Abstract
Exosomes are sub-100 nm extracellular vesicles secreted by normal and cancer cells. We present a high-resolution structure of previously unidentified nanofilaments on glioblastoma-derived exosomes, using nanoscale peak force imaging. These stiff, adhesive, trypsin- and RNAse-resistant surface nanofilaments add a new dimension to the current structural knowledge of exosome-mediated intercellular communication.
Keywords: nanoparticles, glioblastoma exosomes, nanofilaments, intercellular communication, nanotechnology
1. Introduction
The ability of cells to communicate with each other is essential for multicellular organisms. A number of mechanisms, including small nano-sized, cell-derived extracellular vesicles called exosomes [1], have been investigated as mediators of cellular interactions. In particular, the surfaces of exosomes form fascinating interfaces that may mediate their transport between cells, e.g. both as autocrine and paracrine factors in tumour microenvironments, as well as effectors for distant cellular targets, analogous to endocrine signalling. Exosomes are known to play an important role in intercellular communication by extracellular signalling [2] and horizontal mRNA/microRNA transfer [3]. Their roles in tumour proliferation [4,5], pre-metastatic niches [6] and tumour microenvironment modulation, including glioblastoma as a model disease system, have also been proposed [7].
The quantity, surface molecular composition and dynamics of exosomes released vary depending on the cellular origin and physiological state of the cell [8]. Understanding exosome structure at nanometre resolution can provide us detailed insights into the mechanisms of exosome-mediated cell–surface interactions including transport, binding to and uptake by target cells. Electron microscopy has been the gold-standard method [9] to characterize exosomes, but is limited by potential artefacts [10]. No structural details can be resolved with optical imaging as exosomes are smaller than the diffraction limit (200–300 nm) [11]. Recently, atomic force microscopy (AFM) [12] has emerged as a powerful biophysical nanoscale characterization technique that offers three-dimensional imaging of unlabelled single exosomes, and provides additional information about the structure, mechanical properties and their specific biomolecular composition [13,14]. So far, detailed surface characteristics of glioblastoma exosomes at the nanoscale level have not been reported to date.
Here, we provide first evidence of the presence of surface nanofilaments on glioblastoma-derived exosomes and analyse their biophysical characteristics at the nanoscale using AFM peak force imaging [15]. Compared with normal exosomes, glioblastoma exosomes displayed abundant nanofilaments, and the nanofilaments were trypsin- and RNase-resistant. We quantified the distinct structural, biophysical and biochemical properties of the exosomes and surface nanofilaments. Based on in vitro uptake assays, glioblastoma exosomes showed a significantly higher uptake in cells compared with normal exosomes. We discuss how these newly recognized nanofilaments may help in modulating, tethering and transport of exosomes for intercellular communication.
2. Results and discussion
2.1. Glioblastoma cell-line-derived exosomes show abundant networks of surface nanofilaments
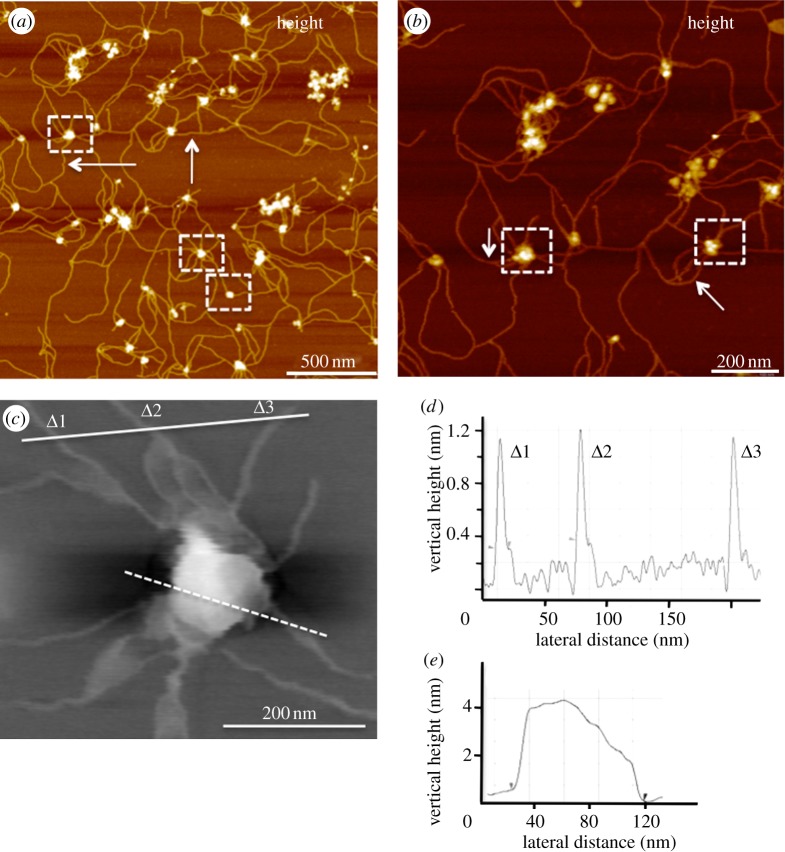
Exosomes derived from several human cell lines were studied, including glioblastoma (U87 and U251), melanoma (SKMEL) and normal human astrocytes (NHA). U87 and U251 are human primary glioblastoma cell lines from grade IV tumours [16]. SKMEL28 is a malignant melanoma cell line. Normal control exosomes were isolated from a primary human astrocyte cell line (NHA). Key structural features not previously reported for exosomes were observed at nanoscale resolution, using the recently developed variation of AFM called peak force microscopy (PFM) [15]. This imaging mode is non-destructive to both tip and sample because it directly controls the peak normal force. PFM operates at low imaging forces (less than 100 pN), permitting subnanometre imaging of soft biological structures (see the electronic supplementary material). The most striking feature of the recorded PF images was the ultra-structural details of exosome surfaces (figure 1). Although ‘microscale’ cellular extensions [17] and nanotubes are known to be exist between cells [18], their functional mechanisms in intercellular communication remain obscure. However, PFM images reveal new and distinct nanofilaments extending from the surface of glioblastoma exosomes (figure 1). Our current work shows that the surface extensions of glioblastoma exosomes form a branched network of nanoscale inter-exosomal connections (figure 1a,b, marked by white arrows). Individual nanofilaments within the network were measured to be 10–20 nm in diameter and up to several micrometres in length (figure 1c,d). Similar, though fewer, nanofilaments were also observed in U251 exosomes. Interestingly, the nanofilaments were rarely observed in the case of normal control NHA exosomes (see electronic supplementary material, figure S1). Sequential ultracentrifugation, both with and without sucrose gradient purification, yielded exosomes with indistinguishable nanofilaments under PFM. Additionally, we found that trypsin–EDTA treatment did not disrupt the network of nanofilaments or exosomes (data not shown), indicating either their non-protein nature or inherent resistance to trypsin digestion.
Figure 1.
PFM reveals glioblastoma exosomes with abundant nanofilamentous surface extensions. (a) Topographic image (z = 10 nm) of glioblastoma U87-derived exosomes showing round bulging vesicles (some shown in dashed boxes) surrounded by a network of nanofilaments. (b) The nanofilaments are shown at a higher resolution (z = 6 nm). Size of U87 exosomes measured from typical high-resolution PFM topographic image (c) is 89.3 nm in diameter and 4 nm in height (e; dashed white line). (d) Cross-section profiles show approximately 1.2 nm in height and 10 nm wide nanofilaments (marked Δ1–3, solid line). Fewer nanofilaments were seen in U251 exosomes, but not seen in NHA-derived exosomes (see the electronic supplementary material, figure S1). The results were confirmed by imaging samples obtained from two independent and commonly used isolations, with and without sucrose gradient purification. Size distributions show an average vesicle size of 89 ± 3.2 nm, 80.8 ± 2.2 nm and 70.9 ± 2.2 nm for U87, U251 and NHA, respectively (n ∼ 100 individual exosomes).
Exosome morphology was measured, using PF topographic images. The findings revealed three-dimensional spherical vesicles, approximately 80–90 nm in diameter for all three cancer cell lines (U87, U251 and SKMEL) and slightly smaller NHA exosomes (see electronic supplementary material, table S1). PFM images provide support to our previous findings showing exosomes as round spherical structures without cup-shaped indentations [19]. As might be expected for the low force of imaging in PFM, the vesicles were measured to be higher (approx. 4 nm) compared with those measured under standard tapping mode imaging owing to minimal compression of the vesicle lipid bilayers [19].
2.2. Mapping of biophysical characteristics (Young's modulus and adhesion strength) of nanofilaments
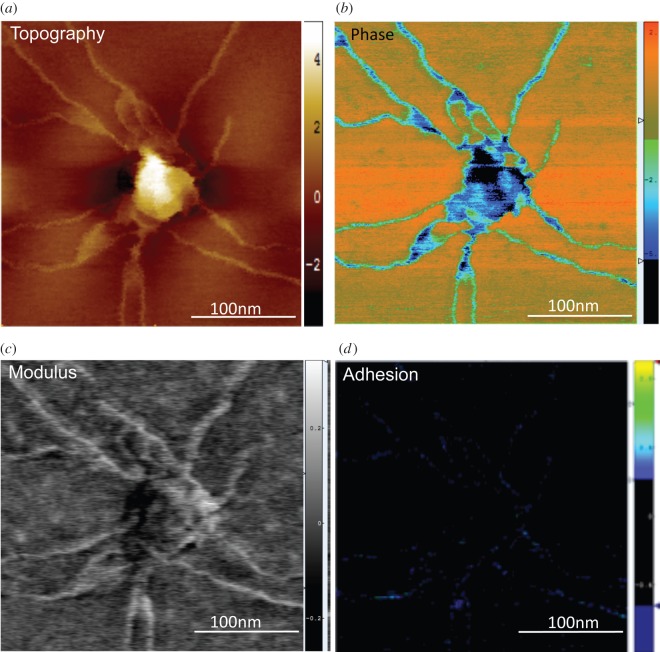
Next, we explored the biophysical properties of glioblastoma exosomes, including their surface nanofilaments. AFM tapping mode height and phase imaging has been widely used to study the structure and morphology of biomolecules [20–22]. Additionally, quantitative mechanical information on surface modulus can be obtained by nanoindentation and force volume AFM imaging [23,24], using appropriate mechanical contact models. However, nanoindentation and force volume imaging are limited in lateral resolution and time extensive. Using PFM, less than 5 nm lateral-resolution maps were typically obtained to determine quantitative information about the topography, change in phase, stiffness (elastic modulus) and adhesion strength (stickiness) of the vesicles (see the electronic supplementary material) as represented in figure 2.
Figure 2.
PFM imaging enabled independent extraction and mapping of biophysical parameters at less than 5 nm lateral resolution, revealing relatively stiffer and more adhesive nanofilaments, in comparison with parent vesicle region. Quantitative mapping of biophysical parameters such as (a) topography (range z = 5 nm), (b) phase (z = 10.6 mV), (c) modulus (z = 2 GPa) and (d) adhesion (z = 1.6 nN) was simultaneously mapped, as represented by U87 exosomes with extending surface nanofilaments.
Figure 2a shows a nanoscale topographic image of a single isolated exosome and extending surface nanofilaments at nanoscale resolution. Complementary to the three-dimensional topography, the phase images can detect inhomogeneous chemical, structural or mechanical properties [25] of the sample. Here, local phase heterogeneities in exosomes (figure 2b) indicated a dense exosomal core relative to surface filaments. Furthermore, the prominent differences in nanofilaments and vesicle regions were revealed in simultaneously recorded elastic modulus and adhesion maps (figure 2c,d). Figure 2c,d corresponds to variations in the elastic modulus and adhesion forces, respectively, both showing significant mechanical heterogeneity between the exosomes and nanofilaments. In PFM, Young's modulus is calculated using a Derjaguin–Muller–Toporov model that is applied to the unloading portion of the force–separation curve, which takes into account the adhesive forces between the tip and the surface [15]. The elastic modulus map obtained by PFM imaging (figure 2c) revealed stiffer (bright) nanofilaments (up to 2 GPa) compared with the exosomal core regions (dark). While surface effects can be expected to affect the obtained modulus values, the contrast between the modulus was also clearly distinct in regions where the stiffer filaments are observed on top of the vesicle regions. Additionally, we performed single force–indentation curve testing over various regions using sharper tips (approx. 1 nm) and thin-film Hertz model [26], which also indicated stiffer nanofilaments. The measured stiffness of exosome nanofilaments was comparable in range with other filamentous biomolecules (see the electronic supplementary material, table S2) including peak force measurements to identify stiffness profiles of nanoscale fibrils such as Tau and alpha-synuclein. Additionally, the non-specific adhesion of the exosomes to bare AFM probes was quantified across the topography. The nanofilaments were found to be significantly more adhesive (p < 0.05) than vesicular regions (figure 2d) with adhesion forces up to 1.6 nN (blue).
2.3. Glioblastoma exosomes show greater cellular uptake compared with normal control exosomes
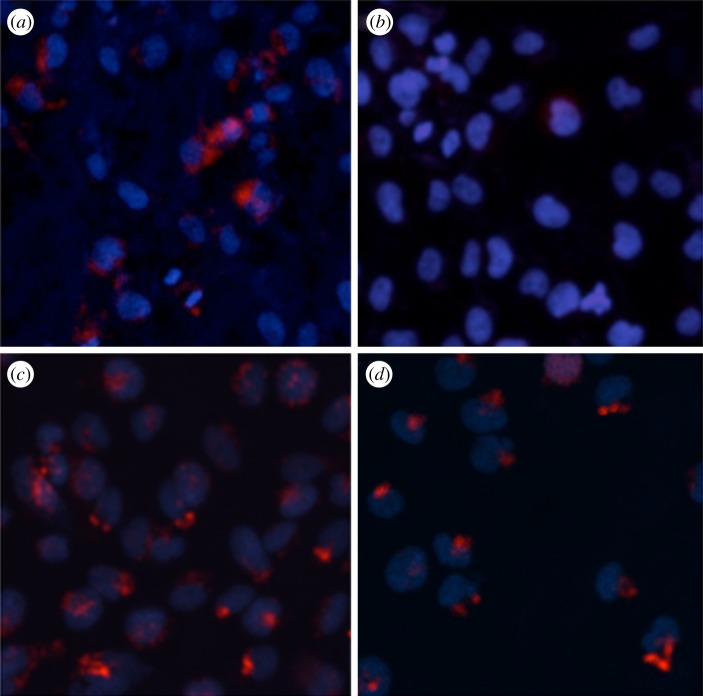
One intriguing question is the functional consequence of exosome nanofilaments in patho-physiological states such as glioblastoma. Our structural data highlight that nanofilaments appear on the surface of glioblastoma exosomes. Consequently, we investigated their potential strength in binding to target cells by assaying the cellular binding and uptake of glioblastoma-derived exosomes compared with normal exosomes. To achieve this, we incubated purified exosomes derived from U251 and NHA cells (labelled with membrane dye CM-DiI) with U251 or NHA cells, respectively (figure 3). We found that in vitro cellular uptake of glioblastoma (U251) exosomes was significantly higher than that of normal control NHA exosomes. Although the exact mechanism of increased uptake of glioblastoma exosomes, which express many fold more surface nanofilaments (with respect to NHA exosomes), still needs to be identified, these preliminary results support our hypothesis that increasing numbers of nanofilaments could play a functional role in cellular binding and uptake of exosomes and may be relevant in intercellular interactions in glioblastoma exosomes.
Figure 3.
In vitro cell assays show greater binding of glioblastoma-derived U251 exosomes compared with normal NHA exosomes (images taken at 200× magnification). (a) NHA cells+ U251 exosomes, (b) NHA cells+ NHA exosomes, (c) U251 cells+ U251 exosomes and (d) U251 cells+ NHA exosomes. Cell nuclei are stained with 4′,6-diamidino-2-phenylindole (blue) and exosomes are stained with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (red).
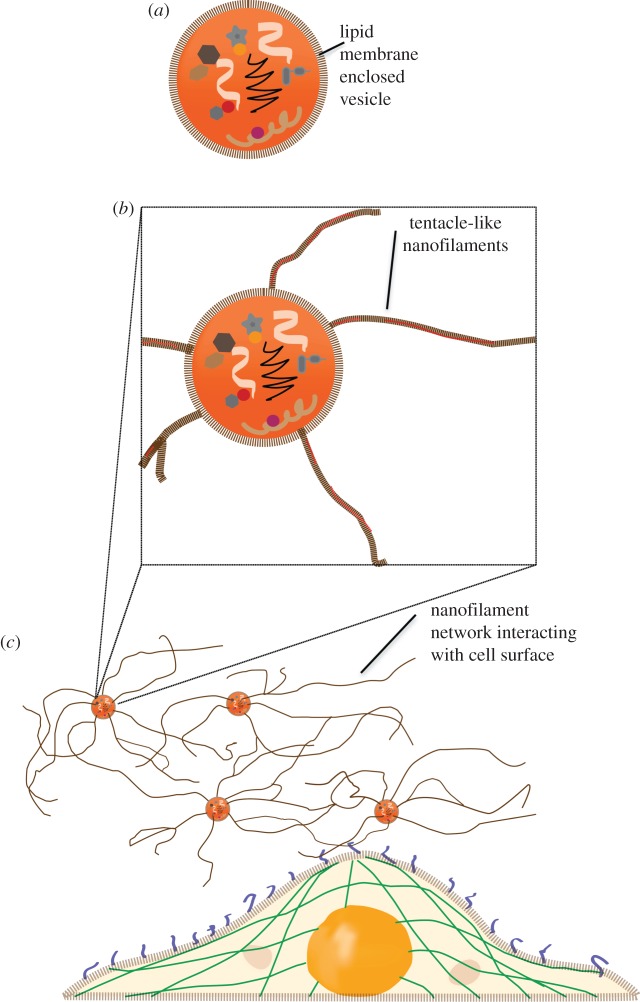
Filamentous connections between cells that allow intercellular communications are typical of multicellular organisms. Recent reports demonstrate the existence of a network of intercellular membrane nanotubes enabling long-distance communication in mammalian cells [17,18]. These tunnelling nanotubes have been shown to facilitate intercellular transfer of cytoplasmic molecules and even organelles and viruses [27,28]. Using correlative field emission scanning electron microscopy (FESEM) and AFM, we previously demonstrated the structure of saliva-derived exosomes and oral cancer exosomes [19,29]. Evidence of associated inter-vesicular structures was observed under FESEM and AFM, but not resolved in detail. Our current findings on structural, nanomechanical and cellular uptake of exosomes provide the first high-resolution nanoscale characteristics of exosome nanofilaments. The adhesive characteristics and cell binding assays suggest their likely role in a direct physical contact with target cells for intercellular communication (figure 4b,c). The nature and function of these observed nanofilaments need to be further investigated, but could well provide a significant advance over existing models of exosome–cell interactions, which are largely based on exosomes being ‘distinct membrane-enclosed vesicles’ (figure 4a) capable of intercellular communication and transport of proteomic and genomic content among distant cells [30].
Figure 4.
Proposed hypothetical schematic model for exosomes with nanofilaments mediating intercellular transport and communication. (a) Current model of exosomes with membrane-enclosed proteomic and genomic content. (b) Glioblastoma exosomes with surface nanofilaments. (c) Nanofilaments of glioblastoma exosomes interact with target cell.
We found a positive correlation between abundance of nanofilament extensions and glioblastoma cell-line-derived exosomes. These nanofilaments were measured to be 10–20 nm wide, compared with the exosome vesicular diameters of around 100 nm. It is known that tip convolution likely results in broadening effect on measured exosome dimensions, yet the images provide good estimates of upper limits of exosome diameters and nanofilament heights and widths. The lengths of nanofilaments extend up to several micrometres, comparable with dimensions of target cells themselves. We tentatively suggest that these structures are composed of lipids and nucleic acids (based on filamentous shape, close to 1.2 nm height in AFM images and similar elastic modulus values ranging from MPa to a few GPa). It is noteworthy that the general physical characteristics and appearance of exosomes, their nucleic acid contents (noted to contain microRNA, mRNA and even potential retro-transposable elements) [3,31], and even their subcellular biological underpinnings suggest their strong relationship to viruses [32]. Within this context, we suggest that the biophysical and potential functional characteristics of these nanofilaments may be analogous to virulence factors for microbes, such as gp120/gp41 for human immunodeficiency virus [33], or haemagglutinin and neuraminidase for influenza virus infection [34,35]. Further exploration of nanofilaments may not only hold significance for exosome biology and their role in intercellular communication, but also for their role as virulence factors in exosome-associated diseases such as cancer, autoimmune diseases and infectious diseases. Further experimental investigations are needed to establish the occurrence and relevance of nanofilaments in a diverse population of exosomes imaged under physiological buffer conditions.
Nanofilaments may serve as long tethers for anchorage, increasing the probability of exosome binding to target cells. They may provide enhanced rigidity or structural integrity to parent exosomes. These rigid and adhesive nanofilaments may also harbour specific recognition motifs for cognate receptors on target cells, perhaps thereby further modulating exosome targeting, uptake and effector functions. Therefore, we propose a tentative model (figure 4) of nanofilament augmented exosome interactions with target cells.
3. Conclusion
Here, we show quantitative differences between the structure and biophysical properties of cancer cell-line-derived and normal control cell-line-derived exosomes, imaged in vitro using PFM. Our results demonstrate that in addition to commonly accepted exosome models of membrane-enclosed vesicles carrying proteomic and genomic cargo, glioblastoma- and malignant melanoma-derived exosomes (and likely other malignant-cell-derived exosomes) also possess surface nanofilaments that have not been identified previously. The PFM method used in these studies allowed the necessary resolution for revealing nanofilaments on exosome surfaces. To the best of our knowledge, this is the first report showing nanofilaments extending from the surface of exosomes, and demonstrates direct physical connections between exosomes. We believe that exosomes with their long tentacle-like nanofilaments may be a fascinating new route for cell–cell communication that warrants further investigations.
Acknowledgements
Support from MEXT WPI Programme: International Center for Materials Nanoarchitectonics (MANA), Japan and CNSI is acknowledged. We acknowledge the use of AFMs at the Nano and Pico Characterization Laboratory at CNSI.
References
- 1.Théry C. 2011. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15 ( 10.3410/B3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calzolari A, et al. 2006. TfR2 localizes in lipid raft domains and is released in exosomes to activate signal transduction along the MAPK pathway. J. Cell Sci. 119, 4486–4498. ( 10.1242/jcs.03228) [DOI] [PubMed] [Google Scholar]
- 3.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. ( 10.1038/ncb1596) [DOI] [PubMed] [Google Scholar]
- 4.Taylor DD, Gercel-Taylor C. 2008. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. ( 10.1016/j.ygyno.2008.04.033) [DOI] [PubMed] [Google Scholar]
- 5.Skog J, et al. 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. ( 10.1038/ncb1800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung T, Castellana D, Klingbeil P, Cuesta Hernández I, Vitacolonna M, Orlicky DJ, Roffler SR, Brodt P, Zöller M. 2009. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 11, 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peinado H, et al. 2012. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891. ( 10.1038/nm.2753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Théry C, Ostrowski M, Segura E. 2009. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593. ( 10.1038/nri2567) [DOI] [PubMed] [Google Scholar]
- 9.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. 1985. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 101, 942–948. ( 10.1083/jcb.101.3.942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raposo G, Stoorvogel W. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. ( 10.1083/jcb.201211138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller G. 2012. Novel tools for the study of cell type-specific exosomes and microvesicles. J. Bioanal. Biomed. 4, 046–060. ( 10.4172/1948-593X.1000063) [DOI] [Google Scholar]
- 12.Binnig G, Quate CF, Gerber C. 1986. Atomic force microscope. Phys. Rev. Lett. 56, 930–933. ( 10.1103/PhysRevLett.56.930) [DOI] [PubMed] [Google Scholar]
- 13.Sharma S, Gimzewski JK. 2012. The quest for characterizing exosomes: circulating nano-sized vesicles. J. Nanomed. Nanotechnol. 3, e115 ( 10.4172/2157-7439.1000e115) [DOI] [Google Scholar]
- 14.Müller DJ, Dufrêne YF. 2011. Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 8, 461–469. ( 10.1016/j.tcb.2011.04.008) [DOI] [PubMed] [Google Scholar]
- 15.Adamcik J, Berquand A, Mezzenga R. 2011. Single-step direct measurement of amyloid fibrils stiffness by peak force quantitative nanomechanical atomic force microscopy. Appl. Phys. Lett. 98, 193701 ( 10.1063/1.3589369) [DOI] [Google Scholar]
- 16.Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF. 2010. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 6, e1000832 ( 10.1371/journal.pgen.1000832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. 2004. Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010. ( 10.1126/Science.1093133) [DOI] [PubMed] [Google Scholar]
- 18.Gerdes HH, Bukoreshtliev NV, Barroso JF. 2007. Tunneling nanotubes: a new route for the exchange of components between animal cells. FEBS Lett. 581, 2194–2201. ( 10.1016/j.febslet.2007.03.071) [DOI] [PubMed] [Google Scholar]
- 19.Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. 2010. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 4, 1921–1926. ( 10.1021/nn901824n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma S, Zhu H, Grintsevich EE, Reisler E, Gimzewski JK. 2013. Correlative nanoscale imaging of actin filaments and their complexes. Nanoscale 5, 5692–5702. ( 10.1039/c3nr01039b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lal R, John SA. 1994. Biological applications of atomic force microscopy. Am. J. Physiol. 266, C1–C21. [DOI] [PubMed] [Google Scholar]
- 22.Brown BP, Picco L, Miles MJ, Faul CF. 2013. Opportunities in high-speed atomic force microscopy. Small 9, 3201–3211. ( 10.1002/smll.201203223) [DOI] [PubMed] [Google Scholar]
- 23.A-Hassan E, Heinz WF, Antonik MD, D'Costa NP, Nageswaran S, Schoenenberger CA, Hoh JH. 1998. Relative microelastic mapping of living cells by atomic force microscopy. Biophys. J. 74, 1564–1578. ( 10.1016/S0006-3495(98)77868-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dufrêne YF, Pelling AE. 2013. Force nanoscopy of cell mechanics and cell adhesion. Nanoscale 5, 4094–4104. ( 10.1039/c3nr00340j) [DOI] [PubMed] [Google Scholar]
- 25.García R, Magerle R, Perez R. 2007. Nanoscale compositional mapping with gentle forces . Nat. Mater. 6, 405–411. ( 10.1038/Nmat1925) [DOI] [PubMed] [Google Scholar]
- 26.Chadwick RS. 2002. Axisymmetric indentation of a thin incompressible elastic layer. SIAM J. Appl. Math. 62, 1520–1530. ( 10.1137/S0036139901388222) [DOI] [Google Scholar]
- 27.Schara K, Jansa V, Sustar V, Dolinar D, Pavlic JI, Lokar M, Kralj-Iglic V, Veranic P, Iglic A. 2009. Mechanisms for the formation of membranous nanostructures in cell-to-cell communication. Cell Mol. Biol. Lett. 14, 636–656. ( 10.2478/s11658-009-0018-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belting M, Wittrup A. 2008. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease . J. Cell Biol. 183, 1187–1191. ( 10.1083/jcb.200810038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma S, Gillespie BM, Palanisamy V, Gimzewski JK. 2011. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir 27, 14 394–14 400. ( 10.1021/la2038763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. 2006. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856. ( 10.1038/Sj.Leu.2404132) [DOI] [PubMed] [Google Scholar]
- 31.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. 2010. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA 107, 6328–6333. ( 10.1073/pnas.0914843107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gould SJ, Booth AM, Hildreth JE. 2013. The Trojan exosome hypothesis. Proc. Natl Acad. Sci. USA 100, 10 592–10 597. ( 10.1073/pnas.1831413100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierson TC, Doms RW. 2003. HIV-1 entry and its inhibition. Curr. Top. Microbiol. Immunol. 281, 1–27. ( 10.1007/978-3-642-19012-4_1) [DOI] [PubMed] [Google Scholar]
- 34.Gerlach T, Kühling L, Uhlendorff J, Laukemper V, Matrosovich T, Czudai-Matwich V, Schwalm F, Klenk HD, Matrosovich M. 2012. Characterization of the neuraminidase of the H1N1/09 pandemic influenza virus. Vaccine 30, 7348–7352. ( 10.1016/j.vaccine.2012.09.078) [DOI] [PubMed] [Google Scholar]
- 35.Kang SM, Song JM, Compans RW. 2011. Novel vaccines against influenza viruses. Virus Res. 162, 31–38. ( 10.1016/j.virusres.2011.09.037) [DOI] [PMC free article] [PubMed] [Google Scholar]