Abstract
The mammalian genome is extensively transcribed, a large fraction of which is divergent transcription from promoters and enhancers that is tightly coupled with active gene transcription. Here we propose that divergent transcription may shape the evolution of the genome by new gene origination.
Widespread divergent transcription
The vast majority of the human genome, including half of the region outside known genes, is transcribed (Djebali et al., 2012). However, most intergenic transcription activity produces short and unstable noncoding transcripts whose abundances are usually an order of magnitude lower than those from typical protein-coding genes. Except for a few well-studied cases (see review in (Guttman and Rinn, 2012; Lee, 2012; Mercer et al., 2009; Ponting et al., 2009; Rinn and Chang, 2012; Ulitsky and Bartel, 2013; Wang and Chang, 2011; Wei et al., 2011; Wilusz et al., 2009), it’s unclear whether most intergenic transcription is regulated or has cellular function.
Recent evidence has shown that most intergenic transcription occurs near or is associated with gene transcription, such as transcription from promoter and enhancer regions (Sigova et al., 2013). The majority of mammalian promoters direct transcription initiation on both sides with opposite orientations, a phenomenon known as divergent transcription (Core et al., 2008; Preker et al., 2008; Seila et al., 2008). Divergent transcription generates upstream antisense RNAs (uaRNAs, or PROMPTs, promoter upstream transcripts) near the 5′ end of genes that are typically short (50–2,000 nucleotides) and relatively unstable (Flynn et al., 2011; Ntini et al., 2013; Preker et al., 2008, 2011). Similar divergent transcription also occurs at distal enhancer regions, giving rise to RNAs termed enhancer RNAs (eRNAs) (Kim et al., 2010; De Santa et al., 2010). In mouse and human embryonic stem (ES) cells most long noncoding RNAs (lncRNAs, longer than 100 nucleotides) are associated with protein-coding genes, including ~50% as uaRNAs and ~20% as eRNAs (Sigova et al., 2013). These observations suggest that divergent transcription from promoters and enhancers of protein-coding genes is the major source of intergenic transcription in ES cells.
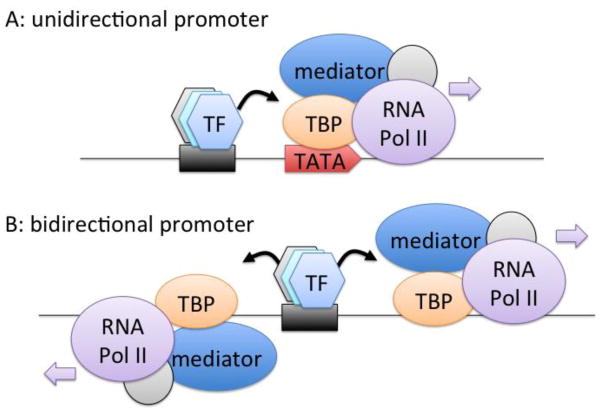
In the textbook model of a eukaryotic promoter, the directionality is set by the arrangement of an upstream cis-element region followed by a core promoter (Fig 1A). The cis-elements are bound by sequence-specific transcription factors whereas the core promoter is bound by TATA-binding protein (TBP) and other factors that recruit the core transcription machinery. Most mammalian promoters lack a TATA element (TATA-less) and are CpG rich (Sandelin et al., 2007). For these promoters, TBP is recruited through sequence specific transcription factors such as Sp1 that bind CpG rich sequences and components of the TFIID complex that have little sequence specificity. Thus, in the absence of strong TATA elements such as for CpG island promoters, TBP-complexes are recruited on both sides of the transcription factors to form pre-initiation complexes in both orientations (Fig 1B). This model is supported by the observation that divergent transcription occurs at most promoters that are associated with CpG islands in mammals, whereas promoters with TATA elements in mammals and worm are associated with unidirectional transcription (Core et al., 2008; Kruesi et al., 2013). In addition, divergent transcription is less common in Drosophila where CpG islands are rare (Core et al., 2012). Since transcription factors with chromatin remodeling potential and transcription activation domains also bind at enhancer sites, it is not surprising that these are also sites of divergent transcription. In fact, promoters and enhancers have many properties in common, and it has been shown recently that many intragenic enhancers can act as alternative promoters producing tissue-specific lncRNAs (Kowalczyk et al., 2012).
Figure 1.
Transcription factors drive divergent transcription. A) Transcription factor (TF) binding helps to recruit TATA-binding protein (TBP) and associated factors, which binds the directional TATA element in the DNA and orientates RNA Pol II to transcribe downstream DNA. B) In the absence of strong TATA elements common of CpG island promoters, TF-recruited TBP and associated factors binds to low specificity sequences and forms initiation complexes at similar frequencies in both directions.
The U1-PAS axis and gene maturation
Promoter-proximal noncoding transcription in both yeast and mammals has been shown to be suppressed at the chromatin level, including nucleosome remodeling (Whitehouse et al., 2007), histone deacetylation (Churchman and Weissman, 2011), and gene loop formation (Tan-Wong et al., 2012). We and others recently found that in mammals promoter upstream antisense transcription is frequently terminated due to cleavage of the nascent RNA by the same process responsible for the generation of the poly A tract at the 3′ ends of genes (Almada et al., 2013; Ntini et al., 2013). In both cases, the primary signal directing this process is the poly (A) signal (PAS) motif, AAUAAA or similar (Proudfoot, 2011). Pol II terminates transcription within several kb after such cleavage (Anamika et al., 2012; Richard and Manley, 2009). Computational analysis showed that relative to the 5′ end of the sense regions, PAS motifs are enriched whereas potential U1 snRNP binding sites, or 5′ splice site-like sequences, are depleted in the upstream antisense regions. The binding of U1 snRNP is known to suppress PAS directed cleavage over regions of thousand nucleotides downstream (Berg et al., 2012; Kaida et al., 2010). Thus, the bias in the distribution of U1 snRNP binding sites and PAS promotes expression of full-length mRNAs by suppressing premature cleavage and polyadenylation but favors early termination of uaRNAs. This conclusion is strongly supported by the finding that inhibition of U1 snRNP dramatically increased termination and polyadenylation of sense-oriented transcripts in the gene region (Almada et al., 2013).
If the U1-PAS axis defines the length of a transcribed region, then it might be expected that for a typical protein-coding gene (~20 kb) to evolve from intergenic noncoding DNA would involve strengthening of the U1-PAS axis by gaining U1 sites and losing PAS in the sense orientation. Examining the distributions of U1 and PAS sites in bidirectional promoters involving UCSC-annotated mRNA-mRNA, mRNA-lncRNA, and mRNA-uaRNA pairs, we found that lncRNAs showed properties resembling intermediates between mRNA genes and uaRNA regions in terms of the density of U1 sites and PAS sites (Almada et al., 2013). That is, the density of PAS decreases from regions producing uaRNA to lncRNA to mRNA, whereas U1 sites show the opposite trend, consistent with the differences in the length and abundance of these transcripts. We also studied the evolution of the U1-PAS axis in vertebrates, and found that older genes exhibit progressive gain of U1 sites and loss of PAS sites at their 5′ ends. Together these observations suggest that strengthening of the U1-PAS axis may be associated with the origination and maturation of genes.
De novo gene origination from divergent transcription
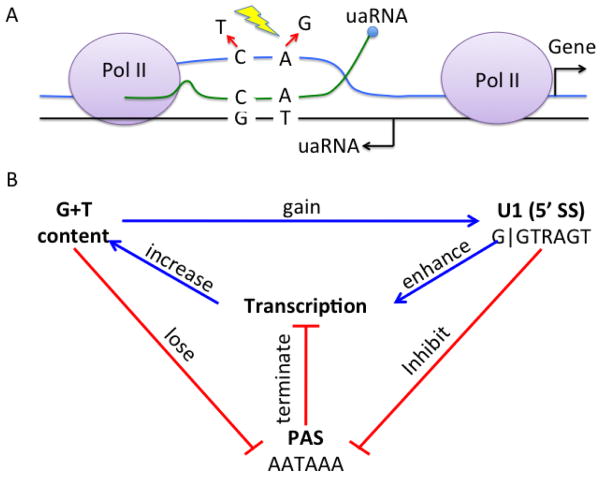
Below we propose a model (Fig 2) arguing that the act of transcription in germ cells strengthens the U1-PAS axis in the upstream antisense region of an active gene, or the associated enhancer regions, creating a feedback loop amplifying transcription activity, which eventually may drive origination of a new antisense-oriented gene (Fig 3).
Figure 2.
Feedback loops between transcription, U1, and PAS signals. (A) Germ cell transcription exposes the coding strand (non-template, which has the same sequence as the RNA) single-stranded and vulnerable to mutations towards G and T bases, (B) which increases the chance of gaining GT-rich sequences such as U1 binding site (5′ splice site (5′ SS)) and also increases the chance of losing A-rich sequences such as PAS, which terminates transcription. U1 binding can enhance transcription through promoting transcription initiation and reinitiation, and also inhibiting the usage of nearby PAS.
Figure 3.
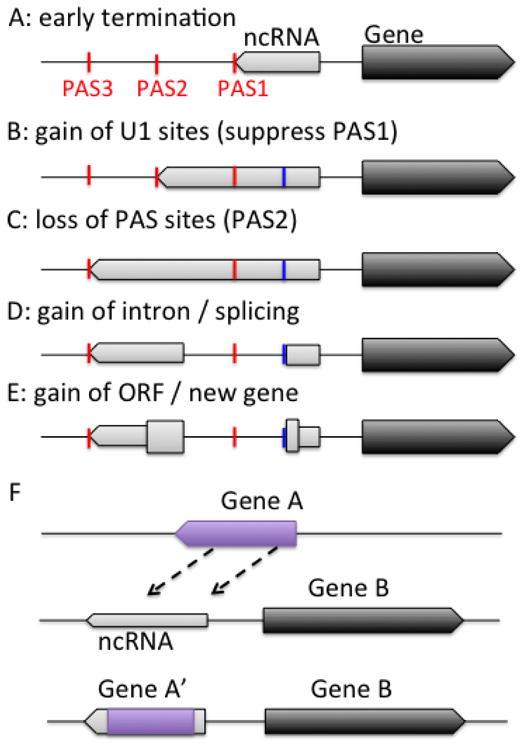
Divergent transcription drives new gene origination. A–E) De novo protein-coding gene origination, and F) gene duplication or translocation. A) Divergent transcription of a gene (right dark block) generates divergent noncoding RNA (ncRNA) in the upstream antisense direction, which is terminated by PAS-dependent mechanism (PAS: red bars). B) Transcription increases G and T frequency on the coding strand, thus increases the chance of encoding a U1 site (blue bar) which suppress a downstream PAS (PAS1), favoring the usage of a downstream PAS (PAS2). C) Increase in G+T content also increases the chance of losing PAS sites (PAS2) which activates a further downstream site (PAS3) and extends the transcribed region. D) The longer transcript acquires splicing signals, which makes it more stable and exported to the cytoplasm. E) The longer transcript encodes a short ORF and the resulting short peptide is selected and fixed in the population and becomes a new protein-coding gene. F) Gene A is translocated or duplicated into the promoter upstream antisense region of gene B, and evolves into a new gene A′. Thin and thick blocks represent transcribed noncoding and coding regions, respectively.
One consequence of transcription is that it can cause mutations, especially on the coding (non-transcribed) strand. During transcription, transient R-loops can be formed behind the transcribing RNA polymerase II, exposing the coding strand as single-stranded DNA whereas the non-coding strand is base-paired with and thus protected by the nascent RNA (Aguilera and García-Muse, 2012). The lack of splicing signals in the divergent transcript also makes it more vulnerable to R-loop formation, as splicing factors have been implicated in suppressing R-loop formation (Li and Manley, 2006, 2005; Paulsen et al., 2009). In addition, divergent transcription generates negative supercoiling at promoters which facilitates DNA unwinding and promotes R-loop formation (Aguilera and García-Muse, 2012; Seila et al., 2009). As a consequence of R-loop formation, the single-stranded coding strand is vulnerable to mutagenic processes, such as cleavage, deamination, and depurination. Genomics studies have shown that during mammalian evolution, transcribed regions accumulate G and T bases on the coding strand, relative to the non-coding strand or non-transcribed regions (Green et al., 2003; Mugal et al., 2009; Park et al., 2012; Polak et al., 2010). Evidence suggests that such strand bias may result from passive effects of deamination, transcription-coupled repair, and somatic hypermutation pathways in germ cell-transcribed genes, in the absence of selection (Green et al., 2003; McVicker and Green, 2010; Polak and Arndt, 2008).
Accumulation of G and T content on the coding strand will strengthen the U1-PAS axis (Fig. 2). A-rich sequences such PAS (AATAAA) is likely to be lost when the genomic DNA accumulates G and T. In contrast, G+T rich sequences, such as U1 snRNP binding sites (e.g., resembling 5′ splice sites, G|GTAAGT and G|GTGAGT), are likely to emerge in these regions. Since promoter-proximal PAS reduces transcriptional activity (Andersen et al., 2012), the loss of PAS and gain of U1 sites should contribute to lengthening of the transcribed region as well as its more robust transcription. The gain of U1 sites could also enhance transcription by recruiting basal transcription initiation factors (Damgaard et al., 2008; Furger et al., 2002; Kwek et al., 2002) or elongation factors (Fong and Zhou, 2001). Therefore a positive feedback loop is formed: active transcription causes the coding strand to accumulate sequence changes favoring higher transcription activity.
As noted above, strengthening of the U1-PAS axis also favors extension of the transcribed region. Being longer gives the transcript several advantages: by chance longer RNAs are more likely to contain additional splicing signals such as a 3′ splice site to become spliced, or binding sites for splicing-independent nuclear export factors, thus escaping nuclear exosome degradation by packaging and exporting to cytoplasm (Nott et al., 2003; Singh et al., 2012). Longer RNAs are also more likely to carry an open reading frame, either generated de novo or by incorporation of gene remnants.
Once in the cytoplasm, the RNA should at some frequency be translated into short polypeptides due to widespread translational activity (Carvunis et al., 2012). Some of the polypeptides may provide advantage to the organism and become fixed in the population, thereby forming a new gene.
Accelerating other new gene origination processes
In addition to de novo gene origination, the model described above also facilitates new gene origination via other mechanisms in regions of divergent transcription. Tandem duplication, retroposition, and recombination of existing genes or gene fragments are the major mechanisms for new gene origination (Chen et al., 2013; Long et al., 2013). Most duplicated genes or gene fragments are silenced due to the lack of required elements such as a promoter. In contrast, genes or gene fragments inserted into regions of divergent transcription, such as upstream of a promoter or flanking an enhancer, will be transcribed, likely under different regulation than prior to their insertion, and thus could evolve to carry out functions different than the original gene. In support of this, a recent survey of human and mouse genes evolved from “domesticated” transposons (Kalitsis and Saffery, 2009) showed that a significant proportion of them are located in bidirectional promoters. Promoter upstream regions also preferentially accumulate transposable elements, which can carry 5′ splice site sequences that may accelerate the process of new gene origination (Gotea et al., 2013).
New gene origination from enhancers
Similar to promoters, enhancers are also divergently transcribed, and as a result, new genes might originate at enhancer regions through the same mechanism described above. The possibility of enhancer derived new genes has not been previously discussed. Manual inspection of a list of 24 hominoid-specific de novo protein-coding genes (Xie et al., 2012) revealed that MYEOV (myeloma overexpressed), a gene implicated in various types of cancer (Janssen et al., 2000, 2002; Leyden et al., 2006; Moss et al., 2006), is likely derived from an intergenic enhancer in mouse. The mouse syntenic region of MYEOV is within a 5 kb region about 100 kb away from any gene, but covered by intensive H3K4me1 marks, diagnostic of an enhancer, and positive for Mediator binding in mouse ES cells, as well as nascent transcription signals (GRO-seq) indicating divergent transcription, all indicating this region is an active enhancer in mouse ES cells. Further analysis is needed to firmly establish the role of enhancer transcription in the origination of the MYEOV gene. For example, it will be interesting to examine the evolutionary dynamics of the spatial and functional relationship between the enhancer/MYEOV locus and the corresponding target gene.
Predictions and supporting evidence
A recent comparative analysis of human-mouse gene annotations detected over a thousand lncRNAs annotated in the upstream antisense region of human genes whereas lncRNAs divergent from the corresponding mouse protein-coding genes could not be detected (Gotea et al., 2013). This observation suggests that promoter divergent transcription could be capable of generating large number of primate-specific transcripts. Another study (Xie et al., 2012) identified 24 hominoid-specific de novo protein-coding genes in human, five of which derive from bidirectional promoters (P < 0.01, compared to shuffled gene positions), confirming promoter divergent transcription as an important source of de novo gene origination, and enhancer transcription may drive other new genes, as noted above.
An important feature of genes originated in the proposed model is that both the new gene and the ancestral gene are likely to be expressed in germ cells. This is because for the transcription-induced G and T bias to accumulate and spread in a population, these mutations should occur in germ cells. A prediction of the model is that new genes are preferentially expressed in germ cells, or tissues with high fraction of germ cells. Consistent with this, previous reports showed that lineage-specific genes in human, fly, and zebrafish genomes are preferentially expressed in reproductive organs or tissues, such as testis (Clark et al., 2007; Levine et al., 2006; Tay et al., 2009; Yang et al., 2013). Moreover, divergent gene pairs in the human genome are enriched for housekeeping genes, such as DNA repair and DNA replication genes (Adachi and Lieber, 2002) that are actively transcribed in germ cells. In addition, the strand bias of G and T content correlates with germ cell but not somatic tissue gene expression levels (Majewski, 2003).
The model could explain the origin of divergent protein-coding gene pairs separated by less than 1 kb (usually less), which account for 10% of human protein-coding genes (Adachi and Lieber, 2002; Li et al., 2006; Piontkivska et al., 2009; Trinklein et al., 2004; Wakano et al., 2012; Xu et al., 2012), far higher proportion than would be expected if genes were randomly distributed in the genome. The model proposed here provides a natural explanation for the evolutionary origin of these gene pairs. It is likely that many more genes originated from divergent transcription, with the bidirectional organization having been disrupted by transposon insertion, recombination, or other genome rearrangement events. The model also predicts that divergent gene pairs commonly have unrelated functions, although they frequently might share co-expression. Except for a few cases, such as histone gene pairs and collagen gene pairs that are likely results of tandem duplication, the majority of divergent gene pairs in the human genome do not share higher functional similarity compared to random gene pairs (Li et al., 2006; Xu et al., 2012). For example, 35 of the 105 annotated DNA repair genes have bidirectional promoters, making DNA repair the most over-represented pathway for genes involved in bidirectional promoters, yet all 35 DNA repair genes are paired with non-DNA repair genes (Xu et al., 2012). Similarly, genes coding subunits of protein complexes are enriched in bidirectional pairs in human, yet none of these pairs code for two subunits of the same complex (Li et al., 2006). A similar observation has been reported for yeast and is consistent with the argument that the bidirectional conformation reduces expression noise and is not strongly selected for share functionality (Wang et al., 2011). The lack of functional relatedness is also illustrated by the parallel evolution of bidirectional promoters of RecQ helicases (Piontkivska et al., 2009). The five RecQ paralogs were duplicated early during metazoan evolution, yet all evolve to have divergent partners in human. However, these partner genes showed no functional or sequence similarity with each other (Piontkivska et al., 2009), suggesting parallel and independent origination of new genes from all five promoters.
Impact on genome organization and evolution
Divergent transcription likely facilitates the rearrangement events that reshape the genome, and also introduces unique features into genome organization, including the sharing of promoters, physical linkage in three-dimensional space, and co-expression of distal genes.
Although vertebrates share most of their genes, the genomic position and orientation of specific genes differ significantly due to genome rearrangement events, such as translocation, recombination, and duplication followed by the loss of the original copy. The survival of the gene or gene fragments at the new position can be facilitated by divergent transcription as discussed above. The role of divergent transcription in preserving the function of the new gene copy is likely significant, given that translocation preferentially occurs near active promoters (Chiarle et al., 2011; Klein et al., 2011). The correlation between transcription and translocation could potentially increase the chance that the translocated gene is still expressed and thus functional, therefore reducing the cost of translocation. For example, although ~40% of human protein coding genes can be traced back to fish, fewer than 7% (83/1262) of human bidirectional gene pairs are also bidirectional in the fish genome (Li et al., 2006), suggesting that most human bidirectional gene pairs formed with young genes, or by bringing together old genes through translocation facilitated by divergent transcription.
In addition to bidirectional organization, spatial and functional coupling between distal gene pairs would be introduced through new gene origination from enhancer transcription. Due to the tight coupling between gene transcription and enhancer transcription, an enhancer derived new gene will share a significant co-expression pattern with the old gene, despite the distance in the linear genome. Such coupled transcription of distal gene pairs brought together by chromatin interactions could contribute to the formation of transcription factories, nuclear foci where multiple genes are transcribed together without the requirement of shared function (Edelman and Fraser, 2012; Sutherland and Bickmore, 2009). The existence of transcription factories has been supported by increasing evidence, including in vivo live imaging (Ghamari et al., 2013) and chromatin interaction mapping (Li et al., 2012). These are probably related to super-enhancers where many genes that are coordinately expressed are associated with a common enhancer region (Lovén et al., 2013; Whyte et al., 2013). Overlaying comparative genomics analysis onto high-throughput chromatin interaction mapping data across multiple species (Dixon et al., 2012; Li et al., 2012) may help to reveal the evolutionary origin of transcription factories.
Conclusions
In conclusion, we propose that divergent transcription at promoters and enhancers results in changes of the transcribed DNA sequences that over evolutionary time drive new gene origination in the transcribed regions. Although the models proposed here are consistent with significant available data, systematic tests of these models await further advances such as in-depth characterization of additional genomes and experiments designed to test specific hypothesis. Over evolutionary times, genes formed through divergent transcription can be shuffled to other locations losing their evolutionary context. We envision future studies will uncover more functional surprises from divergent transcription, and illuminate how intergenic transcription is integrated into the cellular transcriptome.
Acknowledgments
We thank all the Sharp lab members, especially Andrea Kriz, Anthony Chiu, Albert Almada, Mohini Jangi, and Jesse Zamudio for comments, and Christopher Burge, Qifang Liu, Jianrong Wang, and Jeremy Wilusz for critical reading of the manuscript. This work was supported by United States Public Health Service grants RO1-GM34277 and R01-CA133404 from the National Institutes of Health (P.A.S.), partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute, and by Public Health Service research grant (GM-085319) from the National Institute of General Medical Sciences (C.B.B.). X.W. is a Howard Hughes Medical Institute International Student Research fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi N, Lieber MR. Bidirectional gene organization: A common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anamika K, Gyenis À, Poidevin L, Poch O, Tora L. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PloS One. 2012;7:e38769. doi: 10.1371/journal.pone.0038769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PK, Lykke-Andersen S, Jensen TH. Promoter-proximal polyadenylation sites reduce transcription activity. Genes & Development. 2012;26:2169–2179. doi: 10.1101/gad.189126.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. U1 snRNP Determines mRNA Length and Regulates Isoform Expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. Protogenes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M. New genes as drivers of phenotypic evolution. Nature Reviews. Genetics. 2013;14:645–660. doi: 10.1038/nrg3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cell Reports. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Molecular Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Current Opinion in Genetics & Development. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes & Development. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamari A, van de Corput MPC, Thongjuea S, van Cappellen WA, van Ijcken W, van Haren J, Soler E, Eick D, Lenhard B, Grosveld FG. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes & Development. 2013;27:767–777. doi: 10.1101/gad.216200.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Petrykowska HM, Elnitski L. Bidirectional Promoters as Important Drivers for the Emergence of Species-Specific Transcripts. Plos One. 2013;8:e57323. doi: 10.1371/journal.pone.0057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Green ED, Progr NCS. Transcription-associated mutational asymmetry in mammalian evolution. Nature Genetics. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen JWG, Vaandrager JW, Heuser T, Jauch A, Kluin PM, Geelen E, Bergsagel PL, Kuehl WM, Drexler HG, Otsuki T, et al. Concurrent activation of a novel putative transforming gene, myeov, and cyclin D1 in a subset of multiple myeloma cell lines with t(11;14)(q13;q32) Blood. 2000;95:2691–2698. [PubMed] [Google Scholar]
- Janssen JWG, Cuny M, Orsetti B, Rodriguez C, Valles H, Bartram CR, Schuuring E, Theillet C. MYEOV: A candidate gene for DNA amplification events occurring centromeric to CCNDI in breast cancer. International Journal of Cancer. 2002;102:608–614. doi: 10.1002/ijc.10765. [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–U81. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Saffery R. Inherent promoter bidirectionality facilitates maintenance of sequence integrity and transcription of parasitic DNA in mammalian genomes. Bmc Genomics. 2009;10:498. doi: 10.1186/1471-2164-10-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Molecular Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife. 2013;2:e00808–e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O’Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nature Structural Biology. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science (New York, NY) 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden J, Murray D, Moss A, Arumuguma M, Doyle E, McEntee G, O’Keane C, Doran P, MacMathuna P. Net1 and Myeov: computationally identified mediators of gastric cancer. British Journal of Cancer. 2006;94:1204–1212. doi: 10.1038/sj.bjc.6603054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes & Development. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- Li XL, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yu H, Guo ZM, Guo TQ, Tu K, Li YX. Systematic analysis of head-to-head gene organization: Evolutionary conservation and potential biological relevance. Plos Computational Biology. 2006;2:687–697. doi: 10.1371/journal.pcbi.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Vankuren NW, Chen S, Vibranovski MD. New Gene Evolution: Little Did We Know. Annual Review of Genetics. 2013 doi: 10.1146/annurev-genet-111212-133301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. American Journal of Human Genetics. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Green P. Genomic signatures of germline gene expression. Genome Research. 2010;20:1503–1511. doi: 10.1101/gr.106666.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Moss AC, Lawlor G, Murray D, Tighe D, Madden SF, Mulligan AM, Keane CO, Brady HR, Doran PP, MacMathuna P. ETV4 and Myeov knockdown impairs colon cancer cell line proliferation and invasion. Biochemical and Biophysical Research Communications. 2006;345:216–221. doi: 10.1016/j.bbrc.2006.04.094. [DOI] [PubMed] [Google Scholar]
- Mugal CF, von Gruenberg HH, Peifer M. Transcription-Induced Mutational Strand Bias and Its Effect on Substitution Rates in Human Genes. Molecular Biology and Evolution. 2009;26:131–142. doi: 10.1093/molbev/msn245. [DOI] [PubMed] [Google Scholar]
- Nott A, Muslin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. Rna-a Publication of the Rna Society. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nature Structural & Molecular Biology. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Park C, Qian WF, Zhang JZ. Genomic evidence for elevated mutation rates in highly expressed genes. Embo Reports. 2012;13:1123–1129. doi: 10.1038/embor.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A Genome-wide siRNA Screen Reveals Diverse Cellular Processes and Pathways that Mediate Genome Stability. Molecular Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska H, Yang MQ, Larkin DM, Lewin HA, Reecy J, Elnitski L. Cross-species mapping of bidirectional promoters enables prediction of unannotated 5′ UTRs and identification of species-specific transcripts. Bmc Genomics. 2009;10:189. doi: 10.1186/1471-2164-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Arndt PF. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Research. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Querfurth R, Arndt PF. The evolution of transcription-associated biases of mutations across vertebrates. Bmc Evolutionary Biology. 2010;10:187. doi: 10.1186/1471-2148-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA Exosome Depletion Reveals Transcription Upstream of Active Human Promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, Jensen TH. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Research. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes & Development. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes & Development. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual Review of Biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nature Reviews Genetics. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A Large Fraction of Extragenic RNA Pol II Transcription Sites Overlap Enhancers. Plos Biology. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent Transcription from Active Promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle (Georgetown, Tex) 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The Cellular EJC Interactome Reveals Higher-Order mRNP Structure and an EJC-SR Protein Nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nature Reviews Genetics. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. Gene loops enhance transcriptional directionality. Science (New York, NY) 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay SK, Blythe J, Lipovich L. Global discovery of primate-specific genes in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12019–12024. doi: 10.1073/pnas.0904569106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Research. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakano C, Byun JS, Di LJ, Gardner K. The dual lives of bidirectional promoters. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms. 2012;1819:688–693. doi: 10.1016/j.bbagrm.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GZ, Lercher MJ, Hurst LD. Transcriptional Coupling of Neighboring Genes and Gene Expression Noise: Evidence that Gene Orientation and Noncoding Transcripts Are Modulators of Noise. Genome Biology and Evolution. 2011;3:320–331. doi: 10.1093/gbe/evr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Jarvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends in Genetics. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & Development. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang R, Wei L, Li CY. Hominoid-Specific De Novo Protein-Coding Genes Originating from Long Non-Coding RNAs. Plos Genetics. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Chen J, Shen B. The preservation of bidirectional promoter architecture in eukaryotes: what is the driving force? BMC Systems Biology. 2012;6(Suppl 1):S21. doi: 10.1186/1752-0509-6-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zou M, Fu B, He S. Genome-wide identification, characterization, and expression analysis of lineage-specific genes within zebrafish. Bmc Genomics. 2013;14:65. doi: 10.1186/1471-2164-14-65. [DOI] [PMC free article] [PubMed] [Google Scholar]