Summary
Protein conformation dictates a great deal of protein function. A class of naturally unstructured proteins, termed Intrinsically Disordered Proteins (IDPs), demonstrates that flexibility in structure can be as important mechanistically as rigid structure. At the core of the circadian transcription/translation feedback loop in Neurospora crassa is the protein Frequency (FRQ), shown here shown to share many characteristics of IDPs. FRQ in turn binds to Frequency Interacting RNA Helicase (FRH), whose clock function has been assumed to relate to its predicted helicase function. However, mutational analyses reveal that the helicase function of FRH is not essential for the clock, and a region of FRH distinct from the helicase region is essential for stabilizing FRQ against rapid degradation via pathway distinct from its typical ubiquitin-mediated turnover. These data lead to the hypothesis that FRQ is an IDP and that FRH acts nonenzymatically, stabilizing FRQ to enable proper clock circuitry/function.
Introduction
Protein conformation dictates the activity of proteins. Lack of structure or flexibility in structure is thought to be as important mechanistically for some proteins as rigid structure would be for others (Dyson and Wright, 2005; Schlessinger et al., 2011; Tompa, 2012; Uversky and Dunker, 2010). Identification of naturally unstructured proteins is steadily increasing, with a third or more of the total human proteome predicted to be disordered (Fukuchi et al., 2011). Currently, these proteins have been consolidated into a single category, Intrinsically Disordered Proteins (IDPs). Though functionality in IDPs remains highly varied, a commonality is emerging; IDPs are involved in the management of the cell, controlling cellular interactions and biological processes through regulation of activity (Williamson and Potts, 2012).
A well-studied cellular system that relies on perfect coordination of tight activity regulation is the circadian oscillator (Dunlap, 1999). One of the best studied of these clocks is in Neurospora crassa, where the oscillator comprises a transcription/translation loop involving four core proteins, White Collar 1 (WC-1), White Collar 2 (WC-2), Frequency (FRQ) and Frequency Interacting RNA Helicase (FRH). Transcription factors WC-1 and WC-2 form the White Collar Complex (WCC), which drives the rhythmic expression of FRQ. FRQ binds to FRH to form the FRQ/FRH complex (FFC), which then acts on the WCC to inhibit its activity, thus closing the loop. Throughout the circadian cycle, FRQ interacts with many partners that affect the number and location of its post-translational modifications as well as its stability; it is these post-translational modifications of FRQ that set the length of the circadian period (Baker et al., 2012; Diernfellner and Schafmeier, 2011; Liu and Bell-Pedersen, 2006).
Of the four core clock proteins, FRH is the most recently discovered and the protein whose role is least understood. frh is an orthologue of MTR4 which encodes a well-studied cofactor of the Saccharomyces cerevisiae exosome and member of the TRAMP complex with demonstrated RNA helicase activity (Cheng et al., 2005; Jia et al., 2012; LaCava et al., 2005). In Neurospora, FRH is essential for life as well as essential for rhythmicity (Cheng et al., 2005) where it is modeled as regulating the levels of frq message post-transcriptionally via its role in the TRAMP complex (Guo et al., 2009), an essential activity requiring ATPase/helicase enzymatic functions. FRQ association with FRH is important for proper phosphorylation, stability and localization of FRQ (Cha et al., 2011; Guo et al., 2010). Beyond this, FRH is known to play a role in the complex interaction between FRQ and the WCC (Shi et al., 2010); FRH also interacts with VVD to suppress FRQ expression via interaction with the WCC (Hunt et al., 2010).
A FRH mutant, FRHR806H, allows for wild type growth and development but does not support a functional clock (Shi et al., 2010). The observation that this mutation lies outside the highly conserved DEAD box region associated with helicase function and instead resides in the conserved but structurally distinct DSHCT region (Staub et al., 2004), led us to ask whether the role FRH plays in the clock could be unrelated to its putative Dead Box Protein (DBP) helicase and exosome function(s). Strains bearing frhR806H are totally viable and arrhythmic and a second copy of FRH at an exogenous location can rescue the arrhythmicity. Unexpectedly we found that compensatory copies with defects in the conserved helicase-associated ATP binding and RNA unwinding regions were still able to rescue clock function. That is, the helicase functions essential to the health of the organism have no impact on the clock. Because ATP binding and helicase activities are essential for exosome function, this finding makes it unlikely that the role of FRH in the clock is related to its role in the exosome. In a normal circadian cycle, FRQ is slowly and processively phosphorylated leading to its eventual interactions with the ubiquitin ligase, FWD-1 (He et al., 2003). However, without FRH, FRQ is rapidly turned over soon after its synthesis in a manner not dependent on FWD-1, but instead consistent with typical IDP turnover. Moreover, FRQ is highly heat stable, a characteristic in line with typical IDPs. In all, these and other results lead us to hypothesize an independent function for FRH in the clock, that of a stabilizing partner protein for FRQ that interacts with the IDP FRQ to ensure proper folding and interactions within the circadian clock.
Results
Helicase activity is not required for circadian-specific function of FRH
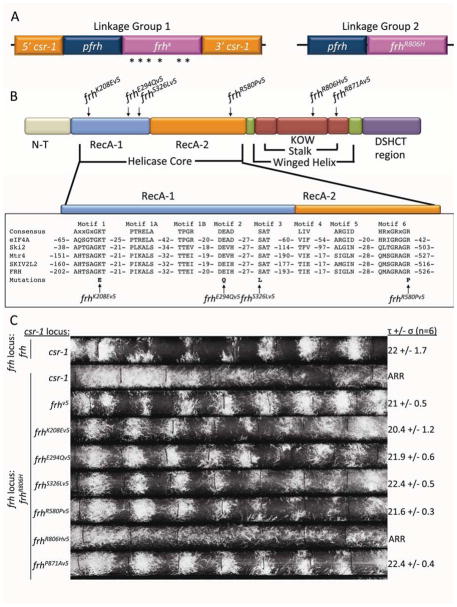
A mutation (R806H) outside the core helicase region (Figure 1) of the DEAD box helicase FRH disables the Neurospora clock while not affecting strain growth or development (Shi et al., 2010; Staub et al., 2004). The position of this mutation, beyond the highly conserved DEAD box region that confers enzymatic functions, suggested that the role of FRH in the clock was unlinked from its exosome function(s) (Guo et al., 2009). frhR806H has been reported to be recessive and we confirmed this by showing that a second copy of frh inserted into the csr-1 locus (Bardiya and Shiu, 2007) could fully rescue rhythmicity. This observation set the stage for a series of experiments in which a variety of alleles of frh, each predicted by sequence identity with known orthologues to be defective in a biochemical activity associated with an essential function, could be assessed for circadian function (Figure 1A and Supplemental Figure 1A).
Figure 1.
Helicase/ATPase activity is not required for the circadian oscillator-specific function of FRH. (A) Strain genotypes: Linkage group 1; at the csr-1 locus: mutant versions of frh driven by the frh promoter. Linkage group 2; at the frh native locus: frhR806H driven by its native promoter. (B) Colored blocks represent the basic structural domains of FRH as predicted from homology with Mtr4p (Jackson et al., 2010; Weir et al., 2010) as well as the core helicase motifs of eIF4A and other members of the DSHCT helicase family. The location of individual point mutations engineered to cripple the enzymatic functions of FRH are shown schematically at the top and the actual sequence context is provided at the bottom. (C) Race tube assay of wild type (rhythmic, top), frhR806H (arrhythmic, next to last) and the frh mutants knocked into the csr-1 locus (τ = period in hours, σ = standard deviation, n = number of race tubes).
The mutations in the frh alleles were designed based on its Saccharomyces orthologue MTR4, which encodes a DEXD box helicase and member of the DSHCT family that has been extensively studied functionally as well as structurally (Jackson et al., 2010; Weir et al., 2010). Because of the high degree of conservation within DEAD BOX helicase regions (comparing Mtr4p to FRH: 51% identical/66% similar; core helicase regions: 67% identical/80% similar; 100% conservation of residues essential for catalytic functions, Figure 1B), frh point mutations could be designed to replicate mutations in Mtr4p and/or other helicases, each with known and well defined effects on the helicase: K208E cripples nucleotide binding and is a loss of function mutation in Mtr4p; E294Q eliminates ATP hydrolysis which has been shown to be essential in helicases other than of Mtr4p; S326L blocks the unwinding function of helicases and is a loss of function mutation in Mtr4p; R580P stops ATPase activity and nucleotide binding and creates a loss-of-function in Mtr4p (Bernstein et al., 2006; Cordin et al., 2006; Liang et al., 1996; Pause et al., 1993; Pause and Sonenberg, 1992). Along with these, mutations were also designed within the DSHCT region reflecting its conservation in multiple species (R806H/clock essential; P871A/elbow bend of KOW region – predicted to play a role in helicase function (Jackson et al., 2010; Weir et al., 2010)) (Figure 1B). All mutant proteins and controls were epitope tagged with V5 at the C-terminus and expression of each was driven by the native frh promoter (Figure 1A) which is strong and constitutive (Cheng et al., 2005; Shi et al., 2010). Strong expression of all FRHX (X= mutations described above) proteins was confirmed by western blotting, and the normal interaction of each mutant protein with FRQ was confirmed by V5 co-IP (Supplemental Figure 2).
The ability of each mutant FRH to function in the circadian oscillator was assessed both by asexual conidial banding on race tubes (Figure 1C) and by luciferase reporter which monitored the rhythmic activation of FRQ expression (Supplemental Figure 2B) (Gooch et al., 2008). In brief, Neurospora was inoculated on one end of a long glass tube containing a bed of agar and, after a day in the light, grows along the tube in the dark. Neurospora strains with functional clocks form daily conidial bands with a periodicity equivalent to the rhythm of the core molecular clock. The frhv5 construct, with no mutation in FRH, served as the positive control and was able to rescue the overt rhythms in conidiation with a period comparable to the control strain (Figure 1C). Similarly but surprisingly, all of the mutations in the helicase region (K208E, E294Q, S326L and R580P) as well as a point mutation outside of the helicase region (in the DSHCT region) that is predicted to play a role in helicase function (P871A) (Jackson et al., 2010; Weir et al., 2010), restore wild type rhythmicity; FRHR806H provided the negative control.
We confirmed that the molecular rhythms in frq expression matched the overt rhythms by examining each frh allele in the frhR806H background that also contained the luciferase gene driven by the frq promoter (Supplemental Figure 2A). As before, all frh alleles designed to cripple essential helicase functions were nonetheless able to rescue wild type rhythmicity (Supplemental Figure 2B). These data strongly suggest that it is not the helicase activity of FRH that is essential to clock function.
FRH helicase activity is required for Neurospora growth
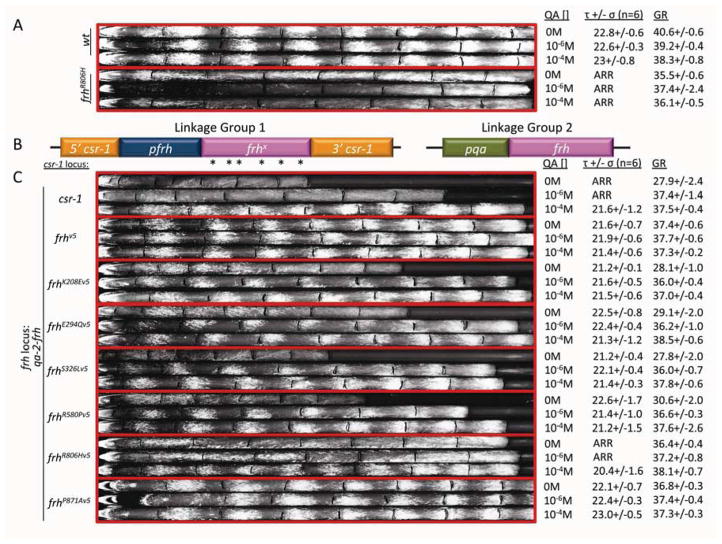
FRH is an essential protein but whether the helicase function is indeed the vital function and is distinct from the clock function is not known (Cheng et al., 2005; Shi et al., 2010). To determine if the helicase function is truly essential, we used a strain in which the regulatable qa-2 promoter drove frh expression at the native frh locus (Figure 2B and C and Supplemental Figure 2D). With no qa-2 inducer (0 M quinic acid - QA), the strain grows slowly (28 mm/day) as just enough FRH is produced from the leaky non-induced promoter to allow the strain to survive. With low levels of inducer (10−6M QA), producing mid range levels of FRH, the strain is able to grow at close to a wild type rate (37 mm/day) but the clock function is not restored as assayed by race tubes. With high levels of inducer (10−4M QA) the strain produces wild type levels of FRH and displays a normal circadian period and growth rate (36 mm/day) on race tubes (Figure 2C) (Shi, 2008; Shi et al., 2010). Into this strain, we transformed the frhX mutants described above, targeting each to the csr-1 locus. The native frh promoter again drove each mutant and in each strain normal levels of FRHX were confirmed by western blot (Supplemental Figure 2D). Thus, in the strain, a regulatable level of essential functions are provided by this qa-2:frh background; these sustain viability but do not allow WT growth. If the frhX mutants that express at normal levels for FRH are able to supply essential functions, then qa-2 induced FRH would not be needed for WT growth.
Figure 2.
The helicase regions provide the FRH functions that are essential for growth. (A) Race tube assay of wild type and FRHR806H shown as three representative race tubes grown with the three concentrations of inducer (QA). (B) Strain genotypes of experimental strains (bottom eight groups of three): Linkage group 1; at the csr-1 locus: mutant versions of frh driven by the frh promoter. Linkage group 2; at the frh native locus: frh driven by the QA-inducible qa-2 promoter. (C) Race tube assay of qa-2 driven FRH and the mutant knock-ins at the csr-1 locus. Each strain contains an inducible copy of wt frh as well as a native promoter-driven but mutated copy of frh at the csr-1 locus and is shown as three representative race tubes grown with the three concentrations of inducer (QA). Less than wild type growth rate indicates a level of functional FRH inadequate for full rescue of essential functions. Restoration of growth indicates that the induced wt FRH was able to supply the essential functions (QA[] = concentration of QA, τ= period in hours, σ = standard deviation, n = number of race tubes, GR = growth rate in mm/day).
The frhv5 construct was able to rescue rhythmicity and normal growth at each level of inducer (0M, 10−6M and 10−4M) (Figure 2C), providing the positive control for rescuing the essential function of FRH. Additionally, the clock mutant FRHR806H also rescued growth at all levels of inducer, consistent with clock function being distinct from essential functions. However, as in the host strain control, clock function is only rescued in the frhR806H background when wild type FRH is driven by the highest level of inducer (10−4M), further confirming that the level of functional FRH needed for the clock to run is higher than that needed to support life (Figure 2C).
The mutations in the helicase region (K208E, E294Q, S326L, R580P) were unable to rescue growth at the lowest inducer concentrations (0M) consistent with these mutations crippling essential enzymatic functions. Only when the wild type copy of FRH was induced at levels sufficient to compensate for this deficiency was growth restored to levels comparable to that seen for the FRHV5 positive control (10−6M and 10−4M) (Figure 2C). Moreover and surprisingly, with no inducer present and growth measurably slowed, expression of the helicase mutant proteins was enough to completely rescue wild type rhythms, highlighting the distinction between clock and essential functions (Figure 2C).
Unlike the mutations in the helicase region, the mutation in the DSHCT region of FRH (P871A) was able to rescue growth at all inducer concentrations (0M, 10−6M and 10−4M) (Figure 2C). This demonstrated that although it is predicted to be involved in helicase activity based on structural similarity (Jackson et al., 2010; Weir et al., 2010), it appears not to play an essential role in helicase activity. As with the other helicase mutations, even with no inducer present, the expression of FRHP871A was enough to rescue wild type rhythms (Figure 2C).
FRQ/FRH interaction demonstrates the need for a clock-functional FRH
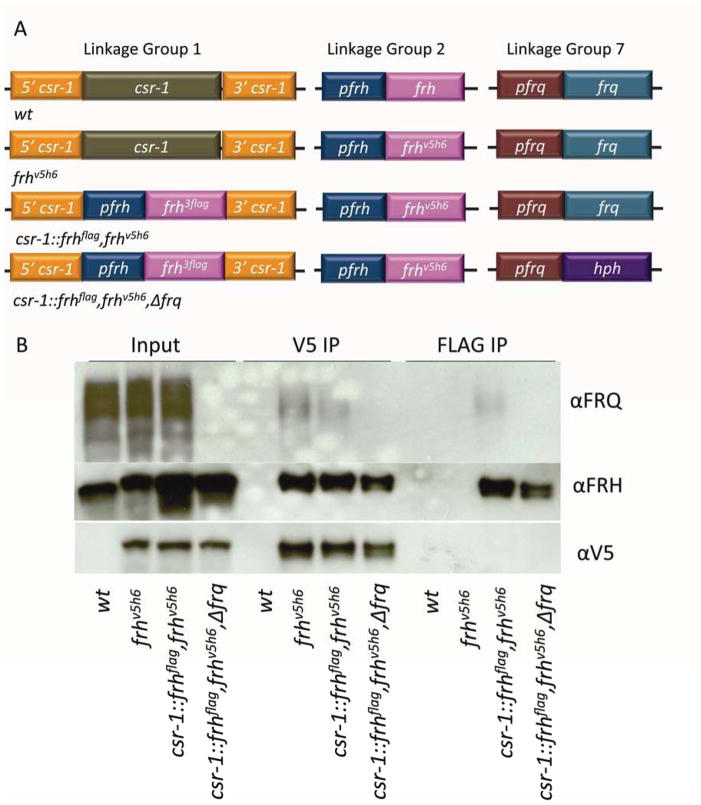
To be conclusive about the distinctive separation between clock function and helicase function in FRH, it was important to demonstrate that FRH does not form a homodimer in its interaction with FRQ dimer. Specifically in our experiment, dimerization of FRH could lead to the formation of a FRQ-FRH complex (FFC) bearing one helicase-null FRH and one clock–null FRH, thereby allowing for cross-complementation in trans within the complex. Although Cheng et al. (2005) estimated based on optical density of protein blots that for every FRQ dimer there was a single FRH bound, a more definitive proof seemed prudent. To determine the stoichiometry of FRH to FRQ, we inserted at the csr-1 locus a second copy of frh (epitope tagged with the 3XFLAG marker) into strains where, at the native locus, FRH had been epitope tagged with V5 (Figure 3A). Extracts of these strains were examined by immunoprecipitation; FRHV5 and FRHFLAG were both detected in the input samples and each tagged version of FRH was pulled down in each respective IP (Figure 3B). In all strains where FRQ was present and FRH was pulled down, FRQ was pulled down as well. However, the αFLAG antibody was not able to pull down V5 tagged FRH (Figure 3B), demonstrating that although FRH is in a complex with FRQ, that complex only involves a single FRH. In the absence of FRQ, FRH monomers still do not interact. The heterotrimeric make up of the FFC further demonstrates that the putative helicase function is not essential for clock function as no compensation in trans could occur in our assay through binding with a second, different copy of FRH.
Figure 3.
A single FRH interacts with the FRQ homodimer. (A) Strain genotypes: Linkage group 1; at the csr-1 locus: native csr-1 or 3XFlag tagged frh driven by the frh native promoter. Linkage group 2; at the frh native locus: frhV5H6 or untagged frh, driven by the frh promoter. Linkage group 7; at the frq locus: native frq, or hph driven by the trpC promoter. (B) IP demonstrating that FRH does not form a dimer in it’s interaction with the FRQ homodimer. V5 and 3Flag antibody were used to pull down tagged FRH. Proteins were western blotted and detected with V5, FRH or FRQ antibody.
FRQ demonstrates the basic characteristics of an IDP
The data above provide strong evidence that the helicase and ATPase functions of FRH are separate from the clock function of FRH, but this leaves the role of FRH in the clock unknown. However, some striking characteristics of FRH’s binding partner, FRQ, suggest a plausible non-enzymatic role for FRH. Unlike FRH, FRQ is predicted to be a largely unstructured protein (Baker et al., 2009; Querfurth et al., 2011; Tang et al., 2009) (Figure 4A and Supplemental Figure 3) and proper co-translational protein folding, influenced by translation rate, has a strong impact on FRQ function (Hurley and Dunlap, 2013; Zhou et al., 2013). Consistent with FRQ being a largely unstructured, mostly surface accessible structure, it is one of the most phosphorylated proteins known (Baker et al., 2009; Tang et al., 2009). FRQ has a large number of binding partners and binds to FRH nearly coincident with translation (Cheng et al., 2005; Guo et al., 2010).
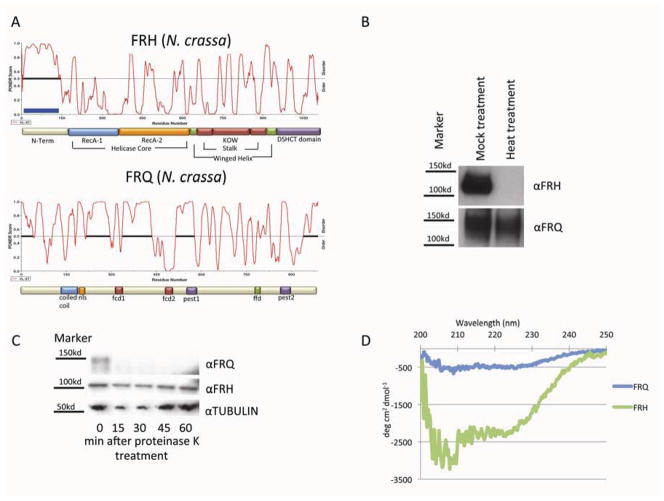
Figure 4.
FRQ demonstrates characteristics of a classic IDP. (A) PONDR analysis of FRH and FRQ from N. crassa. Basic structural domains of each are shown as predicted. Black lines represent distinct and probable regions of low structural complexity (B) Solubility of FRQ and FRH were analyzed after a mock or heat treatment to determine the stability of each protein following heat-induced unfolding. The lack of the protein demonstrates precipitation after heat treatment. (C) Protease sensitivity assay. Comparable levels of FRQ, FRH, and tubulin (see Experimental procedures) were exposed to low levels of the non-specific protease Proteinase K; the globular structured FRH and tubulin proteins were much more stable than FRQ which was rapidly degraded. (D) Far-UV spectra of short-FRQ and FRH. The spectra is an average of three independent acquisitions.
To investigate the structural nature of FRQ, we applied a global test of protein structure. It is well established that IDP’s are highly heat stable and will not aggregate and precipitate out of solution following heat treatment, as globular proteins do (Csizmok et al., 2006; Galea et al., 2009; Hackel et al., 2000; Tantos and Tompa, 2012; Uversky and Dunker, 2010). Lacking structure to unfold, IDP’s see little to no change in structure when heated, unlike globular proteins that will unfold upon heat treatment and then misfold upon returning to physiological temperatures; in fact, solubility following heat treatment is used as a selection to enrich for IDPs in extracts (Tantos and Tompa, 2012). If FRQ is in fact an IDP, then when treated with heat, it should remain in solution while its binding partner FRH becomes insoluble and is precipitated out of solution.
FRQ was enriched using a 6HIS column using a non-denaturing protocol that allowed for the maintenance of the core clock components. The resulting purified clock complex was then subjected to heat treatment (100°C for 10 minutes) with a mock treatment on ice as a control. The soluble proteins were then separated from the aggregated proteins by centrifugation and the supernatant of both the mock and heat treatment were then analyzed by western blotting. As is characteristic for an IDP, FRQ was found in both the mock and heat-treated supernatant while FRH (predicted by both PONDR and other software analyses as well as by sequence homology to be a highly structured protein) (Figure 4A and Supplemental Figure 3), was only found in the mock treated supernatant (Figure 4B). This and other characteristics suggest that FRQ is an IDP – largely unstructured and highly unstable – and that the role of FRH in the clock is to act nonenzymatically, perhaps as a Nanny protein for FRQ (Tsvetkov et al., 2009), to confer both structure and stability.
To further confirm that FRQ satisfies the characteristics of an IDP, limited proteolysis via proteinase K treatment was employed –theory predicts that less structured proteins have more available cleavage sites than globular proteins, and therefore IDPs are degraded at a much faster rate than are the more folded globular proteins (Tsvetkov et al., 2008). As expected, under protease treatment, FRQ was degraded at a much faster rate than the more folded FRH or tubulin controls (Figure 4C).
In a final effort to demonstrate that FRQ shares the classic features of an IDP, we examined in vitro expressed FRQ by circular dichroism (CD). In order to purify protein that was at least 95% pure, we expressed tagged Neurospora proteins short-FRQ and FRH in E. coli and purified them using a 6HIS tag. To ensure that these proteins maintained representative structure when expressed in vitro, short-FRQ and FRH were mixed together and immunoprecipitated with αFRQ antibody. Not only was short-FRQ pulled down, but FRH was also immunoprecipitated (Supplemental Figure 2G). Both proteins were then subjected to CD spectral analysis focusing on wavelengths between 250 and 200 nm that are predictive for structure. The spectra for FRH revealed the significant dip in the short wavelength region characteristic of significant helical structure whereas FRQ showed almost no dip consistent with just a small amount of helical structure, likely arising from its predicted coil-coil region. These data are wholly consistent with FRQ being an IDP (Figure 4D) (Tsvetkov et al., 2008).
The Nanny hypothesis suggests a nonenzymatic circadian function for FRH
Among the explanations for why IDPs persist in cells is the Nanny hypothesis which posits the existence of “nanny” proteins that protect a newly made IDP from birth so that it escapes random degradation by the proteasome (Tsvetkov et al. 2009). If FRH is acting as a Nanny to stabilize FRQ then loss of FRH/FRQ interaction should render FRQ not only less stable but also less stable in an interesting way. In the normal circadian cycle, FRQ is phosphorylated over the course of the day and after about 18 hours, phosphorylation reaches the point where FRQ is bound by FWD-1, the substrate recognition component of an SCF complex, leading to protein turnover (Baker et al., 2012; He and Liu, 2005). As described above, proper phosphorylation of FRQ has been shown to regulate the degradation of FRQ as well as affect the function of the clock. One gene that plays a role in ubiquitination is the F-box/WD40 repeat-containing protein FWD-1. FWD-1 has been shown to directly interact with FRQ, particularly the ubiquitylated phosphorylated form, and is required for the proper degradation of FRQ (He et al., 2003).
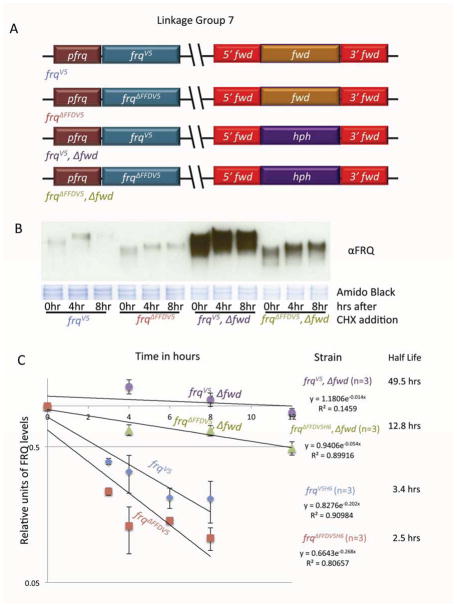
The Nanny hypothesis and similar ideas regarding the loss of nascent protein stabilization predict that the nascent target protein (here FRQ) deprived of its Nanny (FRH) will be rapidly degraded by a mechanism distinct from its normal turnover. To determine if this is true, we tracked the stability of FRQ in three different strains, one in which FRQ no longer interacted with FRH because the FRQ/FRH interacting domain (FFD) (Cheng et al., 2005; Guo et al., 2010) in FRQ had been deleted, one in which the ubiquitin ligase FWD-1 was deleted, and in a double mutant strain in which both FRH-interactions and FWD-1 mediated turnover was lost (Figure 5A).
Figure 5.
FRQ stability is independently determined by interactions with FRH and FWD-1. (A) Strain genotypes: Linkage group 7; at the frq locus: V5H6 tagged frq with the FFD (FRQ-FRH Interaction domain) intact or deleted; at the fwd-1 locus: fwd-1 or hph. (B) Total levels of FRQ during its degradation were followed in epitope tagged strains with or without the FRQ FFD (frq and frq Δ FFD) and/or with or without FWD-1 (frq, Δ fwd-1 and frq Δ FFD, Δ fwd-1) after addition of CHX (to inhibit translation). 10μg of total protein was loaded per lane and membranes (mem) stained with Amido Black were used as loading control. (C) Densitometric analysis of data shown in (B) was used to calculate half-life. For each strain, time zero was set to 100%, allowing degradation rate to be analyzed relative to the other strains. Half-life was calculated by least squares regression of the line through the data points with the resulting equation and goodness of fit (R2) shown. Data are means ± S.E., n = 3.
Both predictions, loss of FRQ stability and a novel mechanism of turnover, are supported by the data. First, FRQ that is bound to FRH (FRQV5) has a half-life that is twice as long as FRQ that is not (FRQΔFFDV5) (Figure 5B and C) (see also (Guo et al., 2010)). Second, in the Δfwd-1 background, which lacks normally regulated turnover of FRQ, again FRQ that is bound to FRH (FRQV5, Δfwd-1) showed an increase in half-life compared to FRQ that cannot bind FRH (FRQΔFFDV5, Δfwd-1) (Figure 5B and C). Although the loss of FWD-1 still stabilizes FRQ even when FRQ cannot interact with FRH (compare FRQΔFFDV5, Δfwd-1 with FRQ ΔFFDV5), perhaps through pleiotropic effects from Δfwd-1, the distinct decrease in steady state FRQ levels in FRQΔFFDV5 as compared to FRQΔFFDV5, Δfwd-1 serves to highlighting the dual pathways to FRQ degradation. In all, these data are consistent with the prediction that FRQ has two paths to degradation, one through the traditionally understood FWD-1-mediated ubiquitination pathway and a second through some other FRH-protected mechanism (Guo et al., 2010).
An unstructured region of FRH mediates interactions with FRQ required for circadian function
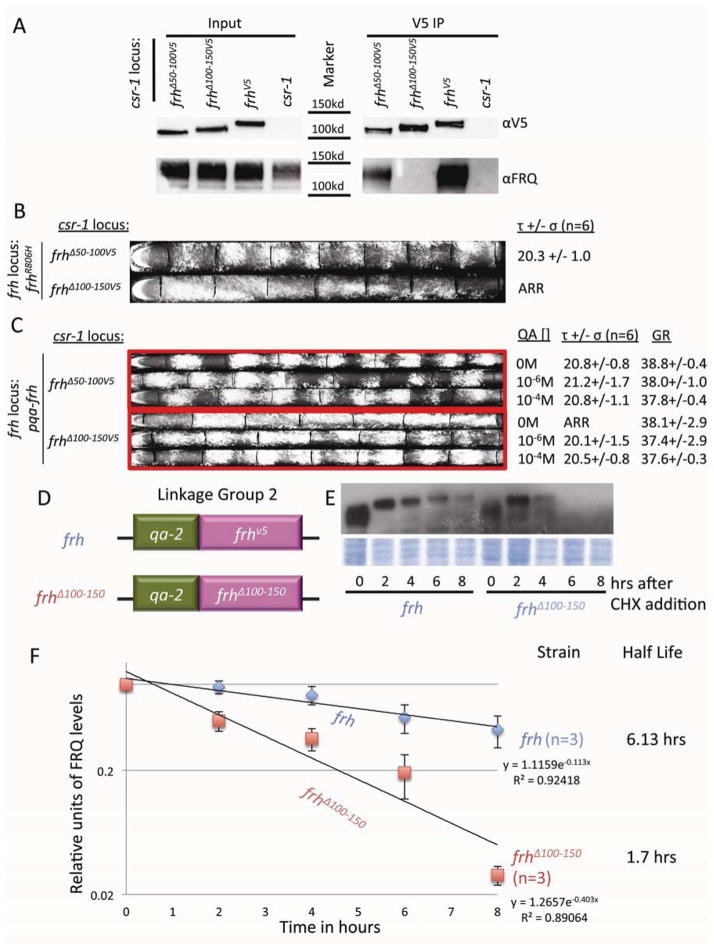
Though the above data strongly suggest that the interaction between FRH and FRQ dictates FRQ stability, the possibility remained that the loss of stability was due to the FRQ deletion in FRQΔFFD rather than the loss of interaction. To eliminate this possibility, we identified the FRQ-FRH interaction domain within FRH. Constructs bearing a series of 50 amino acid deletions within FRH were targeted to the csr-1 locus in both the frhR806H and qa-2p-frh backgrounds (Supplemental Figure 1). Again, the native frh promoter was used to drive each mutant, expression was confirmed by western analysis (Figure 6A and Supplemental Figure 2E), and a V5 IP was performed to test the ability of each stably expressed mutant to interact with FRQ. Most notably, FRHΔ50-100 was able to interact with FRQ while FRHΔ100-150 was not (Figure 6A), localizing the FRQ-FRH interaction domain (FFD) to this interval. The FRHΔ50-100 and FRHΔ100-150 strains were then run on race tubes to test clock function. In the viable but arrhythmic FRHR806H background, the FRHΔ50-100 construct fully rescued the overt rhythm (Figure 1C and 6B), and in the growth-compromised but clock-functional qa-2 driven FRH background, the FRHΔ50-100 rescued growth as well as the clock at all inducer concentrations. This indicated that deletion of amino acids 50-100 impairs neither essential nor circadian function of FRH. In contrast, in the same assay, FRHΔ100-150 was fully able to rescue growth but was not able to rescue rhythmicity. These data identify the FRH interacting region as between amino acids 100 and 150, confirm the distinction between the essential enzymatic and circadian functions within FRH, and complement this distinction by associating separate parts of FRH with each function.
Figure 6.
An N-terminal region of FRH is essential for interaction with FRQ and for FRQ stability, distinguishing oscillator function from essential functions. (A) IP demonstrating that deletions of amino acids 100-150 from FRH eliminated interaction with FRQ. For input, 10ug of protein lysate was loaded and after western blotting proteins were visualized with antisera to V5 or FRQ. For IP, V5 antibody was used to pull down FRH and co-immunoprecipitated FRQ monitored. Proteins were western blotted with V5 and FRQ antibody. (B) Race tube assay of the frh domain deletion mutants at the csr-1 locus in the frhR806H background. (C) Race tube assay of the frh domain-deletion mutants at the csr-1 locus in the qa-2 driven frh background. Each strain contains an inducible copy of frh as well as a native promoter-driven but truncated copy of the gene at the csr-1 locus and is shown as three respective race tubes grown with three concentrations of inducer (QA). Rhythmicity in frhΔ100-150 is not restored until adequate wt frh is expressed from the qa-2 promoter driven copy. (QA[ ] = concentration of QA, τ= period in hours, σ= standard deviation, n = number of race tubes, GR = growth rate in mm/day). (D) Strain genotypes: Linkage group 2; at the frh locus: qa-2 driven frh either wild type or with amino acids 100-150 deleted. (E) Assay of FRQ stability in wt or frh-mutant strains. CHX was added at time 0 and total levels of FRQ were monitored during its degradation in WT and frhΔ100-150 strains. Membranes (mem) stained with Amido Black were used as loading control. (F) Densitometric analysis of data shown in (E) used to calculate half-life. For each strain, time zero was set to 100%, allowing degradation rate to be analyzed relative to the other strains. Half-life was calculated by least squares regression of the line through the data points with the resulting equation and goodness of fit (R2) shown. Data are means ± S.E., n = 3.
Structural predictions by PONDR as well six additional structural prediction programs predict that the N-terminal region of FRH is highly unstructured (amino acids 1-150; encompassing the above described FRQ-FRH Interaction domain) (Li et al., 1999; Romero et al., 1997; Romero et al., 2001) (Supplemental Figure 3). This lack of structure is notable because the closest orthologue to FRH is not predicted to have this unstructured N-terminal region (Supplemental Figure 3 and 4). In fact, in a survey of known FRH orthologues, N-terminal regions of FRH from species having a FRQ orthologue are all predicted to have this unstructured region while the same regions in species lacking FRQ are more variable (Salichos and Rokas, 2010) (Supplemental Figure 4). This conservation is consistent with a need for this unstructured region in interactions with FRQ.
A deletion in FRH affects FRQ stability by affecting interaction
Having identified amino acids 100-150 as a region in FRH required for interactions with FRQ, we predicted that FRQ would be less stable in a frhΔ100-150 background when FRQ-FRH interaction was lost. To determine if that was true, we tracked the stability of FRQ in WT vs. frhΔ100-150 (Figure 6D, E, F and Supplemental Figure 2F). To better control FRH levels, we drove frhX expression using the inducible qa-2 promoter. When comparing FRH to FRHΔ100-150, FRQ that is bound to FRH has a half-life of 6.5 hours while unbound FRQ has a half-life of only 3.6 hours (Figure 6E and F). FRQ stability in FRHΔ100-150 is comparable to the stability seen in strains where the FFD has been deleted in FRQ (Figure 4C).
Discussion
FRH has been linked to the exosome complex and the proper degradation of mRNAs (Guo et al., 2009), and the depletion of FRH from Neurospora (thus removing the active helicase from the system) ablates the clock (Cheng et al., 2005); our data are completely consistent with these prior results. However, we demonstrate here that the role of FRH in the circadian oscillator of Neurospora is not related to its helicase enzymatic function. Indeed, even low levels of helicase-null but clock functional FRH can rescue the clock, and additional data are consistent with a non-traditional role for FRH in the clock. First, although members of the TRAMP complex strongly co-purify with Mtr4p in yeast (LaCava et al., 2005), no members of the TRAMP complex were identified via mass spec among the binding partners of FRH in Neurospora (Baker, 2010). In addition FRH is essential for proper localization of FRQ (Guo et al., 2010), an activity related in no obvious way to exosome function. Further, a mutation outside of the conserved helicase region eliminates the clock [(Shi et al., 2010) and (this work Figure 3)] and FRH mutants lacking amino acids required for core helicase activities are able to rescue clock function (this work Figure 1). Finally, FRH is important for FRQ stability [(Guo et al., 2010) and this work Figure 6] and loss of FRH interaction leads to rapid degradation of FRQ via a non traditional avenue not requiring its known ubiquitin ligase FWD-1. Elimination of an enzymatic role for FRH in the clock of course begs the question, what is the role that FRH is playing and what does this tell us about FRQ?
To answer these questions, we drew upon several pieces of evidence. No free pool of FRQ exists in the cell; instead all FRQ is bound to FRH indicating that FRH must bind FRQ almost immediately after translation (Cheng et al., 2005). FRH interaction with FRQ is essential for clock function [(Guo et al., 2010) and this work]. Moreover, FRQ is predicted to be a largely unstructured protein [(Baker et al., 2009; Tang et al., 2009) and this work]. Most recently, an elegant series of experiments have shown that FRQ must be translated at a slow rate or else the FRQ that is produced is unstable and non-functional, a result that suggests a need for co-translational folding or interaction (Zhou et al., 2013). All of this led us to suggest that a role that FRH could play is to help establish and maintain structure in FRQ. Given the myriad data cited above as well as structural predictions consistent with natively unstructured FRQ and the stabilizing effect of FRH, we theorized that FRQ could be an inherently disordered protein (IDP) and that the principal role of FRH is to stabilize it, perhaps as its cognate Nanny protein.
The lock-and-key and induced fit models (Fischer, 1894; Koshland, 1958) represent classic paradigms for conceptualizing protein-protein interactions, and even more modern models in which proteins exhibit their dynamic personalities by occupying ensembles of conformations (e.g. (Henzler-Wildman and Kern, 2007)) presuppose inherent structural options. However, it is now recognized that there are many types of proteins that do not fit even this dynamic model and those models have been progressively changing. One example of such shifts is that IDPs have emerged as an enormous group of proteins (as many as 45% of eukaryotic proteins may contain significant disorder) (Tompa, 2012). IDP’s are characterized by several specific functional modalities, including adaptability in binding, a large number of functions assigned to the protein and frequent regulation by post-translational modification (Tompa, 2012). FRQ fits many of these characteristics. Extremely highly post-translationally modified, FRQ changes conformation to allow for CK1a binding as it is progressively phosphorylated (Baker et al., 2009; Querfurth et al., 2011; Tang et al., 2009). Indeed FRQ has many different binding partners – CK-1A, WC-1, WC-2, FRH, PRD-4(CHK2) as well as FRQ itself - that associate with specific and distinct regions throughout the protein and these are temporally as well as spatially specific (Baker et al., 2009; Cheng et al., 2001; Denault et al., 2001; Guo et al., 2010; He et al., 2006; Pregueiro et al., 2006; Querfurth et al., 2011). Stability, subcellular localization and function of FRQ all depend on these modifications and binding partners (Baker et al., 2009; Diernfellner et al., 2009; Guo et al., 2010; He and Liu, 2005; Querfurth et al., 2007; Querfurth et al., 2011; Tang et al., 2009) (Figure 5 of this work). PONDR prediction along with a variety of other structural predictors show that roughly 75% of the FRQ amino acid sequence falls into the disordered range (Supplemental Figure 3), a finding consistent with data showing that the longest non-phosphorylated region of mature FRQ is only 129 amino acids out of a total of 998 (Baker et al., 2009; Tang et al., 2009). We demonstrate that FRQ is heat stable, unlike its more structured binding partner FRH, and displays little resistance to limited proteolysis and almost no CD signal. These data taken as a whole provide strong evidence that FRQ is principally comprised of disordered regions.
Flexibility in FRQ structure would allow for flexibility of binding, the observed high levels of post-translational modification, ubiquitination and a variety of protein-protein interactions to occur, all things that are necessary for FRQ function. This promiscuity is important to a protein with so many roles to fill and may be a conserved feature of clock proteins as extensive post translational modifications are a common theme in the fungal/animal transcription-translation feed back loop (FRQ in Neurospora, PER and TIM in Drosophila and the PER and CRY paralogs in mammalian cells) (Mehra et al., 2009). In fact, the PERs of both Drosophila and humans are predicted by PONDR to have multiple regions of disorder (Supplemental Figure 3 and data not shown) (Li et al., 1999; Romero et al., 1997; Romero et al., 2001).
By convention IDP functions must follow one of two paths; either their function directly stems from their disorder, or they bind to a partner molecule at which point they undergo induced folding (Tompa, 2012). The debate between these two models has evidence on both sides and has given rise to a new term as an alternative to IDPs: proteins waiting for partners (PWPs) (Janin and Sternberg, 2013; Uversky and Dunker, 2013). Partners for IDPs have sometimes been termed “Nanny” proteins and the definition of a Nanny is a flexible one (Tsvetkov et al., 2009). It is suggested that IDPs are at risk for degradation by default at birth and that Nanny proteins protect IDPs at birth so that the IDPs can go through an extended, normal and functional life cycle and progress through the proper stages of ubiquitination and degradation (Tsvetkov et al., 2009). In the model, Nanny proteins commonly have functions outside of the Nanny role, often as transcriptional repressors (not unlike the role played by the FFC) (Tsvetkov et al., 2009). Accepting this definition of the IDP/Nanny relationship and with the knowledge that FRQ is indeed an IDP it is plausible that FRH is its cognate Nanny (Figure 7).
Figure 7.
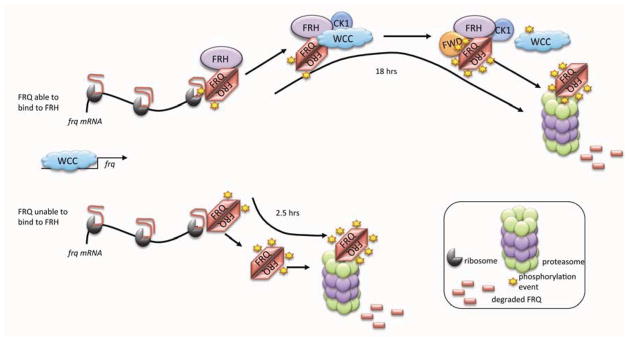
FRH stabilizes the IDP FRQ thereby supporting the normal oscillator cycle. Schematic representation of the role of FRH: If FRQ can bind to FRH, FRH is able to stabilize FRQ throughout its functional cycle of interactions and progressive phosphorylations until it is targeted by FWD-1 for the ubiquitylations that will lead to its degradation in the proteasome. If FRQ cannot bind to FRH, FRQ remains highly unstable and is targeted for degradation as soon as translation is complete.
Given the conserved regulatory architecture of circadian feedback loops in fungi and animals and the predicted unstructured regions in the PERs, we note in passing that two helicases related to FRH, DDX5 and DHX9, are part of the PER complex in mice. Depletion of either protein leads to a shortened period and increased rates of PER transcription, which would be expected if they played a role in PER stability (Padmanabhan et al., 2012). It is possible the role of Drosophila TIM be conceptualized in a manner similar to FRH. Whether the basic IDP concept and the Nanny hypothesis can inform animal clocks is yet to be tested.
Experimental Procedures
Strains, plasmids and reagents
Transformations were performed as described (Bardiya and Shiu, 2007; Colot et al., 2006). Race tube assays, circadian liquid culture experiments, luciferase trace experiments, and FRQ degradation experiments were performed as described with slight modifications (Garceau et al., 1997; Gooch et al., 2008; Hong et al., 2008; Loros et al., 1989; Ruoff et al., 2005).
Short-FRQ and FRH, codon optimized for expression in E. coli, were created by Genscript and cloned into pCOLD1 (a generous gift from the Masayori Inouye lab) with culture conditions and strains used described in the Supplemental Data.
Western blots, protein preparation and immunoprecipitation
Protein lysate extraction, Western blot analyses and immunoprecipitations were performed as described (Chen et al., 2010) with slight modifications (see Supplemental Methods).
Cycloheximide experiments and measurements of half-life and protein stability assay
Mycelial pads grown in at 25°C in LL were treated with 40 μg/mL CHX at indicated times and harvested over 8 hrs with 4-h resolution. Half-lives were measured as in (Hong et al., 2008). Protein stability was performed as described previously (Tsvetkov et al., 2008) with minor changes (see Supplemental Methods).
Heat treatment of FRQ
FRQ was purified with the 6HIS tag as described previously (Baker et al., 2009). Purified protein was then heated to 100 °C for 10 min and centrifuged again for 30,000×g for 30 min at 4 °C as described (Csizmok et al., 2006) and subjected to western blot analysis using SDS-PAGE separation.
Recombinant FRQ and FRH
Recombinant short-FRQ and FRH were purified as described previously with minor differences (Hurley and Woychik, 2009) (see Supplemental Materials).
Circular Dichroism
CD spectra were recorded as in (Audu et al., 2013) with minor changes (see Supplemental Materials).
Supplementary Material
Highlights.
Helicase function of FRH is not essential for Neurospora clock
FFC is in the N-terminal end of FRH
FRQ is an IDP
FRH acts nonenzymatically to support the IDP FRQ
Acknowledgments
We would like to thank Christopher L. Baker providing strains for this project, Josh Weiner and Dale Mierke for assistance with CD analysis and Brian Zoltowski for ideas and discussion. This work was supported by grants from the NIGMS at the NIH to J.M.H. (F32 GM096574), J.J.L. (R01 GM083336), and J.C.D (R01 GM34985 and P01 GM68087). We thank the FGSC at the University of Missouri for supporting our work with Neurospora.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Audu CO, Cochran JC, Pellegrini M, Mierke DF. Recombinant production of TEV cleaved human parathyroid hormone. J Pept Sci. 2013;19:504–510. doi: 10.1002/psc.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. Genetics (PhD thesis) Hanover: Dartmouth College; 2010. Post-translational regulation of the Neurospora crassa circadian system; p. 230. [Google Scholar]
- Baker C, Loros J, Dunlap J. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. 2012;36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiya N, Shiu PK. Cyclosporin A-resistance based gene placement system for Neurospora crassa. Fungal Genet Biol. 2007;44:307–314. doi: 10.1016/j.fgb.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Granneman S, Lee AV, Manickam S, Baserga SJ. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Molecular and Cellular Biology. 2006;26:1195–1208. doi: 10.1128/MCB.26.4.1195-1208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Yuan H, Liu Y. Regulation of the activity and cellular localization of the circadian clock protein FRQ. The Journal of Biological Chemistry. 2011;286:11469–11478. doi: 10.1074/jbc.M111.219782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci U S A. 2010;107:16715–16720. doi: 10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes & Development. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. The EMBO Journal. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Csizmok V, Szollosi E, Friedrich P, Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol Cell Proteomics. 2006;5:265–273. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
- Denault DL, Loros JJ, Dunlap JC. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. The EMBO Journal. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner AC, Querfurth C, Salazar C, Hofer T, Brunner M. Phosphorylation modulates rapid nucleocytoplasmic shuttling and cytoplasmic accumulation of Neurospora clock protein FRQ on a circadian time scale. Genes & Development. 2009;23:2192–2200. doi: 10.1101/gad.538209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner AC, Schafmeier T. Phosphorylations: Making the Neurospora crassa circadian clock tick. FEBS Lett. 2011;585:1461–1466. doi: 10.1016/j.febslet.2011.03.049. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature reviews Molecular cell biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Fischer E. Influence of the configuration on the action of the enzymes. Berichte der deutschen chemischen Gesellschaft. 1894;27:2985–2993. [Google Scholar]
- Fukuchi S, Hosoda K, Homma K, Gojobori T, Nishikawa K. Binary classification of protein molecules into intrinsically disordered and ordered segments. BMC Struct Biol. 2011;11:29. doi: 10.1186/1472-6807-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea CA, High AA, Obenauer JC, Mishra A, Park CG, Punta M, Schlessinger A, Ma J, Rost B, Slaughter CA, et al. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J Proteome Res. 2009;8:211–226. doi: 10.1021/pr800308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell. 2008;7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Cheng P, Liu Y. Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. The Journal of Biological Chemistry. 2010;285:11508–11515. doi: 10.1074/jbc.M109.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel M, Konno T, Hinz H. A new alternative method to quantify residual structure in ‘unfolded’ proteins. Biochim Biophys Acta. 2000;1479:155–165. doi: 10.1016/s0167-4838(00)00051-0. [DOI] [PubMed] [Google Scholar]
- He Q, Cha J, Lee HC, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes & Development. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Yu H, Liu Y. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. The EMBO journal. 2003;22:4421–4430. doi: 10.1093/emboj/cdg425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Liu Y. Degradation of the Neurospora circadian clock protein FREQUENCY through the ubiquitin-proteasome pathway. Biochem Soc Trans. 2005;33:953–956. doi: 10.1042/BST20050953. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Hong CI, Ruoff P, Loros JJ, Dunlap JC. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev. 2008;22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SM, Thompson S, Elvin M, Heintzen C. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc Natl Acad Sci U S A. 2010;107:16709–16714. doi: 10.1073/pnas.1009474107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Dunlap JC. Cell biology: A fable of too much too fast. Nature. 2013;495:57–58. doi: 10.1038/nature11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. The Journal of biological chemistry. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. The EMBO Journal. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J, Sternberg MJE. Protein flexibility, not disorder, is intrinsic to molecular recognition. F1000 Reports Biology. 2013;5 doi: 10.3410/B5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Wang X, Anderson JT, Jankowsky E. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7292–7297. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Molecular and Cellular Biology. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bell-Pedersen D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot Cell. 2006;5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros JJ, Denome SA, Dunlap JC. Molecular cloning of genes under control of the circadian clock in Neurospora. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends in Biochemical Sciences. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Molecular and cellular biology. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. The EMBO journal. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science. 2006;313:644–649. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- Querfurth C, Diernfellner A, Heise F, Lauinger L, Neiss A, Tataroglu O, Brunner M, Schafmeier T. Posttranslational regulation of Neurospora circadian clock by CK1a-dependent phosphorylation. Cold Spring Harb Symp Quant Biol. 2007;72:177–183. doi: 10.1101/sqb.2007.72.025. [DOI] [PubMed] [Google Scholar]
- Querfurth C, Diernfellner AC, Gin E, Malzahn E, Hofer T, Brunner M. Circadian Conformational Change of the Neurospora Clock Protein FREQUENCY Triggered by Clustered Hyperphosphorylation of a Basic Domain. Molecular Cell. 2011;43:713–722. doi: 10.1016/j.molcel.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Romero Obradovic, Dunker K. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform Ser Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Ruoff P, Loros JJ, Dunlap JC. The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17681–17686. doi: 10.1073/pnas.0505137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A. The diversity and evolution of circadian clock proteins in fungi. Mycologia. 2010;102:269–278. doi: 10.3852/09-073. [DOI] [PubMed] [Google Scholar]
- Schlessinger A, Schaefer C, Vicedo E, Schmidberger M, Punta M, Rost B. Protein disorder--a breakthrough invention of evolution? Curr Opin Struct Biol. 2011;21:412–418. doi: 10.1016/j.sbi.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Shi M. Genetics (PhD thesis) Hanover: Dartmouth College; 2008. Circadian and Non-Circadian Oscillators in Neurospora crassa; p. 145. [Google Scholar]
- Shi M, Collett M, Loros JJ, Dunlap JC. FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics. 2010;184:351–361. doi: 10.1534/genetics.109.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E, Fiziev P, Rosenthal A, Hinzmann B. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays. 2004;26:567–581. doi: 10.1002/bies.20032. [DOI] [PubMed] [Google Scholar]
- Tang CT, Li S, Long C, Cha J, Huang G, Li L, Chen S, Liu Y. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10722–10727. doi: 10.1073/pnas.0904898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantos A, Tompa P. Identification of intrinsically disordered proteins by a special 2D electrophoresis. Methods in molecular biology. 2012;896:215–222. doi: 10.1007/978-1-4614-3704-8_13. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends in Biochemical Sciences. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P, Asher G, Paz A, Reuven N, Sussman JL, Silman I, Shaul Y. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins. 2008;70:1357–1366. doi: 10.1002/prot.21614. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. The case for intrinsically disordered proteins playing contributory roles in molecular recognition without a stable 3D structure. F1000 Reports Biology. 2013:5. doi: 10.3410/B5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP, Potts JR. Intrinsically disordered proteins: administration not executive. Biochem Soc Trans. 2012;40:945–949. doi: 10.1042/BST20120188. [DOI] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.