Abstract
Previous studies of the effect of ART on gene expression in HIV-infected individuals have identified small numbers of modulated genes. Since these studies were underpowered or cross-sectional in design, a paired analysis of peripheral blood mononuclear cells (PBMCs), isolated before and after ART, from a robust number of HIV-infected patients (N=32) was performed. Gene expression was assayed by microarray and 4,157 differentially expressed genes (DEGs) were identified following ART using multivariate permutation tests. Pathways and Gene Ontology (GO) terms over-represented for DEGs reflected the transition from a period of active virus replication before ART to one of viral suppression (e.g., repression of JAK-STAT signaling) and possible prolonged drug exposure (e.g. oxidative phosphorylation pathway) following ART. CMYC was the DEG whose product made the greatest number of interactions at the protein level in protein interaction networks (PINs), which has implications for the increased incidence of Hodgkin’s lymphoma (HL) in HIV-infected patients. The differential expression of multiple genes was confirmed by RT-qPCR including well-known drug metabolism genes (e.g., ALOX12 and CYP2S1). Targets not confirmed by RT-qPCR (i.e., GSTM2 and RPL5) were significantly confirmed by droplet digital (ddPCR), which may represent a superior method when confirming DEGs with low fold changes. In conclusion, a paired design revealed that the number of genes modulated following ART was an order of magnitude higher than previously recognized.
Keywords: Antiretroviral therapy, droplet digital PCR, gene expression, HIV, microarray, paired analysis
1. Introduction
Antiretroviral therapy (ART) dramatically reduces the mortality and morbidity of HIV infection (Arts and Hazuda, 2012; Vittinghoff et al., 1999) and commonly refers to the use of multiple nucleoside reverse transcriptase inhibitors (NRTIs) in combination with a protease inhibitor (PI) or a non-nucleoside reverse transcriptase inhibitor (NNRTI). ART induces a sustained effective suppression of viral replication (<50 copies/mL) and optimally leads to an increase in CD4+ T-cell counts. In contrast to the obvious benefits of ART, long-term exposures to this therapy may lead to drug-class specific toxicities. Liver toxicity has been associated with NNRTI treatment (Rivero et al., 2007) and lipodistrophy with PI exposure (Carr et al., 1998), whereas a large number of adverse effects have been attributed to NRTI use: myopathy, pancreatitis, neuropathy, lipodistrophy, bone density loss, hepatic steatosis and lactic acidosis (Grigsby et al., 2010; Montessori et al., 2004; Pacenti et al., 2006). NRTI toxicity is associated with the inhibition of DNA polymerase gamma (Polg), which is responsible for the replication and repair of mitochondrial DNA (mtDNA), and also with the induction of oxidative stress, which further effects mtDNA replication, as well as overall energy production (Desai et al., 2008; Lewis et al., 2003). A review of in vitro studies suggests the following ranking with respect to NRTI effects on mitochondrial Polg: zalcitabine (ddC) ≥ didanosine (ddI) ≥ stavudine (d4T) > lamivudine (3TC) > zidovudine (AZT) > abacavir (ABC) (Kakuda, 2000). Finally, tenofovir (TDF) is a commonly used NRTI thought to have limited toxicity (Venhoff et al., 2007), although long term exposure in mice may be associated with mitochondrial toxicity in the renal proximal tubules (Kohler et al., 2009).
Relatively few studies have examined gene expression in samples from HIV-infected individuals treated with ART using a platform capable of assessing the whole transcriptome (Borjabad et al., 2011; Li et al., 2004; Rotger et al., 2010; Vigneault et al., 2011). Only Li et al. (2004) used a paired design in order to analyze gene expression in lymph node biopsies from 4 HIV-infected individuals before and after 28 days of ART. Paired statistical tests were not used to identify differentially expressed genes (DEGs) between time points but fold changes for genes were calculated for each individual (post-ART/pre-ART) and revealed that 162 genes were consistently differentially expressed across subjects. Although paired samples existed in the study of Rotger et al. (2010), a cross-sectional analysis was performed to identify 204 DEGs between CD4+ T cells isolated from ART-treated (N=67) and untreated (N=52) HIV-infected individuals (personal communication). Therefore, these studies suggest that ART has a limited effect on gene expression in vivo. In order to confirm this finding, gene expression was assayed by microarray in a paired design using PBMCs taken before and after ART from a robust number of HIV-infected subjects (N=32). Remarkably, the current study demonstrated that a much larger number of genes (N = 4,157) than previously known were modulated following ART, which revealed novel findings with respect to the metabolism and toxicities associated with this treatment.
2. Materials and Methods
2.1. Participants
Thirty-six HIV-infected men in the San Diego Primary Infection Cohort (SD PIC) were initially selected retrospectively for this study based on the following criteria: (1) ART-naive before enrollment, (2) continuously adhered to ART, (3) achieved viral suppression, (4) had viably stored PBMC samples both before and 48 weeks after ART. Viral suppression was defined as achieving <50 HIV RNA copies/ml within 48 weeks from the start of ART without evidence of subsequent viral rebound (i.e., no two concurrent measurements of >500 HIV RNA copies/ml). Complete demographic and clinical information for these subjects is presented in Supplementary Table 1.
2.2. Microarray Data Analysis
Microarray data were generated as previously described (Woelk et al., 2010). Briefly, RNA was isolated rom PBMC samples using RNeasy Mini Kits (QIAGEN, Germantown, Maryland, USA) and quality was assessed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA). All samples were deemed of sufficient quality (mean RNA integrity number 8.11 0.91) for hybridization to HumanWG-6 v3 Expression BeadChips (Illumina, San Diego, California, USA). Raw gene expression data were log2 transformed and robust spline normalized using the Bioconductor package lumi (Du et al., 2008) in R (version 2.8.0). The quality of microarray data was examined using plots of inter quartile range (IQR) versus median expression for each array. Four outlier samples were identified and paired array data for subjects 13, 14, 15 and 16, who were all on a PI-based regimen, were removed from analysis to leave a total of 32 subjects (Supplementary Table 1). Unsupervised clustering identified batch effects based on the date of microarray hybridization and these were removed using ComBat (Johnson et al., 2007). Multivariate permutation tests implemented in BRB-Array Tools (Simon et al., 2009) were used in a paired approach to identify genes differentially expressed such that there was 95% confidence of no more than 1% false positives. Gene expression data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE44228.
2.3. Pathway, Gene Ontology and Protein Interaction Network Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, gene ontology terms and protein interaction networks (PINs) associated with DEGs were identified as previously described (Perez-Santiago et al., 2012). Pathway analysis was performed using functional class scoring implemented in BRB-Array Tools (Simon et al., 2009), which analyzes the entire expression data set instead of a list of DEGs and generates a p-value for each KEGG pathway (Kanehisa et al., 2010) as opposed to a p-value for each gene (Datson et al., 2010). The Biological Networks Gene Ontology (BiNGO) tool (Maere et al., 2005) was used to perform gene ontology (GO) analysis and uses a hypergeometric test to identify those GO terms that are significantly overrepresented in the list of DEGs. The protein-protein and protein-DNA interactions of the protein products of DEGs were visualized using MetaCore™ (GeneGo, St. Joseph, MI, USA). These networks reveal hubs, which often represent transcription factors that control the regulation of multiple target genes. In all analyses, the false discovery rate (FDR) associated with multiple testing was corrected using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995) and an FDR-corrected p-value <0.05 was considered significant. In a conservative approach for pathway analysis, a KEGG pathway was only identified as being significant with an FDR-corrected p-value <0.05 for both the Fisher (LS) statistic and the Kolmogorov-Smirnov (KS) test.
2.4. RT-qPCR and ddPCR Analysis
RT-qPCR validation of the gene expression detected by microarray analysis using TaqMan Gene Expression Assays (Life 675 Technologies, Carlsbad, CA) was performed as previously described (Perez-Santiago et al., 2012; Woelk et al., 2012) for 16 targets (Supplementary Table 2). Briefly, cDNA was reverse transcribed from paired RNA samples for 12 randomly selected subjects for each target from a total of 17 subjects (Supplementary Table 1). RT-qPCR was performed with the 7900HT Fast Real-Time PCR System (Life Technologies) using 50 ng of cDNA in a 20 L reaction volume for each target in duplicate. The relative quantitation (RQ) value for each gene in each sample was calculated after assessing the expression of a normalizer (GAPDH, Assay ID: Hs00192713_m1) using the 2−ΔΔCT method. RQ values were log2 transformed and averaged across biological replicates within each time point to calculate fold change differences before and after ART for comparison to microarray data and for significance testing using paired t-tests.
TaqMan Gene Expression Assays were also used to assess differential gene expression by ddPCR using the QX100 system (Bio-Rad, Hercules, CA) for GSTM2, RPL5 and STAT1. ddPCR is thought to be more sensitive than RT-qPCR since target RNA is distributed across thousands of oil emulsion droplets which each undergo reverse transcription and a subsequent end-point PCR reaction. Since some droplets contain no template while others contain one or more copies, the number of target RNA molecules present was calculated from the fraction of positive end-point reactions using Poisson statistics (Hindson et al., 2012). Five nanograms of RNA in a 20 L PCR reaction volume were used for each target in duplicate. The copy numbers for each target were averaged across duplicates and normalized to GAPDH prior to fold change calculations and significance testing using paired t-tests.
3. Results
3.1. Numerous Genes are Differentially Expressed following ART
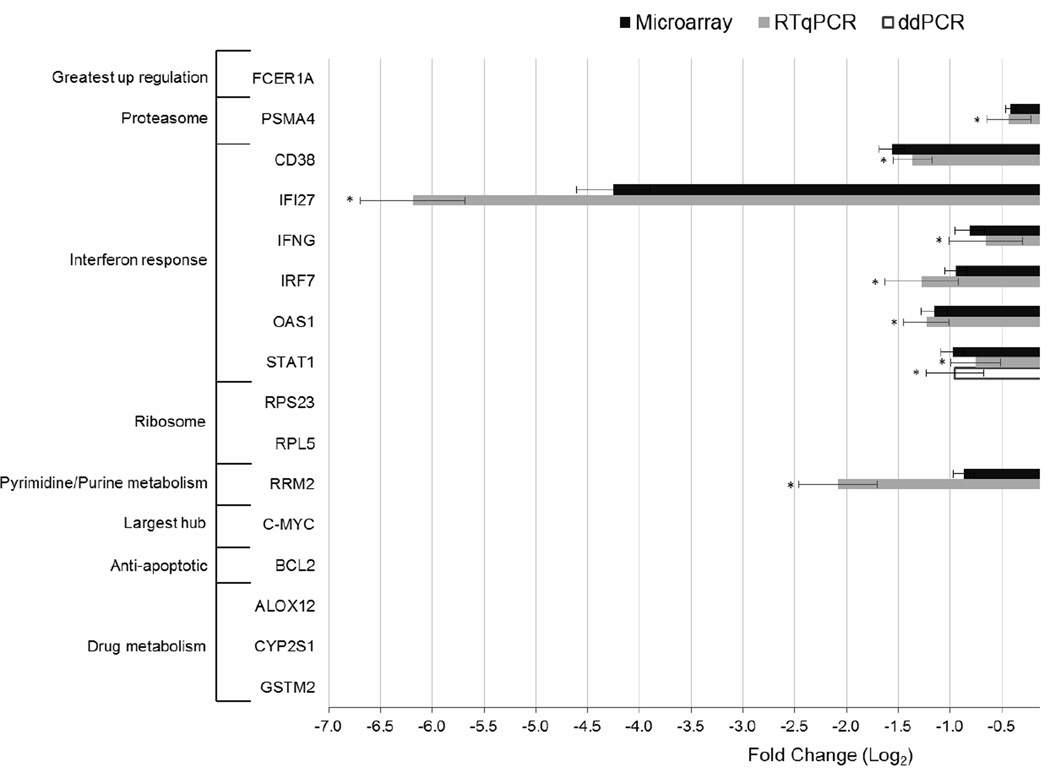
Gene expression was compared in a paired analysis using samples taken before and 48 weeks after ART from 32 HIV-infected individuals (Supplemental Table 1). Multivariate permutation tests identified 4,157 DEGs with 95% confidence that no more than 1% were false positives (Supplemental Table 3). The gene exhibiting the greatest upregulation following ART was the alpha subunit of the immunoglobulin epsilon receptor (FCER1A, fold change of 2.01) and the gene with the greatest downregulation was interferon alpha inducible protein 27 (IFI27, fold change of −19.61). The differential expression of both of these genes was confirmed by RT-qPCR (Figure 1).
Figure 1.
Gene expression analysis of 17 targets by RT-qPCR and a subset of 3 targets by ddPCR in 12 of the patients used for microarray analysis. Fold changes were calculated by dividing expression levels after 48 weeks of ART by those at baseline before ART initiation for microarray (black bars), RT-qPCR (grey bars) and ddPCR (white bars). An asterisk indicates a significant difference in gene expression (p < 0.05) between baseline and 48 weeks of ART as determined using a paired t-test. Error bars depict the standard error of the mean across the 12 samples analyzed and were calculated for fold changes in the RT-qPCR data using the error propagation formula for ratios. For microarray and ddPCR data, error bars were calculated by first converting all values to their natural logarithms and computing the standard error of the mean of the log transformed value such that positive error bars equate to fold change * expSEM-1 and negative error bars equate to fold change * expSEM−1.
3.2. Pathway and Gene Ontology Analysis Reflect the Repression of HIV Replication and Exposure to ART
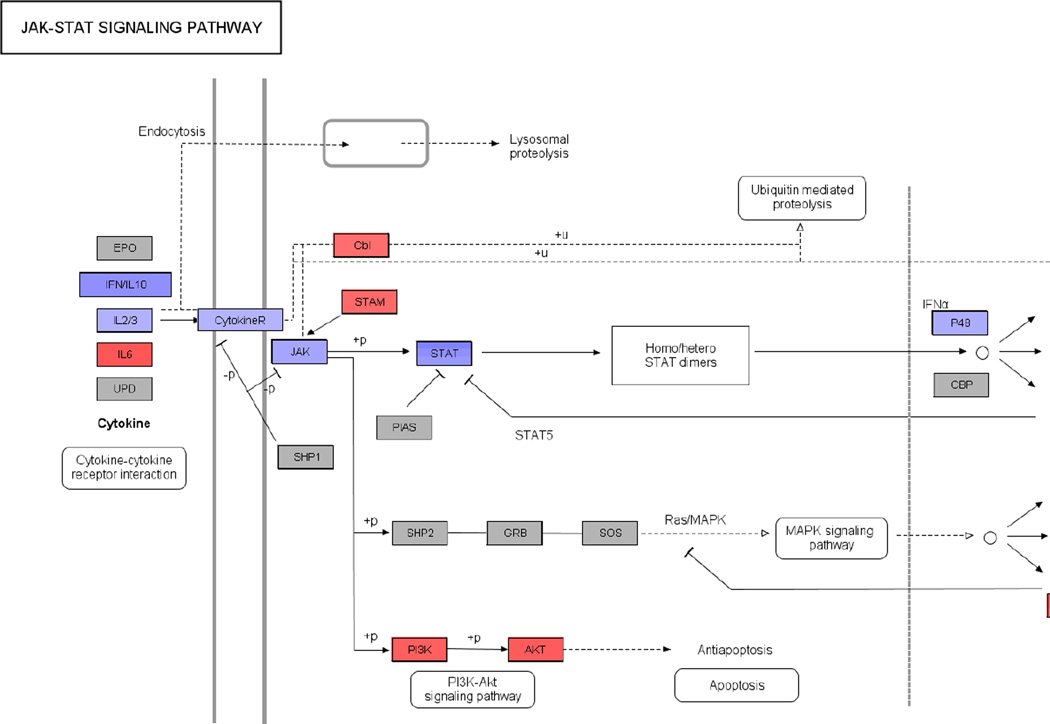
Functional class scoring was used to identify a large number of KEGG pathways significantly enriched for DEGs (Table 1). Pre-ART samples were taken during active HIV replication while post-ART samples were isolated after prolonged drug exposure and the inhibition of virus replication. Fold changes were calculated by dividing post-ART gene expression levels by pre-ART levels. Therefore, DEGs mapping to antiviral pathways originally triggered by HIV replication should be downregulated following ART. This was best exemplified by the Proteasome pathway, where all DEGs mapping to this pathway were downregulated following ART, including those required for conversion of the standard 20S proteasome to the immunoproteasome (Supplementary Figure 1). The proteasome is thought to act as an early intracellular defense against HIV infection since it has been shown to degrade HIV virion components and because proteasome inhibitors increase the efficiency of infection (Schwartz et al., 1998; Wei et al., 2005). The JAK-STAT signaling and Cytokine-cytokine receptor interaction pathways were also significantly over-represented for DEGs. The major components of JAK-STAT signaling were downregulated (i.e., IFNG, JAK2, STAT1, STAT2, and IRF9), thus reflecting the abation of innate immune responses following ART. However, the PI3K-Akt signaling arm of the JAK-STAT signaling pathway was upregulated leading to the upregulation of anti-apoptotic AKT3 (Brunet et al., 1999), suggesting a reversal in HIV-induced apoptosis due to virus suppression (Figure 2). Others have shown that ribosomal genes are downregulated in interferon (IFN) treated PBMCs, which may diminish viral protein synthesis (Stark et al., 1998; Taylor et al., 2004). The cessation of IFN responses following ART resulted in the reversal of this downregulation such that the majority of DEGs mapping to the Ribosome pathway (81%) were upregulated (data not shown). The downregulation following ART of the proteasome subunit PSMA4, components of JAK-STAT signaling (i.e., IFNG and STAT1), and downstream IFN stimulated genes (ISGs, namely OAS1, IFI27, and the activation marker CD38) were confirmed by RT-qPCR, as was the upregulation of ribosomal protein RPS23 but not RPL5 (Figure 1).
Table 1.
KEGG pathways significant for both statistical tests used in functional class scoring analysis implemented in the Gene Set Comparison Tool of BRB-Array Tools.1
| KEGG Pathway Description | KEGG Pathway |
Number of genes |
LS p value |
LS FDR p-value |
KS pvalue |
KS FDR p-value |
|---|---|---|---|---|---|---|
| Purine metabolism | hsa00230 | 195 | 1.00E-05 | 1.67E-04 | 1.01E-03 | 7.33E-03 |
| Pyrimidine metabolism | hsa00240 | 121 | 1.00E-05 | 1.67E-04 | 3.75E-03 | 2.40E-02 |
| One carbon pool by folate | hsa00670 | 18 | 1.00E-05 | 1.67E-04 | 7.70E-04 | 6.43E-03 |
| Reductive carboxylate cycle (CO2 fixation) | hsa00720 | 16 | 1.00E-05 | 1.67E-04 | 1.00E-05 | 1.67E-04 |
| Atrazine degradation | hsa00791 | 12 | 1.00E-05 | 1.67E-04 | 5.21E-03 | 3.11E-02 |
| Ribosome | hsa03010 | 143 | 1.00E-05 | 1.67E-04 | 1.00E-05 | 1.67E-04 |
| Proteasome | hsa03050 | 41 | 1.00E-05 | 1.67E-04 | 1.00E-05 | 1.67E-04 |
| Cytokine-cytokine receptor interaction | hsa04060 | 256 | 1.00E-05 | 1.67E-04 | 1.00E-05 | 1.67E-04 |
| Cell cycle | hsa04110 | 162 | 1.00E-05 | 1.67E-04 | 1.00E-05 | 1.67E-04 |
| Hematopoietic cell lineage | hsa04640 | 106 | 1.00E-05 | 1.67E-04 | 1.00E-05 | 1.67E-04 |
| Folate biosynthesis | hsa00790 | 49 | 2.00E-05 | 2.78E-04 | 2.42E-03 | 1.62E-02 |
| Citrate cycle (TCA cycle) | hsa00020 | 40 | 2.00E-05 | 2.78E-04 | 1.00E-05 | 1.67E-04 |
| Cell adhesion molecules (CAMs) | hsa04514 | 136 | 5.00E-05 | 6.42E-04 | 1.00E-05 | 1.67E-04 |
| Colorectal cancer | hsa05210 | 111 | 1.20E-04 | 1.43E-03 | 3.88E-03 | 2.40E-02 |
| Synthesis and degradation of ketone bodies | hsa00072 | 8 | 2.00E-04 | 2.23E-03 | 8.00E-05 | 1.03E-03 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | hsa00400 | 12 | 2.70E-04 | 2.82E-03 | 1.00E-05 | 1.67E-04 |
| Prion disease | hsa05060 | 14 | 3.10E-04 | 3.05E-03 | 1.70E-04 | 2.03E-03 |
| TGF-beta signaling pathway | hsa04350 | 94 | 4.40E-04 | 4.08E-03 | 2.07E-03 | 1.44E-02 |
| Oxidative phosphorylation | hsa00190 | 130 | 4.70E-04 | 4.09E-03 | 1.00E-05 | 1.67E-04 |
| Tryptophan metabolism | hsa00380 | 107 | 8.70E-04 | 6.68E-03 | 8.00E-05 | 1.03E-03 |
| Butanoate metabolism | hsa00650 | 50 | 9.20E-04 | 6.68E-03 | 2.10E-04 | 2.34E-03 |
| Propanoate metabolism | hsa00640 | 49 | 9.80E-04 | 6.82E-03 | 7.00E-05 | 1.03E-03 |
| Jak-STAT signaling pathway | hsa04630 | 179 | 1.76E-03 | 1.18E-02 | 3.50E-04 | 3.63E-03 |
| Aminoacyl-tRNA biosynthesis | hsa00970 | 34 | 2.49E-03 | 1.60E-02 | 3.70E-04 | 3.63E-03 |
Abbreviations are as follows: KEGG, Kyoto Encyclopedia of Genes and Genomes; LS, refers to Fisher least significance statistic; KS, refers to Kolmogorov-Smirnov statistic; FDR, refers to false discovery rate and was corrected using the Benjamini and method. The Number of genes column refers to the total number of genes assigned to the pathway.
Figure 2.
Jak-STAT Signaling Pathway from KEGG (ID: hsa04630) overlaid with log2 fold change values using PathVisio indicating up (red) or downregulation (blue) following ART. The scale for log2 fold change values is indicated at the bottom of the pathway diagram. Genes not significantly differentially expressed are depicted in grey.
Pathway analysis also identified KEGG pathways significantly enriched for DEGs that may be related to ART exposure. For example, the NRTI components of ART are nucleoside and nucleotide analogs, whose incorporation terminates viral DNA synthesis, and thus it is relevant that the pyrimidine and purine metabolism pathways were significant (Table 1). The downregulation of the ribonucleotide reductase M2 (RRM2) gene, which mapped to both of these pathways, was confirmed by RT-qPCR (Figure 1). The toxicity associated with NRTIs has been attributed, in part, to reduced oxidative phosphorylation activity (Desai et al., 2008). In accordance with this result, the oxidative phosphorylation pathway was significantly enriched for DEGs and the genes that did map complexes I, III and IV were consistently downregulated (data not shown).
The GO terms that were significantly over-represented for DEGs following ART supported the results of pathway analysis. For example, significant GO terms related to biological processes included nucleic acid metabolic process, immune system process, and cellular protein metabolic process (Supplemental Figure 2A). Significant GO terms related to molecular function included purine ribonucleotide binding, chemokine receptor activity, and cytokine binding (Supplemental Figure 2B).
3.3. Interferon Regulatory Factors, Apoptotic Factors, and c-Myc Form Major Hubs in Protein Interaction Networks
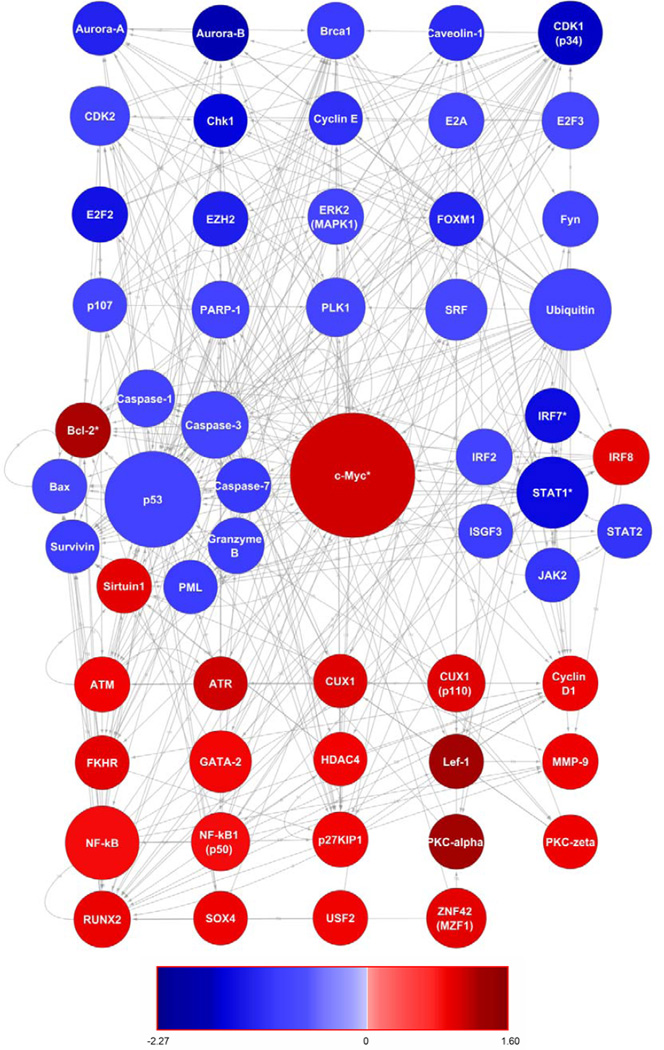
A PIN was generated in MetaCore™ to visualize the protein-protein and protein-DNA interactions of the products of the 4,157 genes that were differentially expressed following ART. For visualization purposes, only those genes whose protein products formed at least 25 interactions were selected (Figure 3). c-Myc formed the most interactions (N=515) and the upregulation (1.46 fold) of this gene was confirmed by RT-qPCR (Figure 1). In support of pathway analysis, which revealed repression of the Jak-Stat signaling pathway following ART, interferon regulatory factors (IRF2, 7, and IRF9 or ISGF3) and signaling molecules (JAK2, STAT1 and 2) were identified as downregulated hubs in the PIN. IRF8 was upregulated factor but this supports the repression of JAK-STAT signaling since IRF8 is a repressor of ISG expression (Weisz et al., 1992). The downregulation of IRF7 was confirmed by RT-qPCR (Figure 1) in addition to other IFN associated genes as previously noted (i.e., CD38, IFNG, STAT1, OAS1, and IFI27). Network analysis also revealed a cluster of apoptosis related hubs with genes encoding pro-apoptotic proteins that were downregulated following ART (e.g., Bax, Granzyme B, PML, and Caspase−1, −2 and −3). Many of these pro-apoptotic proteins are part of p53 dependent apoptotic pathways (Shen and White, 2001) and this was reflected in the protein network, as p53 was also present as a major downregulated hub (Figure 3). In contrast, anti-apoptotic proteins often exhibited upregulation following ART (e.g., BCL2 and Sirtuin1) and the upregulation of BCL2 was confirmed by RT-qPCR (Figure 1).
Figure 3.
Direct protein interaction network constructed using MetaCore™ and visualized in Cytoscape. This network contains only those differentially expressed genes whose protein products had greater than 25 interactions at the protein-protein or protein-DNA level. The size of the node is reflective of the number of interactions and the color key indicates whether the protein coding gene was upregulated (red) or downregulated (blue) following ART. The expression of genes marked with an asterisk was confirmed by RT-qPCR.
3.4. Comparison to Previous Studies of ART-Treated Versus Untreated HIV-Infected Individuals
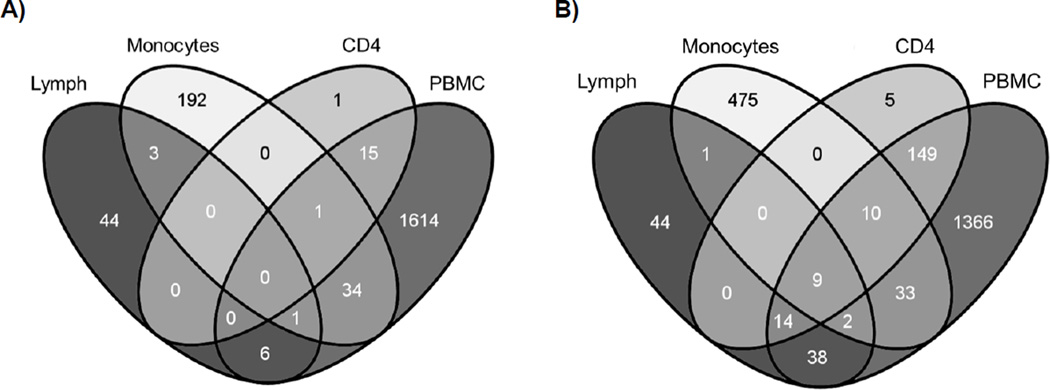
The overlap between the 4,157 genes differentially expressed in PBMCs following ART were compared to those genes identified in similar studies examining tissue from the lymph node (Li et al., 2004), CD4+ T cells (Rotger et al., 2010), and a treatment interruption study in monocytes (Tilton et al., 2006) (Figure 4). Analyzing gene expression in the PBMC compartment captured the vast majority of genes (97%) previously identified as differentially expressed when CD4+ T cells were compared cross-sectionally between ART-treated and untreated HIV-infected individuals. Lesser overlap in DEGs was identified between PBMCs and lymphoid tissue (43%) or monocytes (12%). There were no genes upregulated across all 4 studies but the following 9 ISGs were downregulated in common: IFI6, IFI35, IFIT3, ISG15, OAS2, STAT1, MX2, IFITM1, and GBP1. These genes further emphasize the downregulation of the IFN response following ART regardless of tissue or cell type.
Figure 4.
Venn diagram depicting the overlap in genes differentially expressed between PBMCs isolated from HIV-infected individuals pre- and post-ART (PBMCs), between lymph node samples taken from HIV-infected patients pre- and post-ART (Lymph, Li et al. 2004), between CD4+ T cells from ART-treated or untreated HIV-infected individuals (CD4, Rotger et al. 2010), and between monocytes taken from HIV-infected individuals before and after the interruption of HAART (Monocytes, Tilton et al. 2006). The differentially expressed genes upregulated (A) or downregulated (B) while on ART for all 4 studies are presented.
3.5. Comparison to Previous Studies that Identified Drug Modulated Genes
Microarray studies examining changes in gene expression following NRTI-treatment were identified in order to further investigate the overlap between NRTI-modulated genes and those genes differentially expressed following ART in the current study. NRTI was the primary focus since every HIV-infected individual in this study was treated with NRTIs (Supplemental Table 1). Unfortunately, there appears to be a lack of microarray studies examining gene expression changes in human primary immune cells treated with NRTIs. Instead, Grigsby et al.’s (Grigsby et al., 2010) study examining the effects of TDF on mouse osteoblasts and Desai et al.’s (Desai et al., 2008) investigation of liver cells isolated from mice exposed to AZT in combination with 3TC were selected for comparison purposes. TDF was the most common NRTI used to treat the HIV-infected individuals in our study and AZT/3TC was a common pairing of NRTIs (Supplemental Table 1). In addition to these NRTI-treatment studies, well-known drug metabolism genes comprising the Drug Metabolism PCR Array (SABiosciences) were also selected for comparison.
There was limited overlap between genes differentially expressed in PBMCs following ART, NRTI-modulated genes, and well-known drug metabolism genes (Supplemental Figure 3). The greatest overlap appeared to be with well-characterized drug metabolism genes whereby 9 genes (i.e., ALOX12, CYB5R3, CYP2E1, GPX1 and 4, GSTM2 and 3, GSTT1, and PON2) were upregulated and 5 genes (i.e., BLVRA, GSTZ1, MARCKS, MT2A, and NAT1) downregulated following ART. Although not included on the Drug Metabolism PCR Array, CYP2S1 was also selected for RT-qPCR analysis because it was upregulated (1.30 fold) to a greater extent than CYP2E1 (1.14 fold) and belongs to same family of well-characterized drug metabolism genes. The upregulation of ALOX12 and CYP2S1 but not GSTM2 were confirmed by RT-qPCR (Figure 1) and thus the expression of this latter target was subjected to analysis by ddPCR.
3.6. Confirmation of Gene Expression by RT-qPCR and ddPCR
As discussed throughout the results, the differential expression of a large number of targets was significantly confirmed by RT-qPCR, which validated the conclusions drawn from microarray analysis (Figure 1). However, the significant upregulation of RPL5 and GSTM2 following ART was not confirmed by RT-qPCR. Therefore, the expression of these targets was assessed by ddPCR, which is more sensitive than RT-qPCR because target transcripts are distributed across several thousands of droplets that each undergo an end-point PCR reaction (Baker, 2012; Strain et al., 2013). After confirming the significant downregulation of STAT1 (selected as a positive control), ddPCR was able to identify significant upregulation of both RPL5 and GSTM2 where RT-qPCR had failed (Figure 1). When ddPCR results were substituted for RT-qPCR results, the gene expression for all targets correlated with microarray results with an r2 of 0.93 (data not shown).
4. Discussion
Paired gene expression analysis of 64 PBMC samples taken from 32 HIV-infected individuals before and after ART identified 4,157 genes that were significantly differentially expressed. This number of DEGs is an order of magnitude higher than similar studies that examined expression differences between samples taken from ART-treated and untreated subjects. These previous studies used cross-sectional (Rotger et al., 2010) or underpowered approaches (Li et al., 2004), and our study demonstrates the effectiveness of a sufficiently powered paired analysis. Gene expression differences before and after ART in the PBMC compartment demonstrated near complete overlap with genes identified in the CD4+ T cell subset but much less overlap with the monocyte subset and lymphoid tissue (Figure 4). However, there was a core set of 9 ISGs (IFI6, IFI35, IFIT3, ISG15, OAS2, STAT1, MX2, IFITM1, and GBP1) that were downregulated across compartments following ART. Interestingly, none of the members of this core set of ISGs were detected as differentially expressed in brain tissue between ART-treated and untreated HIV-infected subjects with HAND (Borjabad et al., 2011) suggesting that these findings may not extend across the blood-brain barrier. In summary, these genes have been identified for the first time as biomarkers of the host response to HIV that are relevant across multiple compartments associated with the peripheral blood.
Gene expression was assayed in paired samples that represented a transition from a baseline of active HIV replication to a period of virus suppression and prolonged ART exposure (48 weeks). This transition should be reflected by receding activation of antiviral pathways and the increasing modulation of processes related to drug metabolism. The identification of KEGG pathways and GO terms over-represented for DEGs, as well as the hubs identified in PINs, clearly reflected this transition. Antiviral processes associated with IFN signaling (Figure 1), proteasome degradation (Supplemental Figure 1) and apoptosis (Figure 2) were repressed following ART. Whereas pathways associated with NRTI metabolism and oxidative phosphorylation were enriched for DEGs and may reflect prolonged ART exposure (Table 1). Unfortunately, there was minimal overlap between genes differentially expressed following ART compared with murine osteoblasts (Grigsby et al., 2010) or liver cells (Desai et al., 2008) exposed to NRTIs (Supplemental Figure 3). This may be because these studies were not performed PBMCs, which represent a substrate for HIV replication and are activated during ongoing infection. A number of well-characterized drug metabolism genes were modulated by ART of which the upregulation of ALOX12 and CYP2S1 was confirmed by RT-qPCR (Figure 1). ALOX12 belongs to a family of non-heme iron dioxygenases and CYP2S1 encodes a member of the cytochrome P450 superfamily of enzymes but their roles in ART metabolism are currently not well understood. Encouragingly, Vigneault et al. (Vigneault et al., 2011) also noted that CYP2S1 was expressed to a greater extent in CD4 T cells isolated from HIV-infected ART-treated subjects compared to uninfected controls. Furthermore, Borjabad et al. (Borjabad et al., 2011) demonstrated that CYP46A1 and CYP4x1 were upregulated in the brain tissue of HIV-infected subjects with HAND treated with ART compared to untreated subjects. Finally, many of the pathways identified as significantly over-represented for DEGs in PBMCs (Table 1) were related to similar biological processes identified by Li et al. (2004) who examined gene expression in lymphoid tissue: Immune Activation, IFN Related, Chemokine Receptors and Ligands, Apoptosis, Nucleic Acid and Protein Metabolism, and Extracellular Matrix. This suggests that although the overlap between DEGs was moderate between PBMC and lymphoid tissue (46%) the same higher order biological processes are being affected across tissue types.
Virus suppression resulting from ART clearly prolongs life for HIV-infected individuals but as this population ages it is becoming apparent that they suffer from a higher incidence of several non-AIDS related disorders compared to healthy individuals, which include neurocognitive decline, cancer, as well as heart, liver, kidney and bone disease (Deeks, 2011). With respect to the PBMC samples analyzed in this study, Patel et al. (2008) observed significantly higher rates of Hodgkin’s lymphoma in HIV-infected patients compared to the general population with incidence rates increasing despite the advent of ART. This increase in Hodgkin’s lymphoma (HL) in HIV-infected patients has previously been associated with immunosuppression resulting from co-infection with Epstein-Barr virus (Righetti et al., 2002) but the potential contribution of ART-modulated genes should not be overlooked. Specifically, the current study found that CMYC was upregulated following ART (Figure 1) and was the hub that formed the most protein-protein and protein-DNA interactions (Figure 2). Although this upregulation was modest (1.46 fold by RT-qPCR), c-MYC is believed to regulate 15% of all genes, including genes involved in cell division, cell growth, and apoptosis (Gearhart et al., 2007), and dysregulated expression of c-MYC has been associated with multiple types of cancer (Pelengaris et al., 2002). Increased copy number of CMYC has been noted in HL cell lines (Joos et al., 2003) and downregulation of CMYC by curcumin has been shown to decrease the viability of lymphomas (Mackenzie et al., 2008). Finally, the Oxidative Phosphorylation pathway was significantly over-represented for DEGs (Table 1). Multiple components of complexes I, III and IV, required for oxidative phosphorylation were downregulated in the current study and many cancer cells tend to exhibit reduced oxidative phosphorylation (Solaini et al., 2011). Furthermore, oxidative phosphorylation has been shown to be impaired in HIV-infected children born to seropositive mothers who were exposed to NRTIs (Barret et al., 2003). It has been noted that during acute HIV infection the enzymatic activity of Complexes I, II and III is decreased while those of Complexes IV and V are increased (Ladha et al., 2005; Tripathy and Mitra, 2010). This lends weight to ART resulting in the downregulation of genes in Complexes I and III in this study as opposed to this signal coming from the upregulation of these genes by HIV prior to ART, which might be an argument for Complex IV.
In summary, thousands of genes appear to be differentially expressed in PBMCs when comparing pre- and post-ART samples from HIV-infected individuals. Mapping these genes onto higher order biological processes reflects the transition from active virus replication (pre-ART) to viral suppression and drug metabolism (post-ART). One possible limitation of this study, due to the lack of an uninfected control, is the difficulty in confirming whether a gene is truly upregulated post-ART or was downregulated pre-ART and thus appears upregulated post-ART. Useful controls would be samples taken prior to HIV infection and ART treatment, which are nearly impossible to obtain, or samples taken before and after ART from a control arm of healthy subjects, which is ethically questionable. However, we are confident in our interpretation of the data since they have reconfirmed and extended the findings of previous studies (Li et al., 2004; Rotger et al., 2010) and no evidence in the literature was found for the downregulation of well known drug metabolism genes (i.e., ALOX12, CYP2S1, and GSTM2) by HIV pre-ART suggesting these genes are truly upregulated post-ART. The use of ddPCR to assay gene expression represents a novel aspect of this work and preliminary findings suggest that ddPCR is superior to RT-qPCR when confirming the differential expression of genes with low fold changes. Finally, over the course of this study it became clear that there is currently a lack of studies that have exposed primary immune blood cells to NRTIs in vitro. Future experiments identifying ART-modulated gene expression in blood cells are important since these cells circulate throughout the body and thus may distribute any associated toxicities to a variety of compartments. In addition, studies that examine the difference between different regimen types (e.g. PI vs. NNRTI) should be performed in the future but were underpowered in the current analysis due to the removal of outlier microarray data from paired samples for 4 PI-treated subjects.
Supplementary Material
HIGHLIGHTS.
The number of genes modulated following ART is an order of magnitude higher than previously recognized.
Well characterized drug metabolism genes not previously associated with ART appear upregulated by ART.
A core set of interferon stimulated genes respond to HIV regardless of tissue type and then recede following ART.
Droplet Digital PCR (ddPCR) has been used to confirm gene expression for the first time in an infectious disease setting.
Acknowledgements
Gene expression data were generated via funding through a Developmental Grant from the Center for AIDS Research (CFAR) at the University of California San Diego (UCSD). This work was performed with the support of the Genomics, Flow Cytometry and Translational Virology Cores at the UCSD CFAR (AI036214), equipment from the James B. Pendleton Foundation, the San Diego Veterans Medical Research Foundation, and other National Institutes of Health research grants (AI043638, AI007384, AI074621 and AI087164). Marta Massanella was supported by the Agència de Gestió d’Ajuts Universitaris i de Recerca from the Generalitat de Catalunya and the European Social Fund. Microarray hybridization and scanning was performed at the UCSD Biomedical Genomics (BIOGEM) core facility with the help of Dr. Gary Hardiman (Director) and James Sprague. Class Prediction analysis was performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. We would like to extend our heartfelt appreciation to Drs. Amalio Telenti, Margalida Rotger, Nadine Zangger, Qingsheng Li and Ashley Haase, who provided insights and data from their respective studies. This work is based upon work supported in part by the Department of Veterans Affairs (VA), Veterans Health Administration, Office of Research and Development. The views expressed in this body of work are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arts EJ, Hazuda DJ. HIV-1 Antiretroviral Drug Therapy. Cold Spring Harb Perspect Med. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Digital PCR hits its stride. Nature Methods. 2012;9:541–544. [Google Scholar]
- Barret B, Tardieu M, Rustin P, Lacroix C, Chabrol B, Desguerre I, Dollfus C, Mayaux MJ, Blanche S. Persistent mitochondrial dysfunction in HIV-1-exposed but uninfected infants: clinical screening in a large prospective cohort. AIDS. 2003;17:1769–1785. doi: 10.1097/00002030-200308150-00006. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Borjabad A, Morgello S, Chao W, Kim SY, Brooks AI, Murray J, Potash MJ, Volsky DJ. Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders. PLoS Pathog. 2011;7:e1002213. doi: 10.1371/journal.ppat.1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Carr A, Samaras K, Burton S, Law M, Freund J, Chisholm DJ, Cooper DA. A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. AIDS. 1998;12:F51–F58. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- Datson NA, Speksnijder N, Mayer JL, Steenbergen PJ, Korobko O, Goeman J, de Kloet ER, Joels M, Lucassen PJ. The transcriptional response to chronic stress and glucocorticoid receptor blockade in the hippocampal dentate gyrus. Hippocampus. 2010;22:359–371. doi: 10.1002/hipo.20905. [DOI] [PubMed] [Google Scholar]
- Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai VG, Lee T, Delongchamp RR, Leakey JE, Lewis SM, Lee F, Moland CL, Branham WS, Fuscoe JC. Nucleoside reverse transcriptase inhibitors (NRTIs)-induced expression profile of mitochondria-related genes in the mouse liver. Mitochondrion. 2008;8:181–195. doi: 10.1016/j.mito.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Gearhart J, Pashos EE, Prasad MK. Pluripotency redux--advances in stem-cell research. N Engl J Med. 2007;357:1469–1472. doi: 10.1056/NEJMp078126. [DOI] [PubMed] [Google Scholar]
- Grigsby IF, Pham L, Mansky LM, Gopalakrishnan R, Carlson AE, Mansky KC. Tenofovir treatment of primary osteoblasts alters gene expression profiles: implications for bone mineral density loss. Biochem Biophys Res Commun. 2010;394:48–53. doi: 10.1016/j.bbrc.2010.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2012;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Joos S, Granzow M, Holtgreve-Grez H, Siebert R, Harder L, Martin-Subero JI, Wolf J, Adamowicz M, Barth TF, Lichter P, Jauch A. Hodgkin's lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. International journal of cancer. Journal international du cancer. 2003;103:489–495. doi: 10.1002/ijc.10845. [DOI] [PubMed] [Google Scholar]
- Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clinical therapeutics. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Research. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler JJ, Hosseini SH, Hoying-Brandt A, Green E, Johnson DM, Russ R, Tran D, Raper CM, Santoianni R, Lewis W. Tenofovir renal toxicity targets mitochondria of renal proximal tubules. Laboratory investigation; a journal of technical methods and pathology. 2009;89:513–519. doi: 10.1038/labinvest.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladha JS, Tripathy MK, Mitra D. Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis. Cell death and differentiation. 2005;12:1417–1428. doi: 10.1038/sj.cdd.4401668. [DOI] [PubMed] [Google Scholar]
- Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003;2:812–822. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- Li Q, Schacker T, Carlis J, Beilman G, Nguyen P, Haase AT. Functional genomic analysis of the response of HIV-1-infected lymphatic tissue to antiretroviral therapy. J Infect Dis. 2004;189:572–582. doi: 10.1086/381396. [DOI] [PubMed] [Google Scholar]
- Mackenzie GG, Queisser N, Wolfson ML, Fraga CG, Adamo AM, Oteiza PI. Curcumin induces cell-arrest and apoptosis in association with the inhibition of constitutively active NF-kappaB and STAT3 pathways in Hodgkin's lymphoma cells. International journal of cancer. Journal international du cancer. 2008;123:56–65. doi: 10.1002/ijc.23477. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Montessori V, Press N, Harris M, Akagi L, Montaner JS. Adverse effects of antiretroviral therapy for HIV infection. CMAJ. 2004;170:229–238. [PMC free article] [PubMed] [Google Scholar]
- Pacenti M, Barzon L, Favaretto F, Fincati K, Romano S, Milan G, Vettor R, Palu G. Microarray analysis during adipogenesis identifies new genes altered by antiretroviral drugs. AIDS. 2006;20:1691–1705. doi: 10.1097/01.aids.0000242815.80462.5a. [DOI] [PubMed] [Google Scholar]
- Patel P, Hanson DL, Sullivan PS, Novak RM, Moorman AC, Tong TC, Holmberg SD, Brooks JT. Incidence of types of cancer among HIV-infected persons compared with the general population in the United States, 1992–2003. Ann Intern Med. 2008;148:728–736. doi: 10.7326/0003-4819-148-10-200805200-00005. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Perez-Santiago J, Diez-Alarcia R, Callado LF, Zhang JX, Chana G, White CH, Glatt SJ, Tsuang MT, Everall IP, Meana JJ, Woelk CH. A combined analysis of microarray gene expression studies of the human prefrontal cortex identifies genes implicated in schizophrenia. J Psychiatr Res. 2012;46:1464–1474. doi: 10.1016/j.jpsychires.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Righetti E, Ballon G, Ometto L, Cattelan AM, Menin C, Zanchetta M, Chieco-Bianchi L, De Rossi A. Dynamics of Epstein-Barr virus in HIV-1-infected subjects on highly active antiretroviral therapy. AIDS. 2002;16:63–73. doi: 10.1097/00002030-200201040-00009. [DOI] [PubMed] [Google Scholar]
- Rivero A, Mira JA, Pineda JA. Liver toxicity induced by non-nucleoside reverse transcriptase inhibitors. J Antimicrob Chemother. 2007;59:342–346. doi: 10.1093/jac/dkl524. [DOI] [PubMed] [Google Scholar]
- Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, Shianna KV, Ge D, Gunthard HF, Goldstein DB, Telenti A. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6:e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard JM. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, White E. p53-dependent apoptosis pathways. Advances in cancer research. 2001;82:55–84. doi: 10.1016/s0065-230x(01)82002-9. [DOI] [PubMed] [Google Scholar]
- Simon R. the BRB-Array Tools Development Team, the EMMES Corporation. BRB-Array Tools Version 3.8 User's Manual. 2009 [Google Scholar]
- Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochimica et biophysica acta. 2011;1807:534–542. doi: 10.1016/j.bbabio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Grosse WM, Schaley JE, Sanda C, Wu X, Chien SC, Smith F, Wu TG, Stephens M, Ferris MW, McClintick JN, Jerome RE, Edenberg HJ. Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. J Interferon Cytokine Res. 2004;24:107–118. doi: 10.1089/107999004322813354. [DOI] [PubMed] [Google Scholar]
- Tilton JC, Johnson AJ, Luskin MR, Manion MM, Yang J, Adelsberger JW, Lempicki RA, Hallahan CW, McLaughlin M, Mican JM, Metcalf JA, Iyasere C, Connors M. Diminished production of monocyte proinflammatory cytokines during human immunodeficiency virus viremia is mediated by type I interferons. J Virol. 2006;80:11486–11497. doi: 10.1128/JVI.00324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy MK, Mitra D. Differential modulation of mitochondrial OXPHOS system during HIV-1 induced T-cell apoptosis: up regulation of Complex-IV subunit COX-II and its possible implications. Apoptosis. 2010;15:28–40. doi: 10.1007/s10495-009-0408-9. [DOI] [PubMed] [Google Scholar]
- Venhoff N, Setzer B, Melkaoui K, Walker UA. Mitochondrial toxicity of tenofovir, emtricitabine and abacavir alone and in combination with additional nucleoside reverse transcriptase inhibitors. Antivir Ther. 2007;12:1075–1085. [PubMed] [Google Scholar]
- Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F, Crosby SD, Rychert J, Church G, Martinez-Picado J, Rosenberg ES, Telenti A, Yu XG, Lichterfeld M. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. J Virol. 2011;85:3015–3019. doi: 10.1128/JVI.01846-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittinghoff E, Scheer S, O’Malley P, Colfax G, Holmberg SD, Buchbinder SP. Combination antiretroviral therapy and recent declines in AIDS incidence and mortality. J Infect Dis. 1999;179:717–720. doi: 10.1086/314623. [DOI] [PubMed] [Google Scholar]
- Wei BL, Denton PW, O'Neill E, Luo T, Foster JL, Garcia JV. Inhibition of lysosome and proteasome function enhances human immunodeficiency virus type 1 infection. J Virol. 2005;79:5705–5712. doi: 10.1128/JVI.79.9.5705-5712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz A, Marx P, Sharf R, Appella E, Driggers PH, Ozato K, Levi BZ. Human interferon consensus sequence binding protein is a negative regulator of enhancer elements common to interferon-inducible genes. J Biol Chem. 1992;267:25589–25596. [PubMed] [Google Scholar]
- Woelk CH, Beliakova-Bethell N, Goicoechea M, Zhao Y, Du P, Rought SE, Lozach J, Perez-Santiago J, Richman DD, Smith DM, Little SJ. Gene expression before HAART initiation predicts HIV-infected individuals at risk of poor CD4+ T-cell recovery. AIDS. 2010;24:217–222. doi: 10.1097/QAD.0b013e328334f1f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk CH, Zhang JX, Walls L, Viriyakosol S, Singhania A, Kirkland TN, Fierer J. Factors regulated by interferon gamma and hypoxia-inducible factor 1A contribute to responses that protect mice from Coccidioides immitis infection. BMC Microbiol. 2012;12:218. doi: 10.1186/1471-2180-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.