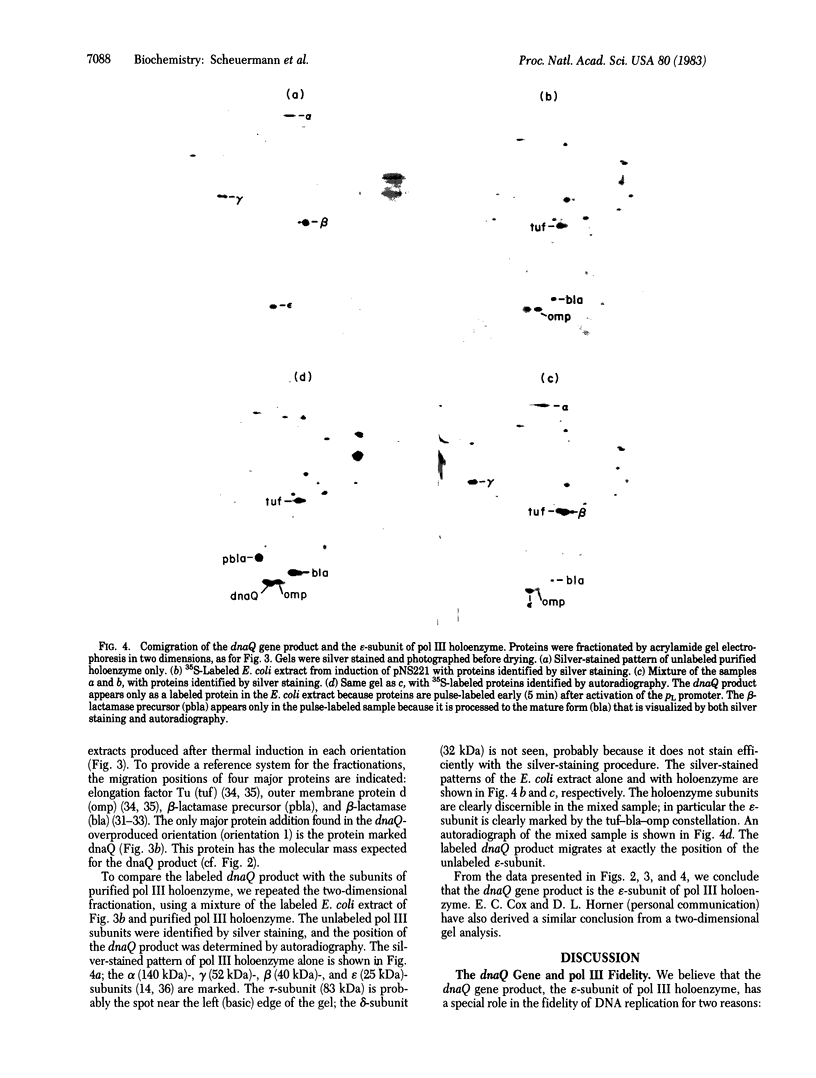
Abstract
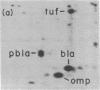
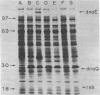
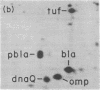
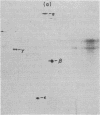
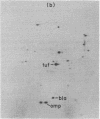
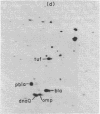
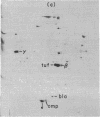
Based on extensive genetic and biochemical studies, the multisubunit DNA polymerase III holoenzyme is considered responsible for the chain-elongation stage in replication of the genome of Escherichia coli and is thus expected to be the major determinant of fidelity as well. Previous experiments have shown that two mutations conferring a very high mutation rate on E. coli, mutD5 and dnaQ49, decrease severely the 3' leads to 5' exonucleolytic editing activity of the polymerase III holoenzyme. To identify more precisely the nature of these mutations, we have carried out genetic mapping and complementation experiments. From these studies and experiments by others, we conclude that the most potent general mutator mutations in E. coli occur in a single gene, dnaQ. To define further the role of the dnaQ gene, we have used two-dimensional gel electrophoresis to compare the labeled dnaQ gene product with purified polymerase III holoenzyme. The dnaQ product comigrates with the epsilon-subunit, a 25-kilodalton protein of the polymerase III "core" enzyme. We conclude that the epsilon-subunit of polymerase III holoenzyme has a special role in defining the accuracy of DNA replication, probably through control of the 3' leads to 5' exonuclease activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Scott G. K. Partial amino acid sequence of penicillinase coded by Escherichia coli plasmid R6K. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3732–3736. doi: 10.1073/pnas.75.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner M., Vapnek D. Versatile cloning vectors derived from the runaway-replication plasmid pKN402. Gene. 1981 Dec;15(4):319–329. doi: 10.1016/0378-1119(81)90175-x. [DOI] [PubMed] [Google Scholar]
- Bloch P. L., Phillips T. A., Neidhardt F. C. Protein identifications of O'Farrell two-dimensional gels: locations of 81 Escherichia coli proteins. J Bacteriol. 1980 Mar;141(3):1409–1420. doi: 10.1128/jb.141.3.1409-1420.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L. Dominant mutators in Escherichia coli. Genetics. 1982 Jan;100(1):7–18. doi: 10.1093/genetics/100.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L. Structure and coding properties of a dominant Escherichia coli mutator gene, mutD. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2295–2299. doi: 10.1073/pnas.80.8.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen G. E., Cox E. C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974 Feb;117(2):477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. Comparative rates of spontaneous mutation. Nature. 1969 Mar 22;221(5186):1132–1132. doi: 10.1038/2211132a0. [DOI] [PubMed] [Google Scholar]
- Echols H., Lu C., Burgers P. M. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. Mutation rate: some biological and biochemical considerations. Biochimie. 1982 Aug-Sep;64(8-9):571–575. doi: 10.1016/s0300-9084(82)80089-8. [DOI] [PubMed] [Google Scholar]
- Echols H. SOS functions, cancer and inducible evolution. Cell. 1981 Jul;25(1):1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Erlich H. A., Cox E. C. Interaction of an Escherichia coli mutator gene with a deoxyribonucleotide effector. Mol Gen Genet. 1980;178(3):703–708. doi: 10.1007/BF00337881. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Maruyama M., Sekiguchi M. Identification of the dnaQ gene product and location of the structural gene for RNase H of Escherichia coli by cloning of the genes. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3770–3774. doi: 10.1073/pnas.78.6.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Sekiguchi M. A new conditional lethal mutator (dnaQ49) in Escherichia coli K12. Mol Gen Genet. 1978 Jul 25;163(3):277–283. doi: 10.1007/BF00271956. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Knight D. M., Das A., Miller H. I., Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982 Dec;31(3 Pt 2):565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- Konrad E. B. Isolation of an Escherichia coli K-12 dnaE mutation as a mutator. J Bacteriol. 1978 Mar;133(3):1197–1202. doi: 10.1128/jb.133.3.1197-1202.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Botstein D. Evidence for posttranslational translocation of beta-lactamase across the bacterial inner membrane. Cell. 1982 Oct;30(3):893–902. doi: 10.1016/0092-8674(82)90294-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lathe R. Fine-structure mapping of the firA gene, a locus involved in the phenotypic expression of rifampin resistance in Escherichia. J Bacteriol. 1977 Sep;131(3):1033–1036. doi: 10.1128/jb.131.3.1033-1036.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. M., Richardson C. C. Deoxyribonucleic acid polymerase III of Escherichia coli. Characterization of associated exonuclease activities. J Biol Chem. 1975 Jan 25;250(2):470–478. [PubMed] [Google Scholar]
- Loeb L. A., Kunkel T. A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Maki H., Horiuchi T., Sekiguchi M. Isolation of conditional lethal mutator mutants of Escherichia coli by localized mutagenesis. J Bacteriol. 1983 Mar;153(3):1361–1367. doi: 10.1128/jb.153.3.1361-1367.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M., Horiuchi T., Maki H., Sekiguchi M. A dominant (mutD5) and a recessive (dnaQ49) mutator of Escherichia coli. J Mol Biol. 1983 Jul 15;167(4):757–771. doi: 10.1016/s0022-2836(83)80109-0. [DOI] [PubMed] [Google Scholar]
- McHenry C. S., Crow W. DNA polymerase III of Escherichia coli. Purification and identification of subunits. J Biol Chem. 1979 Mar 10;254(5):1748–1753. [PubMed] [Google Scholar]
- McHenry C., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem. 1977 Sep 25;252(18):6478–6484. [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978 May;14(1):179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Reyes O., Gottesman M., Adhya S. Formation of lambda lysogens by IS2 recombination: gal operon--lambda pR promoter fusions. Virology. 1979 Apr 30;94(2):400–408. doi: 10.1016/0042-6822(79)90470-7. [DOI] [PubMed] [Google Scholar]
- Sevastopoulos C. G., Glaser D. A. Mutator action by Escherichia coli strains carrying dnaE mutations. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3947–3950. doi: 10.1073/pnas.74.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanos A., Sedgwick S. G., Yarranton G. T., Hübscher U., Banks G. R. Detection of the catalytic activities of DNA polymerases and their associated exonucleases following SDS-polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Apr 24;9(8):1825–1839. doi: 10.1093/nar/9.8.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. M., McHenry C. S. Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol. 1982 Oct;152(1):351–356. doi: 10.1128/jb.152.1.351-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Cox E. C., Horn V. The unusual mutagenic specificity of an E. Coli mutator gene. Proc Natl Acad Sci U S A. 1966 Feb;55(2):274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]