in ciona and vertebrate embryos. PLoS ONE 2, e916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Dorn R., Reuter G. (2003). Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14, 67–75 [DOI] [PubMed] [Google Scholar]
- Scott E. K., Mason L., Arrenberg A. B., Ziv L., Gosse N. J., Xiao T., Chi N. C., Asakawa K., Kawakami K., Baier H. (2007). Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods 4, 323–326 [DOI] [PubMed] [Google Scholar]
- Shkumatava A., Fischer S., Müller F., Strahle U., Neumann C. J. (2004). Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development 131, 3849–3858 [DOI] [PubMed] [Google Scholar]
- Smith M. C., Till R., Brady K., Soultanas P., Thorpe H., Smith M. C. (2004). Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination. Nucleic Acids Res. 32, 2607–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- Thisse B., Heyer V., Lux A., Alunni V., Degrave A., Seiliez I., Kirchner J., Parkhill J. P., Thisse C. (2004). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519 [DOI] [PubMed] [Google Scholar]
- Thorpe H. M., Smith M. C. (1998). In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. USA 95, 5505–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A., Morvan G., Kawakami K. (2006). Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J. (2012). Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and ϕC31 integrase. Methods Mol. Biol. 859, 203–228 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., Spokony R., Wan K. H., Koriabine M., de Jong P. J., White K. P., et al. (2009). Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (1993). The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio), Eugene, OR: M. Westerfield; [Google Scholar]
- White R. M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C. E., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Bellen H. J., Gehring W. J. (1990). Position effects on eukaryotic gene expression. Annu. Rev. Cell Biol. 6, 679–714 [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., Smith S. F., North P., Callaway H., Kelly K., et al. (2005). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyart C., Del Bene F., Warp E., Scott E. K., Trauner D., Baier H., Isacoff E. Y. (2009). Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature 461, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Tong X., Wang Z., Liu D., Pan R., Li Z., Hu Y., Luo Z., Huang P., Wu Q., et al. (2013). TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10, 329–331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





