Abstract
The past decade has brought together substantial advances in human genome analysis and a maturation of understanding of tumor biology. While there is much progress still to be had, there are now several prominent examples in which tumor-associated somatic mutations have been used to identify cellular signaling pathways in tumors. This in turn has led to the development of targeted therapies, with somatic mutations serving as genomic predictors of tumor response and providing new leads for drug development. There is also a realization that germline DNA variants can help optimize cancer drug dosing and predict the susceptibility of patients to the adverse side effects of these drugs; knowledge that ultimately can be used to improve the benefit:risk ratio of cancer treatment for individual patients.
Mechanistic understanding of the biologic pathways regulating human cancers and the normal cells from which they are derived has long influenced the management of cancer. These efforts have shifted from older, cytotoxic therapeutic options toward chemical and biologic therapies that are precisely designed to target a critical gene or pathway. This has delivered a degree of tumor control for common cancers, including breast, lung, colorectal, and extended life and provided cures in some cases of less common cancers, such as testicular cancer and childhood acute lymphoblastic leukemia. Pathway-driven therapeutics has significantly improved the outcomes of chronic myelogenous leukemia and gastrointestinal stromal tumors which may, in the absence of relapse, act as chronic diseases requiring life-long treatment, akin to diabetes or hypertension management.(1) However, these advances have come at a cost, both literally and figuratively, with newer treatments often costing thousands of dollars per month and associated with toxicities that can negatively affect patient quality of life.
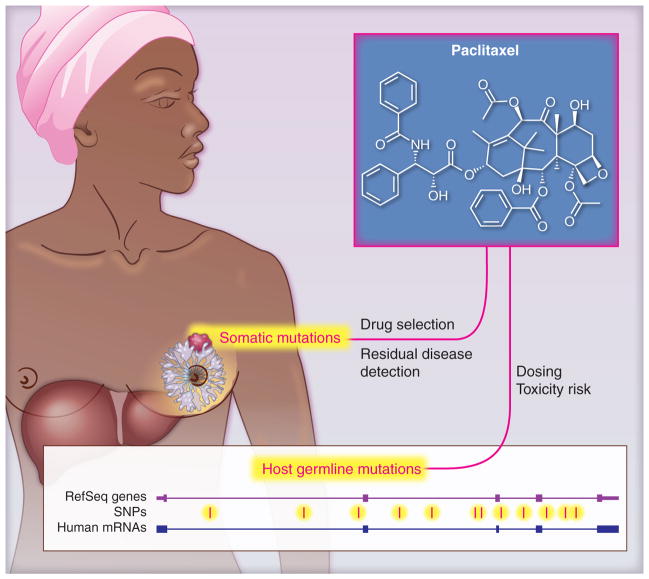
Somatic mutations, variations found within the tumor, and germ-line mutations, heritable variations found within the individual, may influence disease outcome and/or response to therapy (Figure 1). These mutations, or cancer biomarkers, can be broadly classified as prognostic markers, those mainly associated with the course or outcome of a disease, or predictive markers, which can be used to identify subpopulations of patients who are most likely to respond to a given therapy. There is opportunity for genetic information to aid both the selection of effective therapy and the avoidance of treatments with an unacceptable risk of adverse drug reactions (ADR).
Figure 1. Attention must be paid to both tumor and host.
cancer pharmacogenomic variation in both the tumor (somatic changes) and normal tissues (germline variants) influence the treatment of cancer patients.
Inherited differences in drug effects were first documented in terms of drug metabolism in the 1950s,(2, 3) giving rise to the term “pharmacogenetics.” The field has now extended to all aspects of drug disposition, including absorption, distribution, and excretion(4), as well as drug targets and downstream effect mediators. Table 1 outlines some current examples where genotype is used for the selection of cancer chemotherapy.
Table 1.
Pharmacogenomic DNA markers in clinical use for chemotherapy or supportive care of cancer patients
| Germline | Somatic | Drugs | Effect |
|---|---|---|---|
| Thiopurine methyltransferase | -------- | Mercaptopurine, Thioguanine | Neutropenia risk |
| UDP-glucuronosyltrans ferase 1A1 | -------- | Irinotecan, Nilotinib | Neutropenia risk; underdosing risk |
| Glucose-6-phosphate dehydrogenase | -------- | Rasburicase | Anemia |
| Cytochrome P450 2D6 | -------- | Codeine, oxycodone; Tamoxifen | Altered pain control; Altered tumor control |
| -------- | Janus Kinase 2 (JAK2) | Ruxolitinib | Altered drug activity |
| -------- | Human Epidermal Growth Factor Receptor 1 (EGFR) | Cetuximab Erlotinib Gefitinib Panitumumab |
Altered drug activity |
| -------- | Kirsten rat sarcoma viral oncogene homolog (KRAS) | Cetuximab Panitumumab |
Lack of drug activity |
| Abelson murine leukemia viral oncogene homolog 1 (ABL) | Imatinib, Dasatinib, Nilotinib | Altered drug activity | |
| -------- | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT) | Imatinib | Altered drug activity |
| -------- | Human Epidermal Growth Factor Receptor 2 (HER2) | Lapatinib Trastuzumab | Enhanced drug activity |
| -------- | v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) | Vemurafenib | Enhanced drug activity |
| -------- | Anaplastic lymphoma receptor tyrosine kinase (ALK) | Crizotinib | Altered drug activity |
Tumor profiling: from discovery science to patient management
Analysis of tumor DNA to guide patient treatment has been used for over 20 years. An acute lymphoblastic leukemia patient with the presence of a 9:22 chromosomal translocation was once offered bone marrow transplantation, rather then standard cytotoxic chemotherapy; more recently these patients would be offered imatinib or other ABL tyrosine kinase inhibitors. A breast cancer patient with amplification of HER2 might be treated with the antiHER2 monoclonal antibody trastuzumab or the HER2 tyrosine kinase inhibitor lapatinib. Thus, focused profiling is becoming part of routine patient management for select cancers (Table 1) as the lowered costs of high quality DNA sequencing have led to the identification that some somatic mutations are associated with specific benefits (or lack thereof) from targeted therapies.
Somatic DNA mutation assessment has positively impacted patient care for a focused number of cancers. The identification of KRAS mutations in codons 12 or 13 in ~30% of patients with colon cancer suggests that there is no tumor control benefit, but instead some toxicity risk, when patients are treated with expensive antibodies targeting EGFR.(5) Lung cancer, melanoma, and myeloproliferative disorders tend to be sensitive to tyrosine kinase inhibiutors with mutations in the respective genes EGFR, BRAF, and JAK2. However, currently there are molecular predictors of efficacy for less than 10% of the FDA approved cancer drugs.
In addition, cancer cells may mutate and evolve resistance to specific drug treatments resulting in the proliferation of drug resistant tumors. Beyond the initial patient treatment setting, there is a lack of personalized cancer medicine trial data on which to guide patient management decisions. Treatment choices often revert to the use of population average data, where it is difficult to ascertain the value of therapy for an individual patient. We still need definitive discovery and validation/replication efforts for anticancer drugs (old and new). This is particularly true for the older cytotoxic agents, which benefit a meaningful subset of patients, but do not have the diverse scientific and financial advocacy to assure that genomic knowledge is being discovered and deployed in a clinically relevant manner.
As sequencing strategies mature and costs are lowered, there has been an increase in the application of these technologies to tumor profiling.(6, 7) While the current focus is principally directed towards the identification of somatic DNA mutations, cancer may be associated with epigenetic traits including specific miRNAs, variations in RNA expression, methylation patterns and chromatin marks. Currently the most common genetic screening involves performing a targeted DNA capture, focused on a limited number of relevant candidate genes, followed by sequencing. (8) This gives a clinical report that may direct treatment to a signaling pathway that would not be of obvious importance from tumor histology.
Genomic medicine strategies that can identify and clinically annotate the broad assortment of actionable variants are needed to justify these efforts. An initial deep sequencing of 145 genes in colorectal and non-small cell lung cancers found somatic mutations in 39/40 (98%) and 20/24 (83%) of tumors, respectively.(8) More than half (52.5%) of colorectal cancers and 72% of non-small cell lung cancers contained at least one mutation that has been linked to a specific chemotherapy approach.(8) Similar data has come from the NCI/NHGRI Cancer Genome Atlas efforts across tumors from diverse anatomical locations.(6, 7)
Clinical pharmacogenomic efforts to apply deep sequencing to unveil mechanisms of sensitivity or resistance to drug therapy are needed; as we do not know the mechanism of clinical resistance for most anticancer drugs. Sequencing of non-small cell lung cancer which displayed sensitivity and subsequent resistance to EGFR tyrosine kinase inhibitors led to the routine use of EGFR sequencing to guide therapeutic choices.(9) Whole exome sequencing of patients treated with everolimus for advanced bladder cancer revealed that a specific TSC1 mutation correlated with everolimus sensitivity. Patients with TSC1 mutation had a longer time until recurrence of tumor (4.1 versus 1.8 months, 11). This loss of function mutation in TSC1 was subsequently found in 5/96 (5.2%) of advanced bladder cancers, suggesting that there is a subgroup of patients with this disease for whom everolimus treatment might offer substantial benefit.
There are limits to how much tissue can be acquired from a clinical biopsy. The practical issue of low availability of high quality tumor DNA is helping drive analysis from single gene assays to multigene applications, where more knowledge is derived from the existing tissue. Also, quality control issues, resulting in uncertain or erroneous identification of mutations from the use of gene panels or whole genome assessment, may challenge interpretations of molecular diagnostic results across clinical laboratories. Furthermore, we need predictive analyses for the 25–80% of cases where variants of unknown significance are identified in genes that are of interest to a particular tumor. This has been evident in BRCA1/2 testing, associated with breast cancers. We need to generate both laboratory and clinical consensus methods to decide which variants in which genes merit clinical action.(10)
For certain tumor types clinical trial inclusion criteria are starting to focus less on the anatomical origins and more on the somatic mutations identified within a tumor. The focus on “driver” mutations controlling tumor invasiveness and its relative therapeutic response requires screening many patients to find the few that are eligible for a targeted therapy trial.(11) This is not easily implemented at academic centers as currently a disproportionate load of patients are referred for experimental therapy without a previously ascertained molecular profile. This introduces additional time and unsupported expense for somatic sequencing before a transition into a treatment trial of relevance.
There are also unanticipated issues, such as the current preference for fresh tissue, requiring a tissue biopsy for the purposes of the somatic profile. Many clinical practice settings do not have ready access to interventional radiologists for safe biopsy of tumor, nor do they have personnel trained to properly handle tissue to best retain a tumor’s molecular signature (most tissues from community oncologists are placed in a formalin-containing fixative and sent to an outside pathologist). Sequencing efforts targeted at formalin-fixed paraffin embedded tissue and nucleic acid detection in plasma or identification of circulating tumor cells may also provide a means to circumvent some of these barriers. Well-designed practical infrastructure are needed for the application of personalized cancer medicine to ensure that all the right team members will be trained and ready to provide patient support. Commercial and academic efforts that focus on disease-specific gene targets, such as the National Comprehensive Cancer Network or Foundation Medicine, are actively developing the pathways for application in patient management.(12) As the costs decrease and the interpretation ability increases, somatic DNA assessment will become a routine part of the management of cancer.
The role of germline DNA in optimizing dosing and identifying toxicity risk of anti-cancer drugs
While the primary focus of cancer genomic research is on the somatically mutated genes driving tumor growth, inter-patient germline variation can also impact cancer treatment. Indeed, given that most of oncology supportive care is targeted towards mollifying a patient’s adverse effects of cancer treatment while eradicating the cancer, genetic variation can potentially play an important role in the selection and administration of cancer drugs.
There is also an undervalued role of gastrointestinal drug transport and hepatic drug metabolism on the dose, administration schedule, and route of administration of a drug. The use of a highly targeted, effective therapy for chronic myelogenous leukemia can be undone by under dosing, if the drug is metabolized or removed before it encounters the intended cell or molecular target, setting up a milieu for development of drug resistance.(13) Pharmacogenomic variation in drug metabolism has been shown to have a role in the efficacy of certain anticancer therapies such as tamoxifen, an effective anti-estrogen used in the treatment of hormone receptor positive breast cancer. Bioconversion of tamoxifen to several active metabolites including endoxifen, its most abundant active metabolite, is primarily dependent on the highly polymorphic cytochrome P450 2D6 (CYP2D6) enzyme. In the past, tamoxifen was often co-administered with certain antidepressants for treatment of antiestrogen-induced hot flashes until it was discovered that the antidepressants blocked CYP2D6 and the production of endoxifen.(14) Coadministration was abruptly discontinued in clinical practice, because of the risk that the ‘cure’ of hot flashes was compromising the effectiveness of the antiestrogen therapy.(15) Clinical trials of endoxifen as a biomarker for tamoxifen response are now underway.(16)
In addition, approximately 7% of the USA population have a genetic polymorphism or deletion in CYP2D6, resulting in diminished protein levels and/or function. Over 20 published studies have reported an association between CYP2D6 polymorphisms and breast cancer outcomes after tamoxifen treatment, although several recent studies suggest that homozygote variant patients that have no or lower function of CYP2D6 have the poorest outcome.(17) Studies to determine the appropriate dose of tamoxifen for wild-type patients (called extensive metabolizers), and the heterozygous patients (intermediate metabolizers), suggest that the doubling of tamoxifen dose in intermediate metabolizers normalized plasma endoxifen levels to that observed in extensive metabolizers.(18, 19) This level of data is consistent with that usually required for FDA prescribing recommendations for dose-adjustment after organ dysfunction, drug interaction, or age and suggests a relevance to CYP2D6-guided tamoxifen dosing as a routine part of reducing both interpatient variation and the risk of underdosing patients with breast cancer.
In most discussions of clinical management of cancer patients the risk/benefit assessment focuses is on the probability of tumor control from a specific drug, in part because there is a deficiency, of objective data from which one can assess the patients’ risk of developing severe adverse drug effects. This deficiency has prompted the launch of genomic discovery programs that focus on adverse effects of cancer drugs such as sensory peripheral neuropathy, cardiotoxicity, hearing loss, and other toxicities.(2, 20–23) For example, the microtubule inhibitor paclitaxel is used to treat breast, lung, and ovarian cancers, but paclitaxel-induced neuropathy is a common adverse event that often leads to therapeutic disruption and patient discomfort.(24) However, there are currently no mechanisms for prospective identification of patients at heightened risk for neuropathy, for whom choices of drug, administration schedule, and quality of life could be informed.
Both candidate gene and genome-wide association studies have begun to inform the prediction of patient risk for neuropathy. A genome-wide association study involved 855 subjects of European ancestry where paclitaxel was administered as part of therapy for lymph node negative breast cancer. This study identified a single nucleotide polymorphism in the FGD4 gene, which encodes FGD1-related F-actin binding protein, that was associated with the onset of sensory peripheral neuropathy in the discovery cohort [Hazard ratio = 1.57; 95% CI 1.30–1.91] and was also observed in a European and African American replication cohort.(20) As FGD4 is a congenital peripheral neuropathy gene, there is biologic plausibility to further assess the contribution of genetic variation to the development of peripheral neuropathy.
Another study identified, a near doubling of increased risk of paclitaxel-induced neuropathy related to CYP2C8*3 status in breast cancer patients.(21) Although CYP2C8*3 is less common in African-Americans, a significant association was replicated in direction and magnitude of effect. The observation of increased risk of paclitaxel-induced neuropathy in patients who carry the CYP2C8*3 variant across racially distinct patient cohorts suggests that both pharmacokinetic variables (such as CYP2C8) and biologic variation (such as FGD4) contribute to patient risk of neuropathy.
The avoidance of chemotherapy-associated morbidity is critical in the context of adjuvant chemotherapy, where the goal is to kill any stray cancer cells that might have gone undetected. This is especially important in the treatment of childhood malignancies, as many patients will survive their cancer and experience the sequelae from adverse drug events for many years. Cardiomyopathy from anthracycline chemotherapy is a devastating morbidity, with long-term effects on patient productivity and quality of life. Germline variation associated with risk of cardiomyopathy has been identified by means of distinct candidate gene strategies and different patient recruitment strategies. Similar data on germline pharmacogenomics predictors of cisplatin-associated damage to hearing ototoxicity has also been reported, providing a mechanism for prospective identification of patients at risk for this debilitating morbidity.(22)
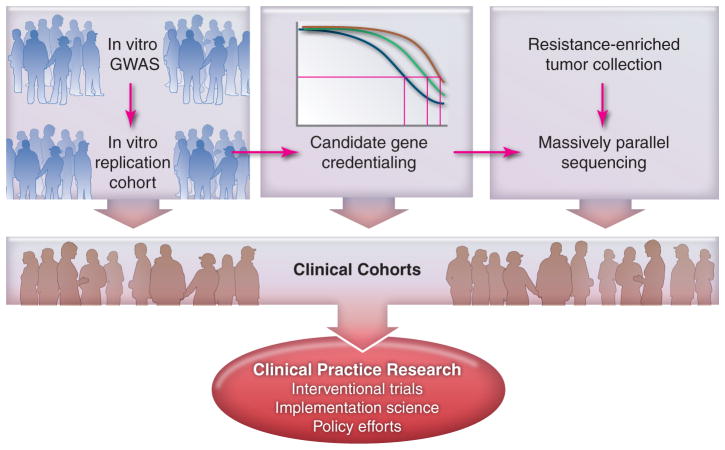
There are significant limitation to pharmacogenomics discovery for anticancer therapies, including the challenges of building large patient cohorts for both discovery and validation purposes. It often takes 7–10 years to construct, conduct, and analyze a clinical trial, which can then be used for pharmacogenomics discovery. The same is true for a validation cohort, which is one of the reasons there are so few discovery and replication studies in the literature. There is also a paucity of information on the heritability of anticancer drug effects, to help justify the quest for genomic solutions to variability in drug effect. One approach is the recent use of cell lines from large, multigeneration families, which have shown a wide variation in heritability of cytotoxicity (10–70%) across 29 commonly prescribed anticancer drugs, with 66% having greater than 30% heritability.(25) This presents an opportunity for both prioritization of drugs for assessment and conduct of ex vivo discovery that will allow precious clinical material to be used for validation studies. The application of bar coding and robotics have allowed the scale up of cell line phenotyping to 500–1000 cell lines per project, followed by ex vivo genome-wide association studies.(25–27) Innovative pharmacogenomics strategies will allow us to more rapidly capture the relevance of genomic information for rational drug therapy selection (Figure 2).
Figure 2. A multidimensional strategy is required.
A blend of in vitro, ex vivo, and in vivo strategies are needed to more rapidly move the promise of pharmacogenomics into application.
Moving into clinical practice
Both replication and validation of pharmacogenomics traits raise challenges. It is often difficult to characterize, uniformly treat, and systematically evaluate patients in order to objectively quantify the drug response phenotype. The standard of care should be to obtain genomic DNA from all patients entered into clinical drug trials, along with appropriate consent to permit pharmacogenetic studies. This is now accomplished in most large trials being conducted by pharmaceutical companies and is routine for some of the NCI clinical trials groups,(20, 28, 29) but has not yet become standard for academic or foundation-supported trials (Figure 2).
The challenge is to balance the desire to apply new information and the need to make sure that there are robust data supporting the idea that acting on a pharmacogenomic marker is in the best interest of the patient. The reliance on prospective, randomized, controlled trials as the only way to justify clinical implementation is not practical and guarantees that new information will have a 5–10 year lag while studies are constructed, conducted, and interpreted. There is also a disconnection between the funding bodies and the prioritization of this type of studies, in terms of financial commitment, clinical trial infrastructure, and ability to rapidly enact new strategies. There have been several efforts to develop ways to gain confidence in early adoption of pharmacogenomics data, on the basis of consensus-building among institutions around the application of genetic information to drug therapy. One such effort is the Clinical Pharmacogenetics Implementation Consortium (CPIC), which includes participants from >80 institutions across 4 continents.(30) A key element to programs such as CPIC is the realization that there are some aspects of the medical decision process, such as drug dosing, that have robust data on which to benefit patients, even as the field waits for the ‘perfect’ studies that definitively guide therapy at a broader level.
Double standards still exist in clinical decision making, in that a drug interaction may be accepted as a credentialed, clinically relevant variable and rapidly integrated into clinical practice, yet the application of genetic data through the exact same mechanism is delayed due to the need for accumulation of large amounts of prospective data. This is occurring for CYP2D6 and tamoxifen, CYP3A4 and taxane chemotherapy, and related interactions for supportive care medications. The standard for drug interaction is influenced by years of familiarity and no additional time or expense to the patient, but has less functional predictability than gene deletion in the same pathway. There is a need to come up with a framework whereby any source of variation in a clinically credentialed pathway can be moved toward clinical implementation.
The endpoints of pharmacogenomics studies have followed a traditional biomarker scheme, trying to explain untoward events, identify low utility, define dose selection, or preemptively predict severe drug reactions. These are important endpoints and should not be neglected in investigational endeavors. However, there are alternate endpoints that are typically considered too mundane for inclusion in NIH grants, but are major drivers of early adoption for new health care modalities. These include avoidance of 30 day readmission rates, economics of ‘bundled care’, and the prioritization of medication access by a health system Pharmacy & Therapeutics committee. These endpoints are often accessible through observational cohorts or electronic health record studies and will likely drive the implementation of pharmacogenomics into practice.
It is time to be more practical as we move forward. Although substantial progress has been made in identifying and characterizing pharmacogenetic phenomena, translation of these data into practical clinical applications remains slow. A variety of factors contribute to this problem, including a lack of clarity on the amount of data needed to prove clinical utility, the paucity of interventional pharmacogenetic studies, and unresolved practical considerations, such as how to establish and implement clear guidelines in departments that manage cancer. There are also societal factors at play, including acceptance of widespread genetic testing as well as implications for insurance coverage and liability. These issues will need to be explored and addressed before the promise of genetically customized medicine can become reality.
Acknowledgments
Supported in part by National Institutes of Health grants UL1 RR025747, P01CA142538, R01 CA161608, and R01HL110380. The author serves on the NIH NHGRI Advisory Council, FDA Clinical Pharmacology Committee, Coriell Institute Pharmacogenomics Advisory Committee, and is a Scientific Advisor to Gentris Corporation. This work is dedicated to my father, whose death from esophageal carcinoma during the writing of this review was a poignant reminder of how far we still have to go in the fight against cancer.
References
- 1.Lee SY, McLeod HL. J Pathol. 2011 Jan;223:15. doi: 10.1002/path.2766. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, McLeod HL, Weinshilboum RM. N Engl J Med. 2011 Mar 24;364:1144. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalow W. Lancet. 1956;2:576. [Google Scholar]
- 4.Meyer UA. Lancet. 2000 Nov 11;356:1667. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 5.Amado RG, et al. J Clin Oncol. 2008 Apr 1;26:1626. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 6.Hammerman PS, et al. Nature. 2012 Sep 27;489:519. [Google Scholar]
- 7.Network CGA. Nature. 2012 Jul 19;487:330. [Google Scholar]
- 8.Lipson D, et al. Nat Med. 2012 Mar;18:382. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paez JG, et al. Science. 2004 Jun 4;304:1497. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 10.Berg JS, et al. Genet Med. 2012 Sep 20; [Google Scholar]
- 11.Kreso A, et al. Science. 2013 Feb 1;339:543. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engstrom PF, et al. J Natl Compr Canc Netw. 2011 Dec;9(Suppl 6):S1. doi: 10.6004/jnccn.2011.0138. [DOI] [PubMed] [Google Scholar]
- 13.Eechoute K, et al. Clin Cancer Res. 2012 Oct 15;18:5780. doi: 10.1158/1078-0432.CCR-12-0490. [DOI] [PubMed] [Google Scholar]
- 14.Stearns V, et al. J Natl Cancer Inst. 2003 Dec 3;95:1758. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 15.Kelly CM, et al. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad A, et al. Clin Pharmacol Ther. 2010 Dec;88:814. doi: 10.1038/clpt.2010.196. [DOI] [PubMed] [Google Scholar]
- 17.Hertz DL, McLeod HL, Irvin WJ., Jr Oncologist. 2012;17:620. doi: 10.1634/theoncologist.2011-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irvin WJ, Jr, et al. J Clin Oncol. 2011 Aug 20;29:3232. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walko CM, McLeod H. Pharmacogenomics. 2012 Apr;13:691. doi: 10.2217/pgs.12.27. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin RM, et al. Clin Cancer Res. 2012 Sep 15;18:5099. doi: 10.1158/1078-0432.CCR-12-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertz DL, et al. Breast Cancer Res Treat. 2012 Jul;134:401. doi: 10.1007/s10549-012-2054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross CJ, et al. Nat Genet. 2009 Dec;41:1345. doi: 10.1038/ng.478. [DOI] [PubMed] [Google Scholar]
- 23.Visscher H, et al. J Clin Oncol. 2012 May 1;30:1422. doi: 10.1200/JCO.2010.34.3467. [DOI] [PubMed] [Google Scholar]
- 24.McWhinney SR, Goldberg RM, McLeod HL. Mol Cancer Ther. 2009 Jan;8:10. doi: 10.1158/1535-7163.MCT-08-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters EJ, et al. Pharmacogenomics. 2011 Oct;12:1407. doi: 10.2217/pgs.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown CC, et al. Pharmacogenet Genomics. 2012 Nov;22:796. doi: 10.1097/FPC.0b013e3283589c50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox NJ, Gamazon ER, Wheeler HE, Dolan ME. Clin Pharmacol Ther. 2012 Oct;92:425. doi: 10.1038/clpt.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratain MJ, et al. Clin Cancer Res. 2006 Jun 1;12:3612s. doi: 10.1158/1078-0432.CCR-06-9008. [DOI] [PubMed] [Google Scholar]
- 29.Innocenti F, et al. Clin Cancer Res. 2012 Jan 15;18:577. doi: 10.1158/1078-0432.CCR-11-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relling MV, Klein TE. Clin Pharmacol Ther. 2011 Mar;89:464. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]