Abstract
Dps proteins, bacterial mini-ferritins that protect DNA from oxidative stress, are implicated in the survival and virulence of pathogenic bacteria. Here we examine the mechanism of E. coli Dps protection of DNA, specifically whether this DNA-binding protein can utilize DNA charge transport through the base pair π-stack to protect the genome from a distance. An intercalating ruthenium photooxidant was employed to generate DNA damage localized to guanine repeats, the sites of lowest potential in DNA. We find that Dps loaded with ferrous iron, in contrast to Apo-Dps and ferric iron-loaded Dps, significantly attenuates the yield of oxidative DNA damage. These data demonstrate that ferrous iron-loaded Dps is selectively oxidized to fill guanine radical holes, thereby restoring the integrity of the DNA. Luminescence studies indicate no direct interaction between the ruthenium photooxidant and Dps, supporting the DNA-mediated oxidation of ferrous iron-loaded Dps. Thus DNA charge transport may be a mechanism by which Dps efficiently protects the genome of pathogenic bacteria from a distance.
Dps proteins are bacterial mini-ferritins that protect DNA under stress conditions. These proteins are thought to protect DNA from oxidative stress by utilizing their ferroxidase activity to deplete ferrous iron and hydrogen peroxide, which can otherwise produce damaging hydroxyl radicals via Fenton chemistry (1). Some Dps proteins also nonspecifically bind DNA, such as that from E. coli which utilizes N-terminal lysines for DNA binding (2). The Dps protein family is involved in the survival of pathogenic bacteria in the oxidizing host environment. Dps is implicated in the virulence of pathogenic bacteria such as Bacteroides fragilis, the most common anaerobic species isolated from clinical infections which is both highly aerotolerant and resistant to oxidative stress (3), and Borrelia burgdorferi, the causative agent of Lyme’s disease (4), among others (5). Moreover, Dps has been shown to protect Salmonella enterica serotype Enteritidis from the Fenton-mediated killing mechanism of bactericidal antibiotics (6). Thus, in the struggle between host and pathogen, oxidative stress is a key factor, and Dps is implicated in bacterial survival when confronted with either the host immune system or antibiotics. What is the mechanism by which Dps is protecting the bacterial genome? Previous experimentation towards elucidating this protection mechanism has shown that Dps protects DNA from DNase cleavage (7), traps hydroxyl radicals, and inhibits DNA nicking by the Fenton reagents Fe2+ and H2O2 (8). We seek to determine more specifically the mechanism of E. coli Dps protection of DNA.
DNA has been shown to conduct charge efficiently through the π-stack of its nucleobases over long molecular distances in a diverse range of systems (9). DNA charge transport (CT) is proposed to be utilized within the cell, both in the long-range activation of redox-sensitive transcription factors and also in facilitating scanning of the genome for damage by DNA-repair enzymes (10). Could ferritins similarly utilize DNA CT to exert their protective effects from a distance? That is, do oxidizing equivalents have to diffuse specifically to the ferroxidase site of Dps, or can Dps become oxidized from a distance through DNA CT, thus protecting the surrounding DNA for potentially hundreds of base pairs?
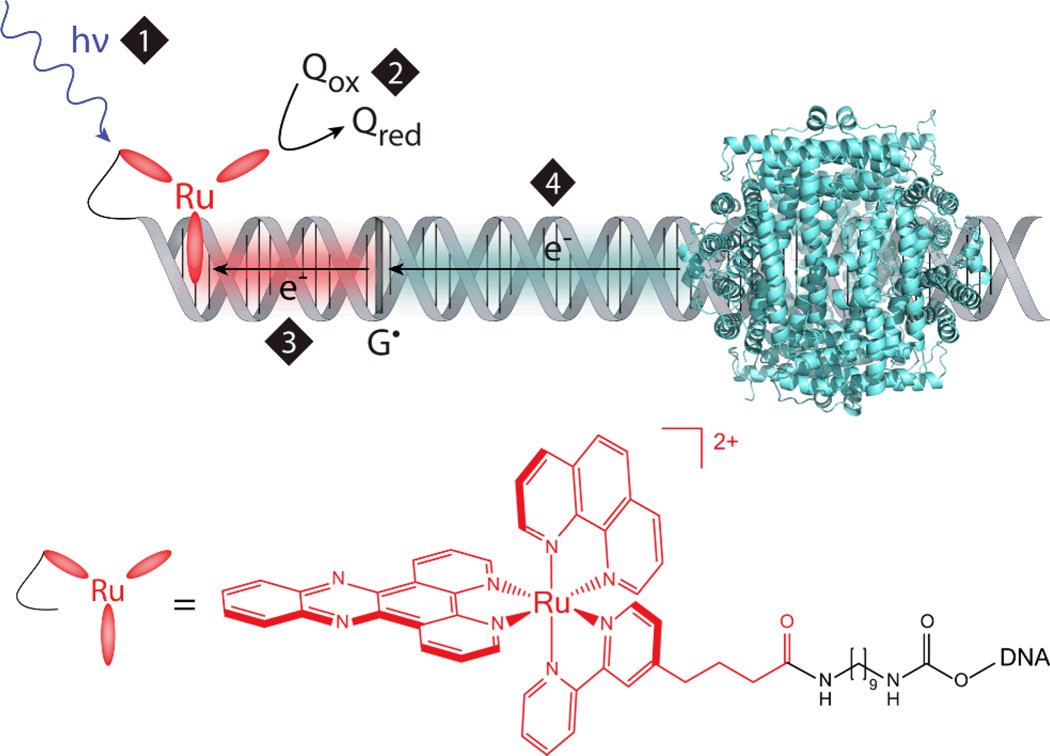
The question of DNA-mediated long distance protection can be answered by generating guanine radicals using ruthenium flash-quench chemistry (11), and investigating if Dps protects the DNA from this oxidative damage (12). The flash-quench technique utilizes dipyridophenazine (dppz) complexes of ruthenium(II) that bind to DNA by intercalation (13). Here, racemic [Ru(phen)(dppz)(bpy’)]2+, where phen is 1,10-phenanthroline and bpy’ is 4-butyric acid-4’-methyl-2,2’-bipyridine, was covalently tethered to amine-modified DNA via the carboxylic acid moiety of the bpy’ ligand (14). In the first step, visible light promotes a t2g → π* metal-to-ligand charge-transfer (MLCT) transition of the Ru(II) complex (15). This Ru(II) excited state is then oxidatively quenched by a diffusing electron acceptor (Q), here [Co(NH3)5Cl]2+, to form a highly oxidizing intercalated Ru(III) complex (1.6 V versus NHE, 16). The in situ generated Ru(III) is competent to abstract an electron from DNA; the hole equilibrates along the DNA π-stack and localizes on guanine, the base with lowest reduction potential (1.3 V versus NHE, 17). The presence of adjacent guanines can further lower the guanine reduction potential, making the 5’-G of guanine doublets and triplets most readily oxidized (18). In this fashion, damage at the 5′-G of guanine repeats is considered a hallmark of one electron oxidative damage created through DNA CT. Further reaction of the guanine radical (G•) with H2O or O2 can form a mixture of irreversible oxidative products (19). These products are analogous to the DNA damage products that can form in vivo as a result of oxidative stress. However, because the lifetime of the guanine radical is long (milliseconds, 13) relative to the timescale of DNA CT (picoseconds, 20), the guanine radical can also interact with DNA-bound, redox-active proteins. Thus electron transfer from Dps through the DNA π-stack could fill the hole on the guanine radical, restoring the integrity of the DNA (Figure 1). Guanine radicals can be monitored by cleaving irreversible guanine oxidation products on radiolabeled oligonucleotides and visualizing them through gel electrophoresis (14). In the presence of E. coli Dps, decreased guanine damage should be observed if Dps is protecting the DNA in this manner.
Figure 1.
Schematic depicting DNA-mediated oxidation of Dps to fill the guanine radical hole generated by flash-quench chemistry. Upper: First, visible light excites an intercalated ruthenium(II) photooxidant (1), which is then oxidatively quenched (2) to a highly oxidizing Ru(III) species by a diffusing quencher (Q). This Ru(III) species is competent to abstract an electron from DNA (3); the hole equilibrates along the π-stack and localizes to the 5’-G of guanine repeats, the most easily oxidized base (G•). DNA CT from Dps to the guanine radical (4) could be a long distance protection mechanism. Note that the location and precise geometry for DNA binding of Dps are unknown. E. coli Dps PDB: 1dps. Lower: Structure of [Ru(phen)(dppz)(bpy’)]2+ covalently tethered to DNA via diaminononane linkage.
It seemed reasonable to consider that any protective effects of Dps would vary as a function of the iron content of the protein. At the intersubunit ferroxidase sites of Dps, ferrous iron is bound, oxidized, and then shuttled to the protein core where it is stored (1). Two conserved histidines together with glutamate and aspartate residues ligate two iron atoms, creating one high affinity iron site and another with much lower affinity. If all of these diiron sites of dodecameric Dps are fully occupied, this would correspond to 24 Fe/Dps. Accordingly, we sought to compare the protective effects of Apo-Dps with Dps in which the ferroxidase site was loaded with either ferrous or ferric iron. As-purified Dps was considered to be Apo-Dps, with reliably ≤ 1 Fe/Dps (based on the absorbance of [Fe(bpy)3]2+, see Supporting Information (SI) for details). The ferroxidase sites of Dps were anaerobically loaded with ferrous iron by incubating the protein with excess ferrous iron and dithionite for 2 hours, and subsequently removing the dithionite and unbound iron using small size-exclusion columns. Despite using excess ferrous iron and long incubation times, reliably only 12 Fe2+/Dps were bound under anaerobic conditions. A linear increase in the quenching of intrinsic protein fluorescence upon anaerobic titration of ferrous iron to Apo-Dps from 0 to 12 Fe/Dps, coupled with minimal quenching past this stoichiometry, supports the observed 12 Fe2+/Dps (Figure S1). In Bacillus anthracis Dps, only one iron per ferroxidase site binds appreciably under anaerobic conditions because of the difference in affinity between the iron sites; a bridging oxidant seems to be required to form the di-iron site (21). Similar results were observed with Listeria innocua Dps (22).
To then confirm specific iron binding to the ferroxidase site of E. coli Dps, a double mutant was prepared where both ligating histidines were changed to glycines: H51G/H63G Dps. Similar secondary structure to the wild-type protein was established by circular dichroism spectroscopy (Figure s2). Under identical conditions, the wild-type protein bound 11.8 ± 0.2 Fe2+/Dps whereas the H51G/H63G mutant bound only 1.0 ± 0.3 Fe2+/Dps, demonstrating that these mutations did indeed abolish iron binding (Figure S3). Therefore we observe specific iron binding only to the higher affinity iron site under anaerobic conditions for E. coli Dps. Ferrous iron-loaded Dps was then oxidized anaerobically using potassium ferricyanide (Figure S4), which was removed in the same fashion as above (23). Iron oxidation was evidenced by a characteristic increase in absorbance at 310 nm (8).
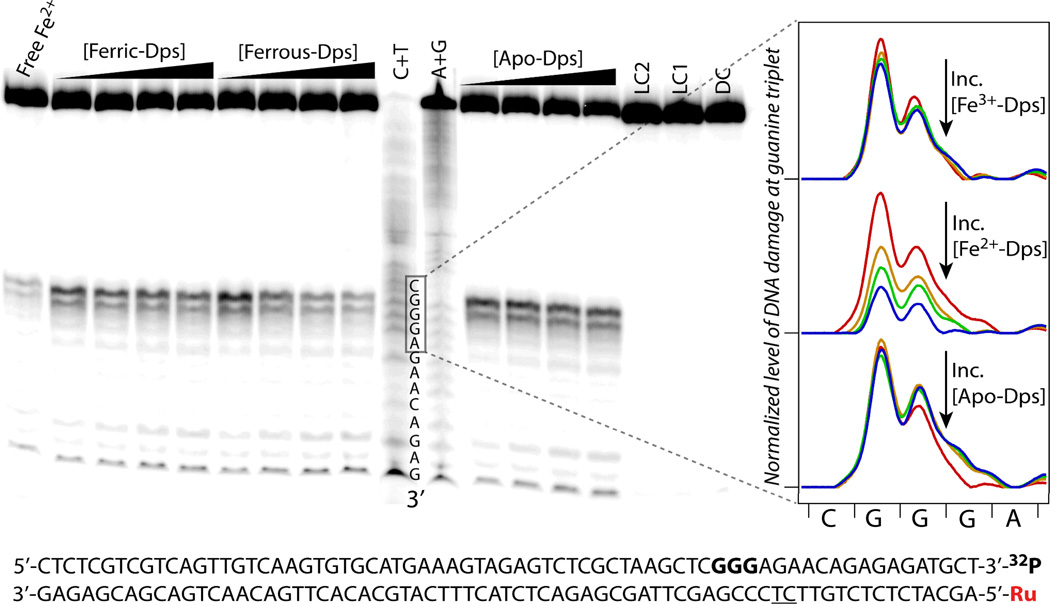
To compare the level of DNA protection by Dps as a function of protein iron loading, samples containing 3’-32P labeled 70 mer duplex DNA appended with 5’-covalently tethered [Ru(phen)(dppz)(bpy’)]2+ (6 µM), the diffusing quencher [Co(NH3)5Cl]2+ (600 µM), and 0–6 µM of Apo-Dps, ferrous iron-loaded Dps, or ferric iron-loaded Dps were irradiated anaerobically at 442 nm to avoid dioxygen oxidation of ferrous iron-loaded Dps. The locations of DNA damage thus generated via flash-quench chemistry were then revealed by treatment with hot piperidine, which cleaves the DNA backbone at sites of oxidative guanine lesions (19), and subsequent visualization by denaturing gel electrophoresis. Figure 2 shows a representative gel featuring the differential effects of Dps with varying iron content. In the absence of Dps, damage is evident predominantly at the 5′-G of the guanine triplet, as expected with one electron DNA oxidation through long range DNA CT (24). When any component is missing, such as in the dark control (DC), which is the full sample (containing DNA, photooxidant, and quencher) but not irradiated, or light control 1 (LC1), which is the sample irradiated but in the absence of quencher, no damage is observed. Ferrous iron-loaded Dps (6 µM) attenuates oxidative DNA damage at the guanine triplet by an average of almost 5-fold (4.9 ± 1.5), whereas equivalent concentrations of Apo-Dps and ferric iron-loaded Dps have relatively little effect (1.1 ± 0.5 and 1.3 ± 0.7 fold attenuation, respectively) (25). Thus increasing equivalents of Dps loaded with ferrous iron, in contrast to Apo-Dps and ferric iron-loaded Dps, significantly decrease the level of DNA damage at guanine repeats, as can be clearly seen in the lane profile comparisons in Figure 2, indicating the importance of both the presence and oxidation state of iron in Dps. It is noteworthy that iron-loaded Dps binds DNA similarly to Apo-Dps, as determined via gel-shift assays (see SI), negating the possibility of a differential binding effect.
Figure 2.
Representative gel comparing ability of Apo-Dps, ferrous-iron loaded Dps, and ferric-iron loaded Dps to protect DNA from damage created via flash-quench chemistry. Left: Autoradiogram of gel showing effect of increasing Dps with varying iron loading. Conditions: 6 µM statistically 3’-32P-labeled 70 mer Ru-DNA, 600 µM [Co(NH3)5Cl]2+, Dps concentrations from 0 to 6 µM, buffer: 50 mM Tris, pH 7.0, 150 mM NaCl. A+G and C+T are Maxam-Gilbert sequencing lanes. Controls: Dark control (DC) contains all components (DNA, photooxidant, and quencher) but is not irradiated; Light control 1 (LC1) is irradiated but lacks quencher; LC2 is irradiated, lacking quencher, but contains 6 µM Dps loaded with ferric iron; Free Fe2+ is irradiated, containing all components but protein, with the addition of free ferrous iron at a concentration equivalent to that in ferrous iron-loaded Dps. The number of Fe/Dps in ferrous iron-loaded Dps was 10.8 ± 0.1. Right: Lane profiles comparing normalized level of DNA damage at the guanine triplet upon titration of ferric iron-loaded Dps (upper), ferrous iron-loaded Dps (middle), or Apo-Dps (lower) from 0 µM protein (red), 2 µM protein (orange), 4 µM protein (green), to 6 µM protein (blue). Lower: DNA sequence, with guanine triplet shown bolded, and the location of a nick in the DNA backbone underlined. For higher synthetic yield, the DNA was made in two pieces (55-mer and Ru-15-mer); breaks in the sugar-phosphate backbone do not affect long-range CT.
Free ferrous iron diminishes damage, as shown by a control which is the full sample with the addition of free ferrous iron at a concentration equivalent to that in ferrous iron-loaded Dps (“Free Fe2+” in Figure 2). To determine if free iron is generated by the irradiation of ferrous iron-loaded Dps, the iron content of a sample was measured before and after irradiation, while ensuring removal of any unbound iron from the irradiated sample by size-exclusion chromatography. The iron content before and after irradiation (12.2 ± 0.3 and 11.8 ± 0.1 Fe/Dps, respectively) was found to be identical, indicating no iron labilization. In view of these controls, the difference in damage attenuation for ferrous iron-loaded Dps compared to Apo-Dps and ferric iron-loaded Dps indicates that Dps containing ferrous iron can become oxidized, perhaps via DNA CT, to fill the hole on guanine radicals.
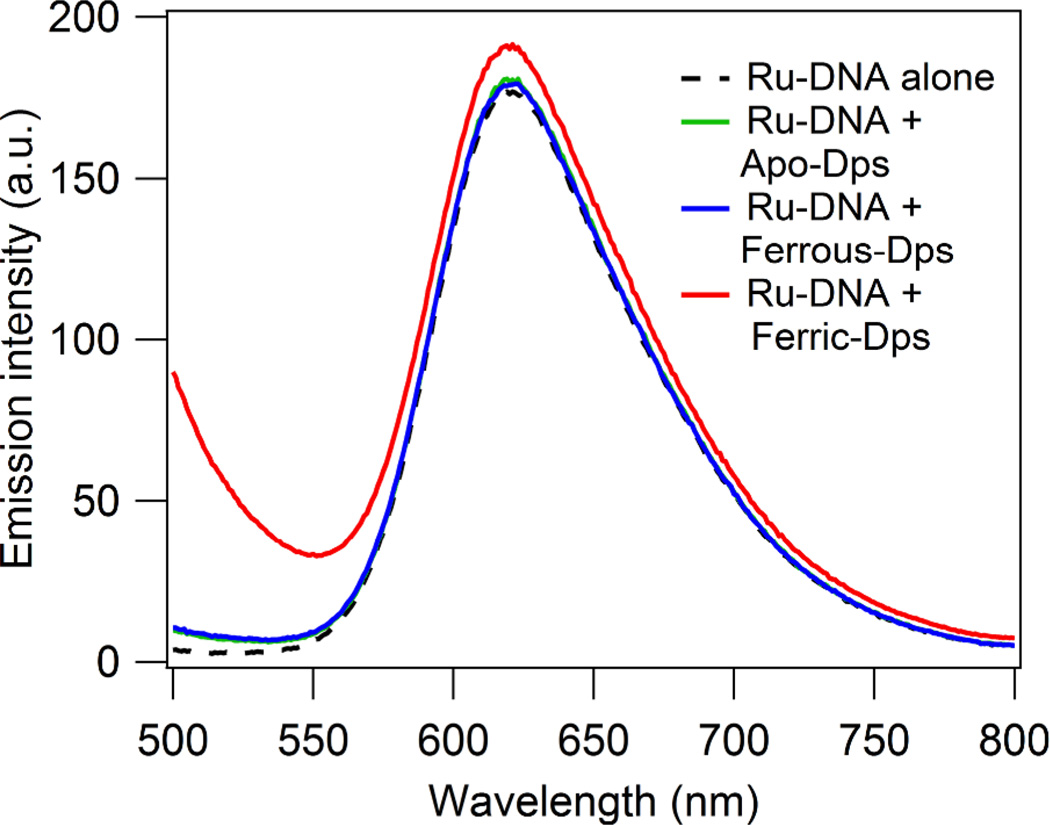
Evidence suggests that Dps does not interact directly with the ruthenium photooxidant. First, the steady-state luminescence of covalently tethered Ru-DNA is not quenched in the presence of Dps, indicating that Dps does not interact with the Ru(II) excited state, Ru(II)* (Figure 3). Further controls for this interaction are samples which were irradiated and lack quencher, but contain 6 µM Dps loaded with ferric iron or ferrous iron (LC2 in Figure 2 and LC3 in Figure S5, respectively). If iron-loaded Dps oxidatively quenched Ru(II)* to form Ru(III), guanine damage would be apparent in these samples.
Figure 3.
Ru-DNA luminescence in absence and presence of Dps. Anaerobically prepared samples containing 8 µM duplexed 70-mer DNA with covalently tethered [Ru(phen)(dppz)(bpy’)]2+ alone or with 8 µM Apo-Dps, ferrous-iron loaded Dps (11.8 ± 0.2 Fe/Dps dodecamer), or ferric-iron loaded Dps in 50 mM Tris, pH 7.0, 150 mM NaCl. Excitation wavelength: 440 nm. Slight precipitation occurs for the sample containing Ru-DNA and ferric iron-loaded Dps, resulting in a raised baseline.
Thus it appears that ferrous iron-loaded Dps, but not Apo-Dps or ferric iron-loaded Dps, can protect DNA from oxidative damage by becoming oxidized to fill guanine radical holes. Coupled with evidence indicating no direct interaction between the photooxidant and Dps, these results support a long-distance protection mechanism for Dps utilizing DNA CT (26). While Dps is highly upregulated in stationary phase, in exponential phase there are approximately 6,000 copies of Dps per E. coli cell (27). Given the size of the E. coli genome (roughly 4,600,000 base pairs), this corresponds to 760 base pairs per Dps, a reasonable distance for DNA CT to occur (28). We would suggest that in the cell, if there was continued availability of ferrous iron, Dps could funnel electrons from the ferrous iron bound to its ferroxidase sites to guanine radical holes until the oxidized iron core of the protein reached capacity, thereby evincing a potentially greater ability to protect DNA from oxidative damage than the 12 Fe2+/Dps used in this study. This possible DNA-mediated protection mechanism of Dps does not obviate direct oxidation by diffusing oxidants but offers a powerful means whereby Dps could effectively protect DNA from a distance. Such a mechanism could contribute to the oxidative stress resistance and virulence of pathogenic bacteria.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Roberto Kolter at Harvard Medical School for his generous donation of the Dps plasmid and strain used for this study. We acknowledge the NIH for funding (GM49216). A.R.A. was supported by the National Institute on Aging of the NIH on a pre-doctoral NRSA (F31AG040954)
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental methods and supporting figures S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Zeth K. Biochem. J. 2012;445:297–311. doi: 10.1042/BJ20120514. [DOI] [PubMed] [Google Scholar]
- 2.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. Nucleic Acids Res. 2004;32:5935–5944. doi: 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sund CJ, Rocha ER, Tzinabos AO, Wells WG, Gee JM, Reott MA, O’Rourke DP, Smith CJ. Mol. Microbiol. 2008;67:129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, Anderson JF, Radolf JD, Fikrig E. Mol. Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- 5.(a) Helicobacter pylori (neutrophil-activating protein, HP-NAP) D’Elios MM, Amedei A, Cappon A, Del Prete G, de Bernard M. FEMS Immunol. Med. Mic. 2007;50:157–164. doi: 10.1111/j.1574-695X.2007.00258.x. (b) Salmonella enterica serovar Typhimurium Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. Infect. Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. and (c) Porphyromonas gingivalis Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. Infect. Immun. 2003;71:1170–1178. doi: 10.1128/IAI.71.3.1170-1178.2003.
- 6.Calhoun LM, Kwon YM. Int. J. Antimicrob. Agents. 2011;37:261–265. doi: 10.1016/j.ijantimicag.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 7.Almiron M, Link AJ, Furlong D, Kolter R. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 8.Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. J. Biol. Chem. 2002;277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 9.Muren NB, Olmon ED, Barton JK. Phys. Chem. Chem. Phys. 2012;14:13754–13771. doi: 10.1039/c2cp41602f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Genereux JC, Boal AK, Barton JK. J. Am. Chem. Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sontz PA, Muren NB, Barton JK. Acc. Chem. Res. 2012;45:1792–1800. doi: 10.1021/ar3001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang I-J, Gray HB, Winkler JR. J. Am. Chem. Soc. 1991;113:7056–7057. [Google Scholar]
- 12.(a) Lee PE, Demple B, Barton JK. Proc. Natl. Acad. Sci. USA. 2009;106:13164–13168. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yavin E, Boal AK, Stemp EDA, Boon EM, Livingston AL, O’Shea VL, David SS, Barton JK. Proc. Natl. Acad. Sci. USA. 2005;102:3546–3551. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stemp EDA, Arkin MR, Barton JK. J. Am. Chem. Soc. 1997;119:2921–2925. [Google Scholar]
- 14.Arkin MR, Stemp EDA, Pulver SC, Barton JK. Chem. Biol. 1997;4:389–400. doi: 10.1016/s1074-5521(97)90129-0. [DOI] [PubMed] [Google Scholar]
- 15.Creutz C, Chou M, Netzel TL, Okumura M, Sutin N. J. Am. Chem. Soc. 1980;102:1309–1319. [Google Scholar]
- 16.Delaney S, Pascaly M, Bhattacharya PK, Han K, Barton JK. Inorg. Chem. 2002;41:1966–1974. doi: 10.1021/ic0111738. [DOI] [PubMed] [Google Scholar]
- 17.Fukuzumi S, Miyao H, Ohkubo K, Suenobu T. J. Phys. Chem. A. 2005;109:3285–3294. doi: 10.1021/jp0459763. [DOI] [PubMed] [Google Scholar]
- 18.Sugiyama H, Saito I. J. Am. Chem. Soc. 1996;118:7063–7068. [Google Scholar]
- 19.Burrows CJ, Muller JG. Chem. Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 20.Wan C, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. USA. 1999;96:6014–6019. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz JK, Liu XS, Tosha T, Adrienne Diebold A, Theil EC, Solomon EI. Biochemistry. 2010;49:10516–10525. doi: 10.1021/bi101346c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su M, Cavallo S, Stefanini S, Chiancone E, Chasteen ND. Biochemistry. 2005;44:5572–5578. doi: 10.1021/bi0472705. [DOI] [PubMed] [Google Scholar]
- 23.While evidence suggests that the preferred oxidant for Dps is hydrogen peroxide (see reference 8), compared to dioxygen for maxi-ferritins, addition of even sub-stoichiometric quantities of hydrogen peroxide led to Dps precipitation. Regardless of the oxidant (hydrogen peroxide or ferricyanide), the important factor is that with oxidized iron, electrons are not available from Dps to fill guanine radical holes.
- 24.Hydroxyl radical damage leads instead to damage at all guanines.
- 25.Experiments were done in triplicate, and oxidative DNA damage was quantified for each lane by the ratio of guanine triplet damage to the undamaged parent band. See SI for further details.
- 26.Since Dps binds DNA nonspecifically, utilizing an intervening mismatch or abasic site between the site of protein binding and the guanine triplet in order to definitively establish that the reaction is long-range via DNA-mediated CT is not possible.
- 27.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Slinker JD, Muren NB, Renfrew SE, Barton JK. Nat. Chem. 2011;3:230–235. doi: 10.1038/nchem.982. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Proc. Natl. Acad. Sci. USA. 2009;106:15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.