Abstract
The evolutionary importance of hybridization as a source of new adaptive genetic variation is rapidly gaining recognition. Hybridization between coyotes and wolves may have introduced adaptive alleles into the coyote gene pool that facilitated an expansion in their geographic range and dietary niche. Furthermore, hybridization between coyotes and domestic dogs may facilitate adaptation to human-dominated environments. We genotyped 63 ancestry-informative single nucleotide polymorphisms in 427 canids in order to examine the prevalence, spatial distribution, and ecology of admixture in eastern coyotes. Using multivariate methods and Bayesian clustering analyses, we estimated the relative contributions of western coyotes, western and eastern wolves, and domestic dogs to the admixed ancestry of Ohio and eastern coyotes. We found that eastern coyotes form an extensive hybrid swarm, with all our samples having varying levels of admixture. Ohio coyotes, previously thought to be free of admixture, are also highly admixed with wolves and dogs. Coyotes in areas of high deer density are genetically more wolf-like, suggesting that natural selection for wolf-like traits may result in local adaptation at a fine geographic scale. Our results, in light of other previously published studies of admixture in Canis, reveal a pattern of sex-biased hybridization, presumably generated by male wolves and dogs mating with female coyotes. This study is the most comprehensive genetic survey of admixture in eastern coyotes and demonstrates that the frequency and scope of hybridization can be quantified with relatively few ancestry-informative markers.
Keywords: admixture, hybridization, Canis, single nucleotide polymorphism, diagnostic markers
Introduction
Hybridization is of immense evolutionary importance as a source of new adaptive genetic variation. Unlike novel mutations, introgressive hybridization with wild species simultaneously introduces many alleles that have already passed through the filter of natural selection. Alternatively, hybridization with domestic species can introduce many alleles that have passed through the filter of artificial selection; this may produce negative consequences in natural populations. Although hybridization has generally been perceived negatively in the conservation and resource management communities (e.g., Rhymer & Simberloff 1996; Allendorf et al. 2001; Wolf et al. 2001; Oliveira et al. 2008), its potential value in enhancing the adaptive potential of parental lineages is gaining recognition (e.g., Seehausen 2004; Kyle et al. 2006; Kays et al. 2010a). Hybridization has played an important evolutionary role in past range expansions and adaptation to changing environments (Willis et al. 2006), and may be vital for the future survival of some taxa under rapidly changing conditions due to anthropogenic land use or climate change. Despite this critical role that hybridization plays in evolution and conservation biology, it remains unclear how habitat variation at the landscape and regional scales affects the flow of introgressed alleles (but see Fitzpatrick & Shaffer 2007). This is particularly true in cases of anthropogenic hybridization, in which stable contact zones may not exist or hybrid swarms may establish.
Modern populations of North American wolf-like canids are known to be admixed in some areas. Coyote-derived DNA was first found in wolf populations of the Great Lakes region in the early 1990s (Lehman et al. 1991). A growing body of evidence indicates that the introgressive hybridization among North American Canis is very complex, with genetic exchange occurring in varying degrees among western gray wolves (Canis lupus), eastern wolves (also known as Great Lakes wolves, C. lupus lycaon, or C. lycaon), Mexican wolves (C. lupus baileyi), red wolves (C. rufus), coyotes (C. latrans), and domestic dogs (C. familiaris) (Kyle et al. 2006 and references therein; Hailer & Leonard 2008; Kyle et al. 2008; Leonard & Wayne 2008; Koblmüller et al. 2009; Wheeldon & White 2009; Wilson et al. 2009; Bohling & Waits 2011; vonHoldt et al. 2011). Although admixture is widely accepted, researchers differ in the interpretations of molecular data and their implications for taxonomic recognition and conservation. Most of the research emphasis has been placed on the wolf side of the admixture story because of ongoing debate regarding the validity of the Great Lakes wolf and red wolf recovery programs, while less attention has been given to the causes and consequences of admixture in eastern coyotes (but see Kays et al. 2010a; vonHoldt et al. 2011). Hybridization with wolves is thought to have aided coyotes in their colonization of eastern forests by allowing them to rapidly evolve larger body size, including wider skulls, which made them more effective deer hunters (Kays et al. 2010a). Hence, we hypothesized that individuals living in areas of high deer density are genetically more wolf-like than those living in areas of lower deer density.
Steadily improving molecular tools and geographic sampling have refined our understanding of this hybridization story. For two decades the extent of the molecular data was limited to restriction fragment length polymorphisms (Lehman et al. 1991), sequences of mitochondrial DNA (mtDNA) usually coupled with genotypes of a few nuclear microsatellites (e.g., Wayne & Lehman 1992; Roy et al. 1994; Koblmüller et al. 2009; Wilson et al. 2009; Rutledge et al. 2010), and sequences of one gene of the major histocompatibility complex (Hedrick et al. 2002). Still, the results of these studies, or more specifically, their interpretations were conflicting. This may be due, in part, to the low resolution offered by a small number of segregating loci in the context of a complex hybridization scenario. Microsatellites accumulate homoplasy quickly and thus have low statistical power for inferring population structure when samples are drawn from an admixed population, especially when admixture proportions are high (Haasl & Payseur 2010). More recently, vonHoldt et al. (2011) published the largest genomic study aimed at addressing the complex evolutionary history of wolf-like canids, taking advantage of the thousands of single nucleotide polymorphisms (SNPs) ascertained from the dog genome project. They used a SNP microarray to assay genomic variation in more than 48,000 loci genotyped in a panel of 277 wolves and coyotes and several hundred dogs. Although this was the most extensive genetic survey of any wild vertebrate group, the geographic sampling of coyotes was limited, with only 13 individuals from northeastern North America and 3 from Ohio, thus limiting inferences about admixture and population subdivision in eastern coyotes.
Here we present data on ancestry-informative SNPs carefully selected and genotyped in a broad geographic sample of 425 eastern coyotes and 2 suspected immigrant wolves, and compare these genotypes to those of 40 western coyotes, 34 western wolves, and 17 eastern wolves from vonHoldt et al. (2011). This represents the largest survey of genomic variation in eastern coyotes to date. Our objectives in this study were to use ancestry-informative SNPs to (1) assess the prevalence and spatial distribution of admixture in eastern coyotes, (2) estimate coyote vs. wolf ancestry of individuals, (3) test for sex-biased hybridization, and (4) investigate the ecological context of admixture.
Methods
Study area and sampling
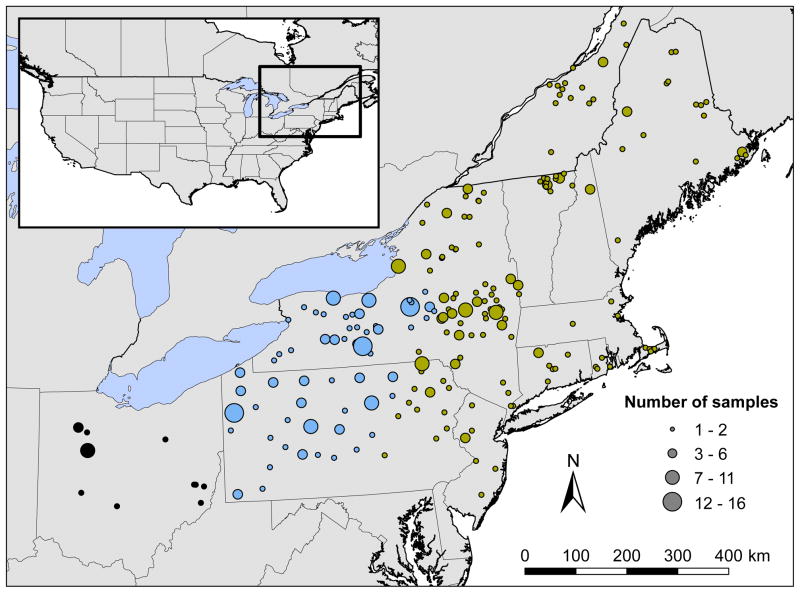
Our study area was located in northeastern North America (Figure 1, Table S1). All samples genotyped in this study (N = 427) are archived and vouchered in the New York State Museum, Albany, NY, and were collected with assistance of local hunters and trappers since 1999. All samples were fresh tissue, except for 10 fecal samples which came from previous scat surveys in New York (Gompper et al. 2006; Kays et al. 2008); the fecal samples generated high-quality DNA as judged by spectrophotometry and mtDNA sequence quality. Two samples (zm14276 from Saratoga County, New York and zm15083 from Orleans County, Vermont) were suspected wolves based on morphology and preliminary genetics (USFWS 2004; 2007); stable isotope data indicate these two wolves were natural immigrants rather than escaped pets (Kays & Feranec 2011). No IACUC ethics review was required for this study because DNA samples came from scat or were salvaged from animals killed for reasons other than research.
Figure 1.
Study area and sampling localities of coyotes in northeastern United States and southeastern Canada. Circle size represents sample size per locality. Circle color represents geographic zone as in Kays et al. (2010a): black, Ohio; blue, contact zone; gold, northeast zone.
Selection of ancestry-informative SNPs
We selected molecular markers based on a previous study that used the Affymetrix Canine Mapping Array to genotype 60,584 autosomal SNP loci in coyotes, western gray wolves, eastern wolves, and dogs (vonHoldt et al. 2010; vonHoldt et al. 2011). We used two independent but complementary tests to select ancestry-informative SNPs. First, we used the program EIGENSTRAT (Patterson et al. 2006; Price et al. 2006) to perform a principal component analysis (PCA) of the genetic variance of western coyote, western wolf, and eastern wolf reference populations (Table S1) at all 60,584 loci; we then ranked all SNPs based on their contributions to the first and second principal components. Second, we computed pairwise FST per locus among the three reference populations and ranked all SNPs based on their degree of differentiation. We selected SNPs that were present both in the top 1% of loci loading the principal component that separates each pair of putative source populations and in the top 1% of an analogous FST comparison (Figure S1).
The goal of our SNP selection process was to obtain a relatively small number of loci with maximum information content to discriminate among three putative parental populations of eastern coyotes: western coyotes, western wolves, and eastern wolves. By choosing SNPs that have a very high FST and PCA score, we genotyped SNPs whose alleles are not shared by eastern wolves and western coyotes or by eastern wolves and western wolves. Although we acknowledge that contemporary eastern wolves are admixed themselves, our approach to selecting SNPs gives the ability to distinguish the relative contributions of eastern and western wolf populations in the genome of eastern coyotes. Despite historic and recent hybridization with both coyotes and western gray wolves, the genomic and geographic distinctiveness of eastern wolves is widely accepted (Rutledge et al. 2010; vonHoldt et al. 2011; Rutledge et al. 2012). Also, the genetic profiles of extant wolves from the western Great Lakes region (i.e., western Ontario, Minnesota, Wisconsin) are similar to those of historic samples, suggesting that current eastern wolves are representative of the historic population (Wheeldon & White 2009; Fain et al. 2010). Further, admixture between wolves and coyotes very likely occurred in the western Great Lakes region (Kays et al. 2010a, b). For these reasons, our sample of eastern wolves included animals from Minnesota, Wisconsin, and Algonquin Park in Ontario (Table S1). Although there is controversy over the systematics of eastern wolves (vonHoldt et al. 2011; Rutledge et al. 2012), our inclusion of eastern wolves as a reference population does not address their taxonomic status.
We designed a custom GoldenGate genotyping assay for the Illumina (San Diego, California) BeadXpress platform. GoldenGate is a medium-throughput, PCR-based method of genotyping many loci in one multiplex reaction, and was recently used to survey genetic variation in wild canids (Sacks et al. 2009; Sacks et al. 2011). We tested in silico the multiplex compatibility of those SNPs that met the abovementioned criteria by downloading from dbSNP (www.ncbi.nlm.nih.gov/SNP) at least 60 bp of flanking sequence on each side of the polymorphism and submitting the sequences to Illumina for processing with Illumina’s Assay Design Tool (ADT). ADT executes an iterative process that evaluates candidate loci and outputs an Illumina score for each SNP that could vary from 0 to 1; SNPs with scores > 0.7 have a high likelihood of being amplified and genotyped successfully in the multiplex assay. In an initial set of 138 submitted SNPs, the ADT score varied from 0.17 to 0.99. We selected SNPs with Illumina scores > 0.7, resulting in a final panel of 63 unlinked SNPs distributed across 25 autosomes. All SNPs are non-genic, except rs22491491, which is located in the intron region of the CASP2 gene. The 63 loci were thus carefully selected to resolve the ancestry of the admixed coyote populations: 21 SNPs diagnostic between western coyote and western wolf, 21 diagnostic between western coyote and eastern wolf, and 21 diagnostic between western wolf and eastern wolf (Table 1).
Table 1.
Ascertainment and FST analysis of 63 ancestry-informative SNPs. SNP rs names correspond to the CanFam2.0 dog genome assembly. Cla = western coyote, Clu = western wolf, Cly = eastern wolf.
| Western coyote – Western wolf SNPs | Western Coyote – Eastern wolf SNPs | Western wolf – Eastern wolf SNPs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Cla allele | Clu allele | FST | SNP | Cla allele | Cly allele | FST | SNP | Clu allele | Cly allele | FST |
| rs24175585 | G | A | 0.97 | rs22927609 | G | C | 0.96 | rs22011433 | C | T | 0.74 |
| rs22333390 | A | G | 0.97 | rs22416514 | G | A | 0.95 | rs23651611 | T | C | 0.72 |
| rs9150379 | G | A | 0.97 | rs23054155 | G | C | 0.94 | rs24207725 | A | G | 0.70 |
| rs22877057 | A | C | 0.97 | rs22691222 | C | T | 0.92 | rs21906101 | T | G | 0.70 |
| rs24471781 | T | G | 0.95 | rs22659787 | C | G | 0.90 | rs22976400 | G | A | 0.69 |
| rs23367849 | A | G | 0.91 | rs22436136 | A | G | 0.90 | rs21962387 | A | G | 0.62 |
| rs24514093 | T | C | 0.91 | rs22491491 | G | A | 0.90 | rs22128776 | A | G | 0.62 |
| rs24543100 | C | T | 0.90 | rs22488932 | C | T | 0.90 | rs21972855 | G | A | 0.62 |
| rs23617324 | T | C | 0.88 | rs22494347 | T | G | 0.89 | rs24617980 | T | C | 0.61 |
| rs22161480 | C | T | 0.88 | rs22582321 | C | T | 0.87 | rs23245491 | G | C | 0.61 |
| rs8612074 | A | G | 0.88 | rs9073720 | C | T | 0.87 | rs23653965 | T | A | 0.60 |
| rs23909187 | A | G | 0.87 | rs24489243 | G | A | 0.86 | rs22817050 | T | C | 0.59 |
| rs23278100 | C | T | 0.86 | rs24373496 | G | A | 0.85 | rs22767921 | A | G | 0.58 |
| rs23037622 | T | C | 0.86 | rs24447332 | C | T | 0.85 | rs23070823 | G | T | 0.57 |
| rs22928481 | C | A | 0.86 | rs9029227 | G | A | 0.85 | rs24401025 | A | G | 0.55 |
| rs23050823 | G | A | 0.86 | rs24218607 | T | C | 0.85 | rs8747831 | T | C | 0.54 |
| rs23006689 | G | A | 0.85 | rs23126832 | T | A | 0.84 | rs24427396 | A | G | 0.54 |
| rs24517393 | T | C | 0.85 | rs8666298 | G | A | 0.84 | rs22521423 | G | A | 0.54 |
| rs22645721 | C | T | 0.85 | rs23410089 | G | T | 0.83 | rs24260906 | A | G | 0.53 |
| rs22409691 | G | A | 0.84 | rs22350704 | C | T | 0.83 | rs23966574 | G | A | 0.53 |
| rs23001750 | C | A | 0.84 | rs23486713 | G | A | 0.83 | rs24312148 | G | C | 0.53 |
Laboratory methods
We extracted total genomic DNA using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California) according to the manufacturer’s instructions or as described in Kays et al. (2010a). We cleaned and concentrated genomic DNA using a modified QIAamp DNA Micro Kit (Qiagen) protocol with SpinSmart PCR Purification columns (Denville Scientific, South Plainfield, New Jersey). We determined final DNA concentrations using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, Delaware) and prepared five 96-well plates with genomic DNA aiming to attain uniform concentration (average: 44 ng/μL).
SNP genotyping was done at the Center for Genomics and Human Genetics in The Feinstein Institute for Medical Research, Manhasset, NY. The GoldenGate assay was performed in accordance with manufacturer’s protocols. We processed the raw data using the genotyping module of Illumina’s GenomeStudio software suite (v2011.1), and followed Illumina’s recommendations for evaluating overall data quality, locus performance, and sample performance. We removed from all downstream analyses low-performing samples with call rates < 90%. GenomeStudio automatically generates genotypes from raw fluorescent signal intensities. All SNPs were biallelic, so there are three possible genotypes per locus - AA, AB, and BB - each forming three distinct clusters when signal intensity is plotted. For any particular locus, some samples did not conform to any of the three possible genotype clusters, so we also manually removed these ambiguous genotypes.
Analyses of admixture
We used two independent approaches to evaluate admixture. First, we used PLINK 1.07 (Purcell et al. 2007; http://pngu.mgh.harvard.edu/purcell/plink) to perform multidimensional scaling analysis (MDS). MDS is a data-agnostic multivariate method that allows one to explore and visualize the variation and dominant relationships in genetic data. MDS in PLINK is based on the pairwise identity-by-state distance matrix and the results are comparable to those of PCA. Second, we used STRUCTURE 2.3 (Pritchard et al. 2000) to quantify admixture and estimate ancestry in our sample in relation to the three reference populations. STRUCTURE implements a Bayesian clustering algorithm to infer the ancestry of admixed individuals by calculating the posterior mean estimates of K proportions of the genome inherited from ancestors in K populations. By employing these two complementary approaches, we address the criticisms of each. For example, STRUCTURE and similar Bayesian clustering methods are powerful analytical tools, but assume a population genetics model that may be violated in natural populations. MDS and similar ordination methods simply provide a scatter-plot which one subjectively examines for interesting patterns, but the data are not required to meet biological assumptions. Therefore, using both complementary approaches strengthens the interpretability of the results (Patterson et al. 2006; Rutledge et al. 2010).
We used a set consisting of 40 western coyotes, 34 western wolves, and 17 eastern wolves from vonHoldt et al. (2011) as reference parental populations to estimate the ancestry proportions of 427 eastern canids genotyped in this study and 10 additional coyotes genotyped by vonHoldt et al. (2011) – nine northeastern coyotes plus one Ohio coyote known to be admixed (Table S1). To ensure that the results of admixture analyses are not largely affected by our choice of priors in STRUCTURE’s Bayesian framework, we first tested the ability of our 63 ancestry-informative SNPs to distinguish among the three reference populations using six combinations of allele frequency and ancestry models available in STRUCTURE 2.3. We also performed these tests using 63 random SNPs; we used the website www.random.org to generate 63 random numbers between 1 and 48,036 in order to extract the genotypes of 63 random SNPs from the phased 48K-SNP dataset of vonHoldt et al. (2011). In these confirmatory tests we utilized 50,000 burn-in and 500,000 MCMC iterations, and set the number of populations (K) to 3.
After validating the diagnostic capability of the 63 ancestry-informative SNPs and their robustness to various modeling settings, we estimated the ancestry proportions of each individual in our sample of eastern coyotes using STRUCTURE and the same parameters as described above, following Bohling and Waits (2011). Specifically, for individuals without prior population information (i.e., the 437 “unknown” samples) we used the admixture model and set STRUCTURE to infer α (a Dirichlet parameter for degree of admixture) using a separate α for each population. Even though the ancestry estimate for each unknown individual depends only on the reference set and not on the other unknown hybrid individuals, Vähä and Primmer (2006) concluded that STRUCTURE may be somewhat sensitive to the proportion of hybrids in the overall sample. We tested whether the posterior ancestry estimates would be affected by this proportion by analyzing only the Ohio coyotes (N = 24) with the reference set (N = 91), and comparing their ancestry estimates with those generated when the full sample (N = 437) is analyzed with the reference set.
Although domestic dogs were not included in our SNP ascertainment panel or in the initial STRUCTURE reference set, vonHoldt et al. (2011) discovered substantial levels of admixture with dogs, especially in Ohio and eastern coyotes. To test for the degree of dog admixture in our samples of Ohio and eastern coyotes, we conducted a post hoc analysis by adding a fourth reference population of domestic dogs, choosing ten modern dog breeds based on their size, presence in North America, and potential to mate with coyotes and wolves (Table S1). We carried out this analysis in STRUCTURE using the same parameters as above, but setting K to 4. For all STRUCTURE analyses, we conducted five replicate runs and used CLUMPP (Jakobsson & Rosenberg 2007) to align and average the five replicate cluster membership coefficient matrices.
In order to investigate the broader ecological context of admixture, we tested the hypothesis that individuals living in areas of high deer density are more wolf-like genetically than those living in areas of low deer density. We obtained white-tailed deer (Odocoileus virginianus) densities from a Quality Deer Management Association 2008 map which depicts deer densities across management units or counties and summarizes data provided by state wildlife agencies (www.qdma.com/shop/qdma-white-tailed-deer-density-map). Deer density was treated as a categorical ordinal variable with four density bins (< 15, 15–30, 30–45, and > 45 deer per square mile) because that is how Quality Deer Management Association standardized data across wildlife agencies. We assigned to each individual canid the deer density that corresponds to its sampling locality. We analyzed the association of wolf ancestry to deer density using a non-parametric Kruskal-Wallis test in R (R Development Core Team 2012). We used the combined western + eastern wolf ancestry component estimated when dogs were included in the analysis, thus eliminating the latrans and familiaris components of individuals’ ancestry.
Results
Genotyping results
We attempted to genotype 480 samples at 63 SNP loci. Our quality control measures filtered out 53 samples: 51 that performed poorly with call rates < 90% and two that were mislabeled. Thus we report results for the remaining 427 samples. Out of 26,901 expected genotypes, 1372 were not called or were manually removed due to ambiguity, for a total genotyping call rate of 94.9%. Across all 63 SNPs, call rates ranged from 83% to 100%, and averaged 95.5% (± 3.5% SD). Ten of the thirteen scat samples we attempted to genotype amplified successfully with high call rates. Genotyping success did not depend on source of DNA, whether fecal or tissue sample (chi-squared test for independence: χ2 = 2.158, df = 1, P = 0.142).
Admixture
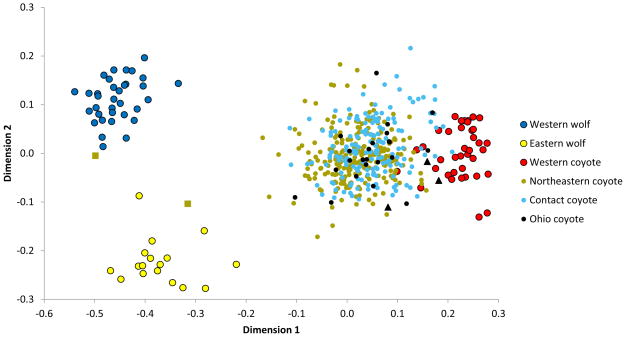
The distribution of genetic variation among genotyped individuals in relation to the reference populations can be visualized with MDS (Figure 2). In this analysis using only the 63 ancestry-informative SNPs, the three reference populations formed their own distinct clusters in a manner similar to when the ordination is done on all 60,000+ loci (Figure 2, Figure S1). MDS axis 1 separated coyotes from wolves, and axis 2 separated the two wolf reference populations (western vs. eastern). Most of the 425 eastern coyote samples formed their own cluster that has little overlap with the western coyote cluster. As expected, eastern coyotes were situated between western coyotes and wolves in MDS-space, with large variability in genomic contributions from each reference population; i.e., some individuals were genetically more similar to western coyotes while others were much more wolf-like. Most individuals from Ohio were far from western coyotes in MDS-space. Instead, all Ohio coyotes were fully within the distribution of contact zone and northeast zone coyotes (Figure 2), even in higher MDS dimensions (data not shown). The three Ohio coyotes genotyped by vonHoldt et al. (2011) - graphed as black triangles in Figure 2 - were outliers relative to other Ohio coyotes genotyped in this study. In addition, one of the suspected wolf immigrants (zm15083 from Orleans County, VT) clearly clustered with western wolves and the other (zm14276 from Saratoga County, NY) with eastern wolves (Figure 2).
Figure 2.
Multidimensional scaling plot of the three reference populations and the 427 canids genotyped in this study. Data for all samples are only from 63 ancestry-informative SNPs. Samples genotyped in this study were partitioned into three geographic zones as in Kays et al. (2010a): Ohio, contact zone, and northeast zone. Black triangles represent the three Ohio coyotes genotyped by vonHoldt et al. (2011). Gold squares represent two immigrant wolves from Vermont (left) and New York (right).
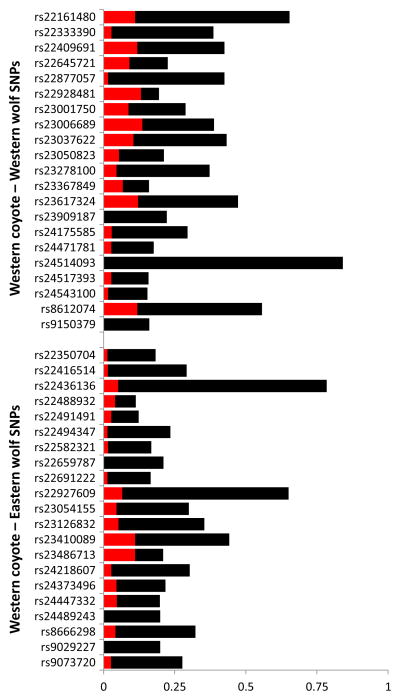
In all 42 SNPs that distinguish western coyotes from western and eastern wolves (Table 1), the frequency of the rare wolf allele was much higher in eastern coyotes than in western coyotes (Figure 3). The average frequency of the wolf allele in western coyotes was 0.05, but increased to 0.31 in eastern coyotes. In five loci, the wolf allele became the major allele in eastern coyotes, attaining frequencies in excess of 0.5. In five loci that were monomorphic in western coyotes (i.e., the wolf allele was absent in a sample of 40 diploid individuals), the wolf allele appeared in moderate to high frequencies in eastern coyotes (Figure 3). In these same 42 SNPs, eastern coyotes had moderate to high dosage of coyote alleles and an average heterozygosity of 0.33 (range: 0.02–0.73; Figure S2).
Figure 3.
Frequency of the wolf allele in 42 SNPs that distinguish western coyotes from western and eastern wolves. The length of the red bar is the frequency of the rare wolf allele in the western coyote reference population. The total length of the bar is the frequency of the wolf allele in the full sample of eastern coyotes, thus the black portion is the increase in frequency due to introgression.
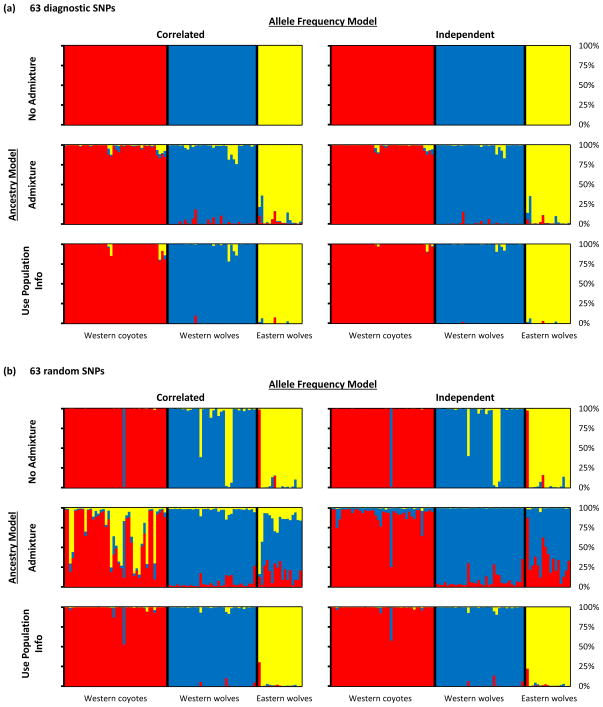
We conducted preliminary tests in STRUCTURE with six model combinations of ancestry and allele frequency priors, using only the 91 reference individuals. The 63 ancestry-informative SNPs performed well in all simulations, clearly discriminating the three reference populations (Figure 4a). Thus the “cleanness” of the bar plots in Figure 4a is not a statistical artifact and the parameters used as priors in STRUCTURE’s Bayesian approach do not strongly affect the results. With 63 randomly chosen SNPs, individuals from all three reference populations were misassigned in the No Admixture simulations, and the reference populations were not clearly partitioned in the Admixture simulations (Figure 4b). The random SNPs did not perform well, except in the Use Population Info simulations (Figure 4b).
Figure 4.
Ability of 63 ancestry-informative SNPs (a) versus 63 random SNPs (b) to distinguish among 40 western coyotes (red), 34 western wolves (blue), and 17 eastern wolves (yellow) using six combinations of three ancestry models and two allele frequency models available in STRUCTURE 2.3. The purpose of this analysis was to validate the diagnostic capability of our carefully selected SNPs and to test the sensitivity of the results to the priors in STRUCTURE’s Bayesian framework.
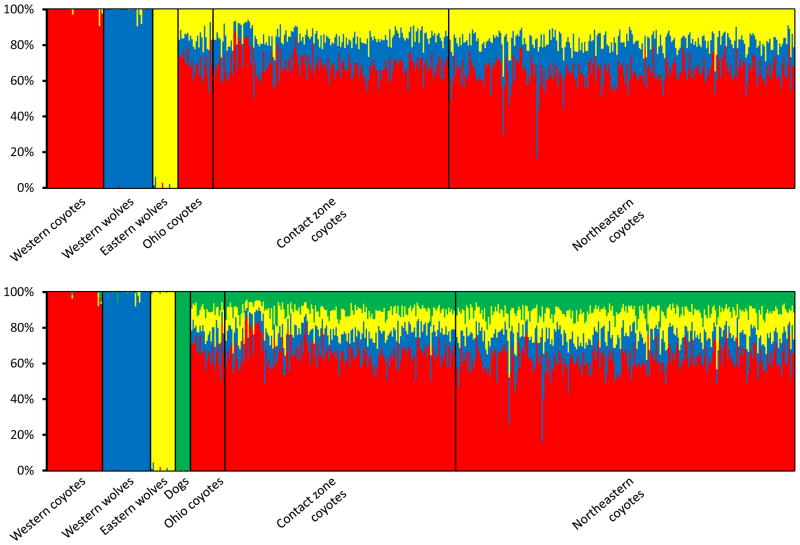
After determining that the 63 ancestry-informative SNPs successfully distinguished among western coyotes, western wolves, and eastern wolves, and that the results are not much affected by modeling decisions, we used them to assess the ancestry of 437 eastern canids in relation to the three parental populations. Although the 63 ancestry-informative SNPs were not initially selected to distinguish dogs from wild Canis, they did so remarkably well (Table 2). This allowed us to also estimate the proportion of dog ancestry in eastern coyotes. All coyotes examined showed a signal of admixture, but there were some subtle geographic differences in the degree of wolf and dog introgression (Figure 5, Table 2). Coyotes from the northeast zone were significantly more wolf-like than Ohio and contact zone coyotes (Kruskal-Wallis chi-squared = 26.45, P < 0.001 with K = 3; Kruskal-Wallis chi-squared = 22.54, P < 0.001 with K = 4). There was a widespread signal of dog ancestry, with about 10% of eastern coyotes’ genome assigned to dog, but northeastern coyotes had significantly higher dog ancestry (Kruskal-Wallis chi-squared = 6.66, P = 0.010). Ohio coyotes had, on average, 31% wolf ancestry (western + eastern), which is not significantly different from contact and northeast zone coyotes (Kruskal-Wallis chi-squared = 3.26, P = 0.071). When we compared the ancestry estimates of the Ohio coyotes when analyzed alone versus when analyzed with the other 413 canids, the differences were negligible, thus the STRUCTURE analyses were not sensitive to the proportion of hybrids in the overall sample.
Table 2.
Ancestry analysis of 437 admixed canids in relation to three or four parental populations (in bold): Cla = western coyotes, Clu = western wolves, Cly = eastern wolves, and Cfa = domestic dogs. Ohio, contact, and northeastern subdivisions correspond to geographic zones in Figure 1. Values indicate mean ancestry estimate of N samples.
| N | Three parental populations | Four parental populations | ||||||
|---|---|---|---|---|---|---|---|---|
| % Cla | % Clu | % Cly | % Cla | % Clu | % Cly | % Cfa | ||
| Western coyotes | 40 | 99.59 | 0.05 | 0.37 | 99.44 | 0.03 | 0.27 | 0.26 |
| Western wolves | 34 | 0.05 | 99.32 | 0.63 | 0.03 | 99.30 | 0.46 | 0.21 |
| Eastern wolves | 17 | 0.20 | 0.58 | 99.22 | 0.13 | 0.40 | 99.30 | 0.17 |
| Dogs | 10 | - | - | - | 0.01 | 0.06 | 0.08 | 99.85 |
| Ohio coyotes | 24 | 68.94 | 14.62 | 16.43 | 66.23 | 11.67 | 12.36 | 9.74 |
| Contact coyotes | 167 | 68.64 | 15.56 | 15.80 | 65.77 | 12.31 | 11.66 | 10.26 |
| Northeastern coyotes | 244 | 64.69 | 17.10 | 18.21 | 61.73 | 13.62 | 13.58 | 11.07 |
| Northeastern wolves | 2 | 23.22 | 49.80 | 26.99 | 20.99 | 44.98 | 19.88 | 14.16 |
Figure 5.
Ancestry analyses of 437 admixed canids in relation to the three (top) or four (bottom) parental populations. STRUCTURE bar plots depict each individual as a vertical bar divided into three or four posterior mean estimates of its admixed ancestry, i.e., the estimated proportion of its genome inherited from western coyote, western wolf, eastern wolf, or dog ancestors. Ancestry-informative genetic markers were selected to make the parental populations as distinct as possible. The two northeastern individuals with high wolf ancestry are immigrant wolves sampled in New York and Vermont. Average ancestry estimates per group are given in Table 2.
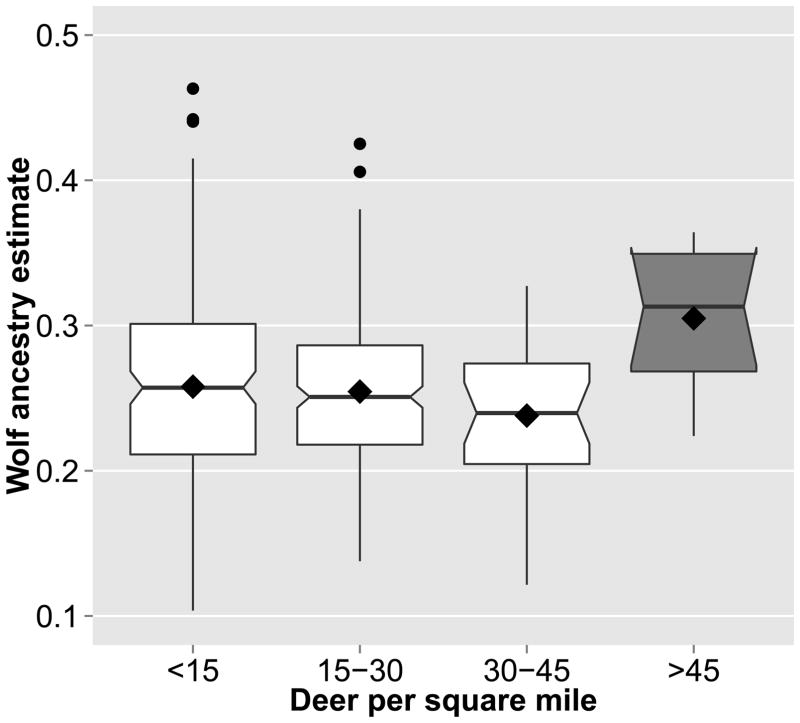
Ten coyotes sampled from the highest deer density habitats (> 45 deer/mile2) were genetically more wolf-like than coyotes sampled from habitats with lower deer density (Figure 6, Kruskal-Wallis chi-squared = 7.21, P = 0.007). The average pairwise relatedness of these ten individuals was r = 0.13 (2× Lynch and Ritland 1999 estimator). We note here that the Kruskal-Wallis statistic and corresponding P-value test the equality of medians and not a causal relationship; i.e., the statistical hypothesis was that wolf ancestry varies in areas of different deer density, not that deer density “shapes” wolf ancestry.
Figure 6.
Wolf ancestry plotted against deer density. Canids in areas of very high deer density are significantly more wolf-like than in lower density classes, as denoted by the dark gray box. Wolf ancestry on the vertical axis is the combined western + eastern wolf ancestry proportion estimated when dogs were included in the STRUCTURE analysis, thus eliminating the latrans and familiaris components. The lower and upper boundaries of each box correspond to the first and third quartiles, respectively; black line is the median, black diamond is the mean, circles are outliers. Non-overlapping notches on the sides of boxes indicate strong evidence that the medians differ.
Discussion
Admixture in North American canids
We analyzed 63 ancestry-informative autosomal SNPs in 437 northeastern canids and found that admixture is pervasive across the region. The ancestry of all coyotes we sampled showed a clear signal of hybridization with various Canis groups: western wolves, eastern wolves, and domestic dogs. Recent evidence from mitochondrial and Y-chromosome DNA support our finding of extensive hybridization among western gray wolves, eastern wolves, coyotes, and dogs in eastern North America (Wilson et al. 2012; Wheeldon et al. 2013). This coyote-wolf-dog hybrid swarm extends into the Midwestern United States. Contrary to our expectations from an earlier finding of no wolf mtDNA in Ohio coyotes (Kays et al. 2010a), these same individuals were, on average, 66% coyote and 24% wolf in their nuclear genome (Table 2). The extension of wolf introgression into Ohio was unexpected because vonHoldt et al. (2011) found that midwestern/southern coyotes were genetically distinct from hybrid northeastern coyotes, and that admixture in midwestern/southern coyotes was primarily with dogs. In their analyses, midwestern/southern coyotes had, on average, 7.5% dog ancestry and 2.4% wolf ancestry. However, their inference came from a limited sample of 13 northeastern and 19 midwestern/southern coyotes, only three of which were from Ohio. Those three Ohio coyote samples are on the periphery of the statistical distribution of other Ohio coyotes genotyped in this study (Figure 2). This is not surprising since those three coyotes were selected for genotyping with the SNP microarray because they were morphologically peculiar, having unusual pelage and craniodental phenotypes.
How did wolf-derived DNA arrive in Ohio? We propose three hypotheses that require further investigation: (1) coyote-wolf hybrids, descendants of the northern expansion front, circled around the Great Lakes and back westward into Ohio; (2) coyote-wolf hybridization occurred in Minnesota or western Ontario (Kays et al. 2010b) and the initial colonizers of Ohio were admixed; (3) coyote-wolf hybrids from southern Ontario moved into the southern peninsula of Michigan and then south into Ohio. These three and any other hypotheses must be able to account for the disparate patterns in mitochondrial and nuclear DNA.
In theory, it is possible that northeastern coyotes evolved to be more wolf-like genetically due to natural selection, genetic drift, or both, thus appearing admixed in the absence of actual hybridization. We reject this hypothesis on several grounds. First, early studies were highly suggestive of a hybrid origin for northeastern coyotes, long before the availability of any molecular data needed to confirm this (Monzón 2012). The hybridization hypothesis was proposed by various authors entirely on the basis of morphology (Lawrence & Bossert 1969) and captive rearing experiments (Silver & Silver 1969; Kolenosky 1971; Mengel 1971). Second, molecular evidence has unequivocally confirmed coyote-wolf admixture in the Great Lakes region and further east since the early 1990s (Lehman et al. 1991; Wayne & Lehman 1992), including recent evidence of wolf mitochondrial DNA introgressing northeastern coyotes (Koblmüller et al. 2009; Kays et al. 2010a). Third, genetic drift alone would make some rare wolf alleles become more frequent at a few loci but become extinct at most other loci, and selection alone would make rare wolf alleles more frequent at a few loci but remain constant at most other loci. However, our results show the rare wolf alleles universally became more frequent in eastern coyotes than in western coyotes, even if they were absent in the western coyote parental population (Figure 3), demonstrating the rapid influx of wolf DNA from introgressive hybridization.
During the design phase of our study there was little evidence that hybridization with domestic dogs is prevalent in the Northeast. Way et al. (2010) found no dog mtDNA in 67 coyotes from eastern Massachusetts, and Kays et al. (2010a) found only one dog mtDNA haplotype in a region-wide sample of 715 eastern coyotes. Consequently, we did not select any SNPs to be diagnostic of dog ancestry when designing our study, but were able to consider this using a post hoc analysis. Our findings are consistent with those of vonHoldt et al. (2011), who found that northeastern coyotes have on average 9.1% dog ancestry; we found that region-wide (including Ohio) coyotes have on average 10.7% (± 3.3% SD) dog ancestry. Using twelve autosomal microsatellites, Wheeldon et al. (2013) recently found that coyotes in southeastern Ontario have on average 2.3% dog ancestry. Together, these results suggest a limited, but appreciable, amount of coyote-dog hybridization in the recent past (11 to 24 generations, estimated by vonHoldt et al. 2011). Since then, the dog components of the genome have been diluted and integrated into the wild gene pool through generations of backcrossing with eastern coyotes. We found no evidence for ongoing coyote-dog hybridization; the homogeneity and low proportion of the dog component in our large sample of wild eastern coyotes suggest that coyote-dog hybridization is infrequent, although the wild population is so abundant that coyote-dog F1 hybrids may appear at a frequency below the detection power of our sample. On the other hand, the dosage of coyote vs. wolf alleles and the fraction of heterozygous loci (Figure S2) suggest that at least some individuals are first- or second-generation coyote-wolf hybrids backcrossed to coyote (vonHoldt et al. 2013).
Our data reveal a complex pattern of admixture among coyotes, dogs, and two distinct wolf populations. We do not believe the common name “Coywolf,” proposed for northeastern coyotes by Way et al. (2010), captures this complexity. Similar patterns of three- and four-way hybridization have been observed in North American Canis. Hailer and Leonard (2008) found some degree of hybridization among sympatric coyotes, Mexican wolves, and red wolves in Texas; Bohling and Waits (2011) detected frequent admixture among coyotes, gray wolves, red wolves, and domestic dogs in North Carolina; and Rutledge et al. (2010) showed that eastern wolves in Ontario act as a conduit of gene flow between coyotes and western wolves by hybridizing with both. Hybridization in Canis extends outside North America: domestic dog genes have introgressed into the wild Australian dingo, European gray wolf, and Ethiopian wolf (Canis simensis) populations (Gottelli et al. 1994; Elledge et al. 2008; Godinho et al. 2011).
Sex-biased hybridization
The observation that wolf DNA is present in the nuclear genome but absent in the mitochondrial genome of Ohio coyotes is clear evidence of sex-biased hybridization between male wolves and female coyotes. The first genetic evidence of coyote-wolf interbreeding suggested that hybridization is unidirectional and occurs only with male wolves and female coyotes (Lehman et al. 1991). Additional mitochondrial, Y-chromosome, and autosomal microsatellite data revealed that male western wolves tend to cross with female eastern wolves, and that male eastern wolves tend to cross with female coyotes across Ontario (Rutledge et al. 2010). A recent survey of several populations across eastern North America found a surprisingly high frequency of western wolf Y-chromosome haplotypes in eastern coyotes, despite an absence of western wolf mtDNA (Wilson et al. 2012). Evidence for interspecific crosses between male coyotes and female wolves is much scarcer. Only 1 in 70 Texas coyotes surveyed carried maternal gray wolf DNA (Hailer & Leonard 2008); and although hundreds of eastern coyotes carried maternal eastern wolf DNA (Kays et al. 2010a), nearly all carried the same haplotype, possibly the result of a single hybridization event. However, it should be noted that the genetic ancestry of wolf is expected to be lost much faster from the mitochondrial genome compared to the nuclear genome if hybridization is followed by subsequent backcrosses with coyote. For example, if the F1 hybrid son of a male coyote and female wolf backcrosses with a female coyote, his offspring will have 25% wolf ancestry in the nuclear genome but 0% wolf ancestry in the mitochondrial genome.
Similarly, the observation that dog DNA is present in the nuclear genome but absent in the mitochondrial genome of eastern coyotes reveals that male dogs mated with female coyotes, but not vice versa. A recent analysis of maternally inherited mtDNA and paternally inherited Y-chromosome markers revealed that the asymmetric introgression of dog genes into northeastern coyotes is mediated by male dogs (Wheeldon et al. 2013). Hybridization of European and African wolves with dogs is also consistently mediated by male dogs and female wolves (Gottelli et al. 1994; Vilà et al. 2003; Godinho et al. 2011; but see Hindrikson et al. 2012 for a rare exception). It is conceivable that our coyote study and the Old World wolf studies failed to sample the hybrid progeny of wild males and domestic females as these pups would likely be reared by bitches in domestic settings or eliminated by dog owners. In contrast to this general pattern of sex-biased hybridization, Adams et al. (2003) documented about 10% of southeastern coyotes with a dog mtDNA sequence, but they postulated a more unnatural cause: young male coyotes from Texas were periodically trapped and released in the Southeast for sport hunting before the main front of coyotes colonized the region; a male coyote that escaped had no female conspecifics and mated with a local dog instead. Our data are consistent with other observations that sexual interactions between wild and domestic canids generally involve male dogs. It is not uncommon for males of certain dog breeds to be as large, or larger, than female wolves or coyotes.
Overall, our data support the hypothesis that the directionality of coyote-wolf-dog sexual interactions is largely determined by body size, with the males of the larger species mating with females of the smaller, as some dog breeds are larger than coyotes. But at least two other hypotheses may account for the apparent directionality of hybridization in Canis. First, wolf populations, usually being sparse, may be subject to Allee effects. One such effect may be the perception by male wolves of heterospecific females as potential mates. Male wolves may encounter lone female coyotes much more frequently than male coyotes encounter lone female wolves. Second, strong maternal effects in Canis may preclude F1 progeny of domestic mothers and wild fathers to enter the wild population, as mentioned earlier. The body size, Allee effects, and maternal effects hypotheses are not mutually exclusive, but one way to start testing them is to look for coyote and wolf introgression in rural and feral dogs. For example, the size hypothesis predicts greater introgression of wild alleles in litters of smaller female dogs than in litters of larger females. Alternatively, the maternal effects hypothesis predicts the presence of wild diagnostic alleles in hybrid sons of domestic mothers, but dog diagnostic alleles in hybrid sons of wild mothers.
Ecological context of hybridization
Coyotes living in areas of high deer density are more wolf-like genetically (Figure 6), supporting the idea that introgressive hybridization with wolves facilitated the colonization of eastern forests and introduced adaptive genetic variation that allowed coyotes to exploit a prey base rich with ungulates (Kays et al. 2010a). Broadly comparing regional populations, northeastern coyotes eat more deer than their western and southern counterparts. Many studies (reviewed by Parker 1995 and Gompper 2002) from western, midwestern, and southern areas of its range have shown the coyote to be an insignificant predator of deer. On the other hand, deer are the most important component of the diet of northeastern coyotes, especially for reproductive adults and coyotes in areas with dense forest and severe winters (Harrison & Harrison 1984; Major & Sherburne 1987; Brundige 1993). At a more local scale, predation of white-tailed deer, especially of fawns, is much greater in areas of high deer density versus areas of low deer density (Blanton & Hill 1989). Wolf ancestry of admixed canids in southern Ontario also is positively associated with moose density (Benson et al. 2012). The strong dependence of northeastern coyotes on ungulate prey presents the possibility that heterogeneity in prey density exerts differential selection pressures on coyotes’ ability to hunt them. Wolves are better adapted to kill ungulates; thus, habitats rich in deer and moose may be favorable to hybrid individuals with a greater degree of wolf ancestry, resulting in fine-scaled local adaptation.
The positive association of wolf-likeness with deer density observed in this study is driven solely by a small sample of 10 individuals from two areas of very high deer density in New Jersey; these two areas measure 1380 and 842 km2. The association is not clinal and disappears when the two highest levels of deer density are compared with the two lowest. Further research is needed to confirm the local adaptation hypothesis with more sampling sites of high deer density, or better, finer-grained, continuous deer density data and genetic markers linked to traits of known function, such as genes related to body size and skull morphology. Further, an analysis of skulls may reveal that coyotes living in areas of high deer density appear more wolf-like morphologically. Coyote-wolf hybrids in undisturbed landscapes of southeastern Ontario indeed have a more wolf-like morphology and diet, while those in nearby fragmented and disturbed landscapes have a more coyote-like form and diet (Sears et al. 2003). These and our results preliminarily indicate that natural selection for wolf-like versus coyote-like traits may be occurring at a fine geographic scale based on landscape characteristics, such as prey availability and human land use.
Methodological matters
Several methodological issues are noteworthy. First, our use of ancestry-informative markers allowed us to quantify the relative genomic contributions of four putative parental populations to eastern coyotes. Fitzpatrick and Shaffer (2004, 2007) employed a similar approach using 8 diagnostic restriction-fragment-length polymorphisms to ascertain the degree of introgression from an introduced salamander’s genes into the gene pool of the threatened California tiger salamander (Ambystoma californiense). Talbot et al. (2011) likewise developed an assay to distinguish among four poplar (Populus) species and their hybrids using 26 diagnostic SNPs. vonHoldt et al. (2013) developed an assay to detect recent wolf-dog hybridization using a panel of 24 diagnostic SNPs with divergent allele frequency distributions. These studies demonstrate how readily hybridization can be quantified with just a few carefully chosen fixed or nearly fixed diagnostic markers. Indeed, a simulation study suggested that 12 loci with an average FST of 0.21 have sufficient power to detect hybrids of two parental populations, although the hypothetical loci were multiallelic (Vähä & Primmer 2006). The 63 markers we used in this study have very high FST values (average: 0.79, range: 0.53–0.97; Table 1) compared to genome-wide FST (western coyote-western wolf: 0.14, western coyote-eastern wolf: 0.11, western wolf-eastern wolf: 0.05; vonHoldt et al. 2011). As a result we were able to still use the admixed eastern wolves as a reference population in order to assess their relative contribution to eastern coyotes. Even though none of the loci investigated in this study were in coding regions, it is possible that they are linked to functional genes; this is likely because of their high divergence across parental populations. If wolf alleles are favorably selected in northeastern coyotes as we propose, then the admixture proportions may be somewhat overestimated.
Second, several recent studies have employed STRUCTURE or similar Bayesian clustering programs to assess hybridization (e.g., Oliveira et al. 2008; Bohling & Waits 2011; Godinho et al. 2011; Sacks et al. 2011; vonHoldt et al. 2011). Most of these studies avoid using prior population information (the usepopinfo ancestry model in STRUCTURE) that may bias the posterior probabilities of assignment. However, the usepopinfo model is necessary when using a sample of reference populations to assess admixture in another population. In STRUCTURE’s user manual, Pritchard et al. (2010) caution the user to also run the program without population information to ensure that the pre-defined populations are in rough agreement with the genetic information because this model assumes that the predefined populations are usually correct. We ran our reference samples through the program with and without the usepopinfo model and observed that it is unwise to use prior population information if the genetic markers are polymorphic but not diagnostic. If, on the other hand, diagnostic markers are chosen to maximize differentiation among groups, user decisions concerning biological assumptions are less influential (Figure 4).
Third, using fecal samples for population-level genetic analyses became popular in the 1990s, but has some technical complications because scat-derived DNA tends to be highly fragmented and degraded (Kohn & Wayne 1997; Kohn et al. 1999). The Illumina GoldenGate assay we used worked as well with scat samples as with tissue samples, most likely because the PCR amplicons are short (100–120 bp). Sacks et al. (2011) obtained similar positive results using the same method. Thus, we recommend that researchers working with fecal DNA use a method that amplifies similarly short sequences, such as assays of SNPs and small indels or next-generation sequencing procedures that produce short reads.
Fourth, SNPs are the new vogue in population and conservation genetics (Morin et al. 2004; Kohn et al. 2006). The completion of the dog genome project enabled researchers to discover and interrogate many SNPs in wild canids. Despite the technological advances, several pioneering studies were fraught with laborious SNP discovery and genotyping methods, small sample sizes, and low genotyping rates (Seddon et al. 2005; Andersen et al. 2006; Sacks & Louie 2008). As in all molecular studies, researchers must balance the costs of genotyping many individuals versus many loci. We navigated these issues by choosing a mid-throughput genotyping method and judiciously selecting SNPs with the highest information content for admixture analyses. This inclined us to genotype many samples versus many SNPs. The resulting dense geographic sampling gives unparalleled resolution to understand admixture dynamics and facilitates future investigations of cryptic population structure and local adaptation (Sacks et al. 2004; Sacks et al. 2005; Monzón 2012; Monzón, in review).
Philosophical matters and implications for coyote and wolf management
Although hybridization is increasingly accepted as a natural phenomenon, even among vertebrate groups, it is often problematic for conservation practitioners. Many conservation policies have the biological species concept as their foundation, thus assuming that “species” are reproductively isolated. Yet, one million years of divergence from a common ancestor, and tens of thousands of years of intense artificial selection in dogs have been insufficient for reproductive isolation to fully evolve in Canis. In many cases, even when reproductive isolation has not fully evolved, there tends to be some outbreeding depression from the loss of locally adaptive genotypes (gene × environment interactions) or the disruption of coadapted gene complexes (gene × gene interactions) (Edmands 1999). The opposite outcome, hybrid vigor, appears to be the case in North American Canis. A more diverse genome, with genes from both wolves and dogs, likely allowed northeastern coyotes to survive in new habitats, both forested and human-dominated. The introgression of adaptive genetic variation via hybridization with wolves presumably permitted admixed coyotes to rapidly colonize the Northeast (Kays et al. 2010a). In fact, the movement of coyotes into the Northeast did not occur until after they began hybridizing with wolves about 154–190 years ago or 86 ± 9 coyote generations ago, assuming a 2-yr generation time (vonHoldt et al. 2011). Admixed northeastern coyotes have higher genome-wide heterozygosity than non-admixed populations (vonHoldt et al. 2011), and modern admixed Great Lakes wolves have more mitochondrial and Y-chromosome haplotypes than western coyotes and gray wolves (Koblmüller et al. 2009).
One phenomenon that is of particular concern is hybridization between a domestic species and its wild relatives. Some examples among vertebrates include bison (Halbert & Derr 2007), wolves (Godinho et al. 2011), and wildcats (Oliveira et al. 2008). It is worrisome that some wildlife may lose its wildness and untamed nature by hybridizing with a domestic species. Indeed, genetic evidence from a domesticated line of foxes (Vulpes vulpes) reveals that domestication leads to marked differences of gene expression in brain regions that modulate emotions and behavior (Lindberg et al. 2005). The possibility that introgression of dog alleles has made eastern coyotes more adapted to human-dominated environs warrants further research.
The type of hybridization documented in this study may be perceived in a negative or a positive way. Seehausen et al. (2008) noted that hybridization may result in a net loss of species numbers, effectively reversing speciation; they used eastern coyote-wolf hybrids to exemplify how two species may coexist in sympatry in some parts of their range, but merge as a hybrid swarm in another more disturbed area. But they could have also used eastern coyote-wolf hybrids to exemplify the rescuing of local biota or the colonization of a new niche via hybridization. As mentioned above, admixed coyote and wolf populations in the Northeast are more genetically diverse than their parental populations, and this enhanced genetic diversity may be adaptive. In a way, hybridization between coyotes and wolves and the subsequent colonization of eastern forests have yielded a net increase in local species diversity by restoring a large wolf-like canid into the Northeast United States. It is unclear whether this admixed canid will prevent the recolonization of true, full-sized wolves. The two wolves genotyped in this study were likely natural dispersers from Canada (Kays & Feranec 2011). One of them had a genetic profile of western Canis lupus, and likely dispersed from northern Quebec where wolves remain distinct from Great Lakes wolf populations.
Wolf and coyote management policies should consider the ecological importance of large predators. With 16–20 million white-tailed deer in the United States, the direct and indirect socioeconomic costs of overpopulated deer are staggering: annual estimates of deer damage are reported to exceed $2 billion nationwide, including $1 billion/year in car damages (Rondeau & Conrad 2003). There is no question that we need to restore natural predator-prey dynamics, lest we allow deer populations to be regulated by cars. Because eastern North American canids form a taxonomically complex group characterized by reticulate evolution, we argue that management policies in the region should aim at conserving natural ecological and evolutionary processes (Ennos et al. 2005; Kyle et al. 2006), such as trophic dynamics and local adaptation.
Supplementary Material
Acknowledgments
We thank Robert Wayne and Bridgett vonHoldt for helpful comments in the early phases of this study and for sharing the 61K- and 48K-SNP “Magic” datasets. We thank Bridgett vonHoldt, John Pollinger, Heather Lynch, Kevin Shoemaker, Matthew Aiello-Lammens, and Leone Brown for generous advice on data analyses. We thank Christian Roccanova for help in the laboratory and Joseph Bopp for help in collecting samples. Staffan Bensch and six anonymous reviewers provided constructive comments to earlier drafts. Research reported in this publication was supported by the National Institute of General Medical Science of the National Institutes of Health IRACDA grant K12GM102778 to Jorge Benach, National Institutes of Health grant GM-6073102 to D. Dykhuizen, the New York State Museum, and grants to J. Monzón from the American Museum of Natural History, American Society of Mammalogists, Stony Brook University’s Turner Fellowship, and National Science Foundation’s Alliance for Graduate Education and the Professoriate.
Footnotes
Author contributions
JM, RK, and DED conceived and designed the research; RK collected the samples; JM performed the laboratory work and analyzed the data; DED contributed laboratory materials and reagents; JM wrote the paper.
Data accessibility
SNP genotypes, sample locations, PLINK and STRUCTURE input/output files are deposited in the Dryad Digital Repository doi:10.5061/dryad.1bh5q/1.
References
- Adams J, Leonard J, Waits L. Widespread occurrence of a domestic dog mitochondrial DNA haplotype in southeastern US coyotes. Molecular Ecology. 2003;12:541–546. doi: 10.1046/j.1365-294x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: setting conservation guidelines. Trends in Ecology & Evolution. 2001;16:613–622. [Google Scholar]
- Andersen DH, Fabbri E, Santini A, et al. Characterization of 59 canine single nucleotide polymorphisms in the Italian wolf (Canis lupus) population. Molecular Ecology Notes. 2006;6:1184–1187. [Google Scholar]
- Benson JF, Patterson BR, Wheeldon TJ. Spatial genetic and morphologic structure of wolves and coyotes in relation to environmental heterogeneity in a Canis hybrid zone. Molecular Ecology. 2012;21:5934–5954. doi: 10.1111/mec.12045. [DOI] [PubMed] [Google Scholar]
- Blanton K, Hill E. Coyote use of white-tailed deer fawns in relation to deer density. Proceedings of the Annual Conference of the Southeastern Association of Fish and Wildlife Agencies. 1989;43:470–478. [Google Scholar]
- Bohling JH, Waits LP. Assessing the prevalence of hybridization between sympatric Canis species surrounding the red wolf (Canis rufus) recovery area in North Carolina. Molecular Ecology. 2011;20:2142–2156. doi: 10.1111/j.1365-294X.2011.05084.x. [DOI] [PubMed] [Google Scholar]
- Brundige GC. Thesis, SUNY College of Environmental Science and Forestry. New York: Syracuse; New York, USA: 1993. Predation ecology of the eastern coyote, Canis latrans var., in the Adirondacks. [Google Scholar]
- Edmands S. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999;53:1757–1768. doi: 10.1111/j.1558-5646.1999.tb04560.x. [DOI] [PubMed] [Google Scholar]
- Elledge AE, Allen LR, Carlsson BL, Wilton AN, Leung LKP. An evaluation of genetic analyses, skull morphology and visual appearance for assessing dingo purity: implications for dingo conservation. Wildlife Research. 2008;35:812–820. [Google Scholar]
- Ennos RA, French GC, Hollingsworth PM. Conserving taxonomic complexity. Trends in Ecology & Evolution. 2005;20:164–168. doi: 10.1016/j.tree.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Fain SR, Straughan DJ, Taylor BF. Genetic outcomes of wolf recovery in the western Great Lakes states. Conservation Genetics. 2010;11:1747–1765. [Google Scholar]
- Fitzpatrick B, Shaffer H. Environment-dependent admixture dynamics in a tiger salamander hybrid zone. Evolution. 2004;58:1282–1293. doi: 10.1111/j.0014-3820.2004.tb01707.x. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick B, Shaffer H. Introduction history and habitat variation explain the landscape genetics of hybrid tiger salamanders. Ecological Applications. 2007;17:598–608. doi: 10.1890/06-0369. [DOI] [PubMed] [Google Scholar]
- Godinho R, Llaneza L, Blanco JC, et al. Genetic evidence for multiple events of hybridization between wolves and domestic dogs in the Iberian Peninsula. Molecular Ecology. 2011;20:5154–5166. doi: 10.1111/j.1365-294X.2011.05345.x. [DOI] [PubMed] [Google Scholar]
- Gompper ME. WCS Working Paper No 17. Wildlife Conservation Society; Bronx, New York: 2002. The ecology of northeast coyotes: current knowledge and priorities for future research. [Google Scholar]
- Gompper ME, Kays RW, Ray JC, et al. A comparison of noninvasive techniques to survey carnivore communities in northeastern North America. Wildlife Society Bulletin. 2006;34:1142–1151. [Google Scholar]
- Gottelli D, Sillero-Zubiri C, Applebaum GD, et al. Molecular genetics of the most endangered canid: the Ethiopian wolf Canis simensis. Molecular Ecology. 1994;3:301–312. doi: 10.1111/j.1365-294x.1994.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity. 2010;106:158–171. doi: 10.1038/hdy.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer F, Leonard J. Hybridization among three native North American Canis species in a region of natural sympatry. PLoS one. 2008;3:e3333. doi: 10.1371/journal.pone.0003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert ND, Derr JN. A comprehensive evaluation of cattle introgression into US federal bison herds. Journal of Heredity. 2007;98:1–12. doi: 10.1093/jhered/esl051. [DOI] [PubMed] [Google Scholar]
- Harrison DJ, Harrison JA. Foods of adult Maine coyotes and their known-aged pups. Journal of Wildlife Management. 1984;48:922–926. [Google Scholar]
- Hedrick PW, Lee RN, Garrigan D. Major histocompatibility complex variation in red wolves: evidence for common ancestry with coyotes and balancing selection. Molecular Ecology. 2002;11:1905–1913. doi: 10.1046/j.1365-294x.2002.01579.x. [DOI] [PubMed] [Google Scholar]
- Hindrikson M, Männil P, Ozolins J, Krzywinski A, Saarma U. Bucking the trend in wolf-dog hybridization: first evidence from Europe of hybridization between female dogs and male wolves. PLoS one. 2012;7:e46465. doi: 10.1371/journal.pone.0046465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kays R, Curtis A, Kirchman J. Rapid adaptive evolution of northeastern coyotes via hybridization with wolves. Biology Letters. 2010a;6:89–93. doi: 10.1098/rsbl.2009.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays R, Curtis A, Kirchman J. Reply to Wheeldon et al. ‘Colonization history and ancestry of northeastern coyotes’. Biology Letters. 2010b;6:248–249. doi: 10.1098/rsbl.2009.0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays R, Feranec RS. Using stable carbon isotopes to distinguish wild from captive wolves. Northeastern Naturalist. 2011;18:253–264. [Google Scholar]
- Kays RW, Gompper ME, Ray JC. Landscape ecology of eastern coyotes based on large-scale estimates of abundance. Ecological Applications. 2008;18:1014–1027. doi: 10.1890/07-0298.1. [DOI] [PubMed] [Google Scholar]
- Koblmüller S, Nord M, Wayne RK, Leonard JA. Origin and status of the Great Lakes wolf. Molecular Ecology. 2009;18:2313–2326. doi: 10.1111/j.1365-294X.2009.04176.x. [DOI] [PubMed] [Google Scholar]
- Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. Genomics and conservation genetics. Trends in Ecology & Evolution. 2006;21:629–637. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Kohn MH, Wayne RK. Facts from feces revisited. Trends in Ecology & Evolution. 1997;12:223–227. doi: 10.1016/s0169-5347(97)01050-1. [DOI] [PubMed] [Google Scholar]
- Kohn MH, York EC, Kamradt DA, et al. Estimating population size by genotyping faeces. Proceedings of the Royal Society of London Series B-Biological Sciences. 1999;266:657–663. doi: 10.1098/rspb.1999.0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenosky GB. Hybridization between wolf and coyote. Journal of Mammalogy. 1971;52:446–449. [PubMed] [Google Scholar]
- Kyle C, Johnson A, Patterson B, Wilson P, White B. The conspecific nature of eastern and red wolves: conservation and management implications. Conservation Genetics. 2008;9:699–701. [Google Scholar]
- Kyle CJ, Johnson AR, Patterson BR, et al. Genetic nature of eastern wolves: past, present and future. Conservation Genetics. 2006;7:273–287. [Google Scholar]
- Lawrence B, Bossert WH. The cranial evidence for hybridization in New England Canis. Breviora. 1969;330:1–13. [Google Scholar]
- Lehman N, Eisenhawer A, Hansen K, et al. Introgression of coyote mitochondrial DNA into sympatric North American gray wolf populations. Evolution. 1991;45:104–119. doi: 10.1111/j.1558-5646.1991.tb05270.x. [DOI] [PubMed] [Google Scholar]
- Leonard J, Wayne R. Native Great Lakes wolves were not restored. Biology Letters. 2008;4:95. doi: 10.1098/rsbl.2007.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg J, Björnerfeldt S, Saetre P, et al. Selection for tameness has changed brain gene expression in silver foxes. Current Biology. 2005;15:R915–R916. doi: 10.1016/j.cub.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major JT, Sherburne JA. Interspecific relationships of coyotes, bobcats, and red foxes in western Maine. Journal of Wildlife Management. 1987;51:606–616. [Google Scholar]
- Mengel RM. A study of dog-coyote hybrids and implications concerning hybridization in Canis. Journal of Mammalogy. 1971;52:316–336. [PubMed] [Google Scholar]
- Monzón J. PhD Thesis, Stony Brook University. Stony Brook; New York, USA: 2012. Rapid evolution of northeastern coyotes. [Google Scholar]
- Monzón J. First regional evaluation of nuclear genetic diversity and population structure in northeastern coyotes (Canis latrans) doi: 10.12688/f1000research.3567.1. (In review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, Luikart G, Wayne RK. SNPs in ecology, evolution and conservation. Trends in Ecology & Evolution. 2004;19:208–216. [Google Scholar]
- Oliveira R, Godinho R, Randi E, Alves PC. Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Philosophical Transactions of the Royal Society B-Biological Sciences. 2008;363:2953–2961. doi: 10.1098/rstb.2008.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G. Eastern coyote: the story of its success. Nimbus Publishing; Halifax, Canada: 1995. [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen W, Falush D. Documentation for STRUCTURE software: version 2.3. 2010. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. http://www.R-project.org. [Google Scholar]
- Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annual Review of Ecology and Systematics. 1996;27:83–109. [Google Scholar]
- Rondeau D, Conrad JM. Managing urban deer. American Journal of Agricultural Economics. 2003;85:266–281. [Google Scholar]
- Roy MS, Geffen E, Smith D, Ostrander EA, Wayne RK. Patterns of differentiation and hybridization in North American wolflike canids, revealed by analysis of microsatellite loci. Molecular Biology and Evolution. 1994;11:553–570. doi: 10.1093/oxfordjournals.molbev.a040137. [DOI] [PubMed] [Google Scholar]
- Rutledge L, Garroway C, Loveless K, Patterson B. Genetic differentiation of eastern wolves in Algonquin Park despite bridging gene flow between coyotes and grey wolves. Heredity. 2010;105:520–531. doi: 10.1038/hdy.2010.6. [DOI] [PubMed] [Google Scholar]
- Rutledge LY, Wilson PJ, Klütsch CFC, Patterson BR, White BN. Conservation genomics in perspective: a holistic approach to understanding Canis evolution in North America. Biological Conservation. 2012;155:186–192. [Google Scholar]
- Sacks B, Louie S. Using the dog genome to find single nucleotide polymorphisms in red foxes and other distantly related members of the Canidae. Molecular Ecology Resources. 2008;8:35–49. doi: 10.1111/j.1471-8286.2007.01830.x. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Brown SK, Ernest HB. Population structure of California coyotes corresponds to habitat-specific breaks and illuminates species history. Molecular Ecology. 2004;13:1265–1275. doi: 10.1111/j.1365-294X.2004.02110.x. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Mitchell BR, Williams CL, Ernest HB. Coyote movements and social structure along a cryptic population genetic subdivision. Molecular Ecology. 2005;14:1241–1249. doi: 10.1111/j.1365-294X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Moore M, Statham MJ, Wittmer HU. A restricted hybrid zone between native and introduced red fox (Vulpes vulpes) populations suggests reproductive barriers and competitive exclusion. Molecular Ecology. 2011;20:326–341. doi: 10.1111/j.1365-294X.2010.04943.x. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Våge DI, Statham MJ. A medium-throughput SNP assay for detecting genetic variation in coding and non-coding portions of the red fox genome. Conservation Genetics Resources. 2009;1:459–463. [Google Scholar]
- Sears HJ, Theberge JB, Theberge MT, Thornton I, Campbell GD. Landscape influence on Canis morphological and ecological variation in a Coyote-Wolf C. lupus × latrans hybrid zone, southeastern Ontario. Canadian Field-Naturalist. 2003;117:589–600. [Google Scholar]
- Seddon JM, Parker HG, Ostrander EA, Ellegren H. SNPs in ecological and conservation studies: a test in the Scandinavian wolf population. Molecular Ecology. 2005;14:503–511. doi: 10.1111/j.1365-294X.2005.02435.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends in Ecology & Evolution. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Takimoto G, Roy D, Jokela J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Molecular Ecology. 2008;17:30–44. doi: 10.1111/j.1365-294X.2007.03529.x. [DOI] [PubMed] [Google Scholar]
- Silver H, Silver WT. Growth and behavior of the coyote-like canid of northern New England with observations on canid hybrids. Wildlife Monographs. 1969;17:3–41. [Google Scholar]
- Talbot P, Thompson S, Schroeder W, Isabel N. An efficient single nucleotide polymorphism assay to diagnose the genomic identity of poplar species and hybrids on the Canadian prairies. Canadian Journal of Forest Research. 2011;41:1102–1111. [Google Scholar]
- US Fish and Wildlife Service [USFWS] Saratoga County, NY canid. Ashland, OR: 2004. [Google Scholar]
- USFWS. Orleans County, VT canid shooting: Agency case 06-000413. Ashland, OR: 2007. [Google Scholar]
- Vähä JP, Primmer CR. Efficiency of model based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Vilà C, Walker C, Sundqvist A, et al. Combined use of maternal, paternal and bi-parental genetic markers for the identification of wolf-dog hybrids. Heredity. 2003;90:17–24. doi: 10.1038/sj.hdy.6800175. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Earl DA, et al. Identification of recent hybridization between gray wolves and domesticated dogs by SNP genotyping. Mammalian Genome. 2013;24:80–88. doi: 10.1007/s00335-012-9432-0. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Earl DA, et al. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Research. 2011;21:1294–1305. doi: 10.1101/gr.116301.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Lohmueller KE, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JG, Rutledge L, Wheeldon T, White BN. Genetic characterization of eastern “coyotes” in eastern Massachusetts. Northeastern Naturalist. 2010;17:189–204. [Google Scholar]
- Wayne RK, Lehman N. Mitochondrial DNA analysis of the eastern coyote: origins and hybridization. In: Boer A, editor. Ecology and management of the eastern coyote. Wildlife Research Unit, University of New Brunswick; Fredericton: 1992. pp. 9–22. [Google Scholar]
- Wheeldon TJ, Rutledge LY, Patterson BR, White BN, Wilson PJ. Y-chromosome evidence supports asymmetric dog introgression into eastern coyotes. Ecology and Evolution. 2013;3:3005–3020. doi: 10.1002/ece3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeldon T, White B. Genetic analysis of historic western Great Lakes region wolf samples reveals early Canis lupus/lycaon hybridization. Biology Letters. 2009;5:101–104. doi: 10.1098/rsbl.2008.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis BL, van Oppen MJH, Miller DJ, Vollmer SV, Ayre DJ. The role of hybridization in the evolution of reef corals. Annual Review of Ecology, Evolution, and Systematics. 2006;37:489–517. [Google Scholar]
- Wilson PJ, Grewal SK, Mallory FF, White BN. Genetic characterization of hybrid wolves across Ontario. Journal of Heredity. 2009;100:S80–S89. [Google Scholar]
- Wilson PJ, Rutledge LY, Wheeldon TJ, Patterson BR, White BN. Y-chromosome evidence supports widespread signatures of three-species Canis hybridization in eastern North America. Ecology and Evolution. 2012;2:2325–2332. doi: 10.1002/ece3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Takebayashi N, Rieseberg L. Predicting the risk of extinction through hybridization. Conservation Biology. 2001;15:1039–1053. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.