Abstract
Using a Drosophila model of Alzheimer's disease (AD), we systematically evaluated 67 candidate genes based on AD-associated genomic loci (P < 10−4) from published human genome-wide association studies (GWAS). Genetic manipulation of 87 homologous fly genes was tested for modulation of neurotoxicity caused by human Tau, which forms neurofibrillary tangle pathology in AD. RNA interference (RNAi) targeting 9 genes enhanced Tau neurotoxicity, and in most cases reciprocal activation of gene expression suppressed Tau toxicity. Our screen implicates cindr, the fly ortholog of the human CD2AP AD susceptibility gene, as a modulator of Tau-mediated disease mechanisms. Importantly, we also identify the fly orthologs of FERMT2 and CELF1 as Tau modifiers, and these loci have been independently validated as AD susceptibility loci in the latest GWAS meta-analysis. Both CD2AP and FERMT2 have been previously implicated with roles in cell adhesion, and our screen additionally identifies a fly homolog of the human integrin adhesion receptors, ITGAM and ITGA9, as a modifier of Tau neurotoxicity. Our results highlight cell adhesion pathways as important in Tau toxicity and AD susceptibility and demonstrate the power of model organism genetic screens for the functional follow-up of human GWAS.
INTRODUCTION
Advances in human genetics have generated rapid progress in our understanding of the genetic risk underlying complex diseases, including common neurodegenerative disorders such as Alzheimer's disease (AD). Emerging data suggest that a large collection of genetic variants impact disease risk, and the critical next step will be to understand the function of the implicated genomic loci in disease pathogenesis. Accomplishing this goal will require numerous complementary approaches, including bioinformatics, intermediate traits, in vitro systems, cell-based strategies, as well as model organism studies. These efforts will be essential to confirm the genes responsible for association signals (fine mapping), to understand how genetic variants affect gene function (gain- or loss-of-function), and to identify the relevant cellular pathway(s) that mediate disease susceptibility.
At autopsy, AD pathology is characterized by extracellular amyloid plaques and intracellular neurofibrillary tangles, predominantly composed of the amyloid-beta peptide and Tau protein, respectively (1). Tau neurotoxicity is central in current models of AD pathogenesis and may mediate the effects of amyloid-beta (2). Rare mutations in either the amyloid precursor protein or microtubule-associated protein Tau genes cause familial dementia syndromes. Besides risk alleles at APOE, genome-wide association studies (GWAS) have identified common genetic variation at numerous other loci in association with AD susceptibility (3–6). Further, numerous studies directly link genetic risk factors to changes in AD pathology (7–10) or related biomarkers (11). Therefore, direct modulation of AD neuropathology is likely an important mechanism of disease susceptibility, and this realization also suggests a basis for successful functional screening strategies.
Expression of human Tau in the nervous system of the fruit fly, Drosophila melanogaster, recapitulates several features of AD and provides a useful experimental model for functional genetic dissection of mechanisms (12,13). In addition, large-scale and unbiased genetic modifier screens relevant to AD have been successfully performed in Drosophila (14–16), including for validation of results from human GWAS (17,18). Here, we extend this proven strategy for functional screening of results from two published AD GWAS (3,19), systematically considering candidate genes with both significant (P < 5 × 10−8) and suggestive (5 × 10−8 < P < 10−4) evidence of association. The latter group of associated loci, though falling short of genome-wide significance criteria, is likely enriched for true positive signals (20,21). Our Drosophila screening strategy successfully identifies genes within such susceptibility loci likely to influence AD. Specifically, we highlight 10 genes with fly orthologs that modulate Tau toxicity, including CD2AP, FERMT2 and CELF1. Importantly, while CD2AP was an established AD risk locus when our screen was undertaken, FERMT2 and CELF1 have been only recently and independently validated in association with AD susceptibility in the latest international GWAS meta-analysis (6). Thus, our results demonstrate how model organism studies can complement GWAS to identify AD susceptibility genes and further suggest that a number of these loci may modulate mechanisms of Tau-mediated neuronal injury.
RESULTS
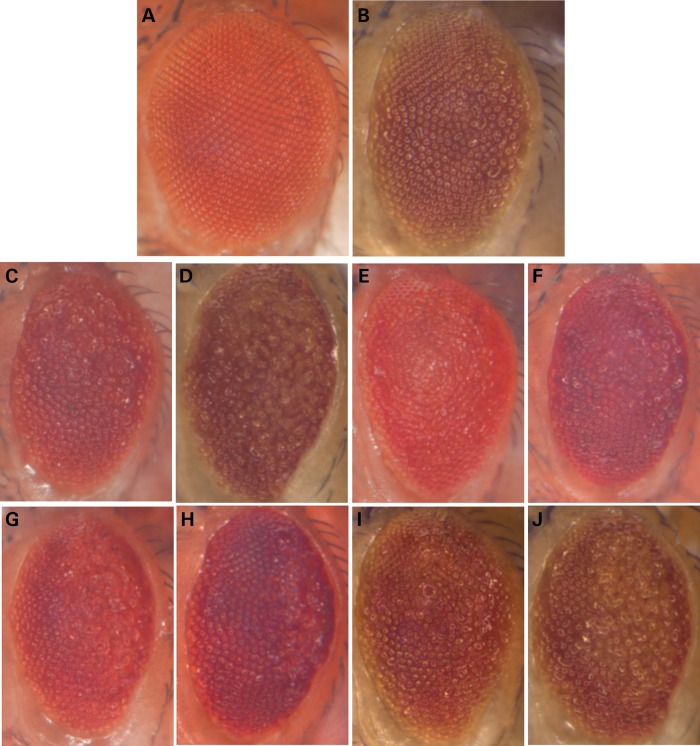
At the time our study was undertaken, 10 genetic loci had demonstrated significant associations with AD susceptibility (P < 5 × 10−8), and 5 of the implicated genes (PICALM, CD2AP, ABCA7, EPHA1 and AMPH) have conserved orthologs (22) in the Drosophila genome (Supplementary Material, Table S1). We utilized FlyBase (23) to identify all available RNA-interference (RNAi) transgenic stocks to facilitate a systematic gene disruption screen (24,25). In addition, we obtained lines predicted to activate gene expression. All reagents were crossed to transgenic flies allowing tissue-specific expression of human TauV337M, a mutant form of Tau associated with familial frontotemporal dementia (12). When expressed in the fly eye, under the control of GMR-GAL4, TauV337M causes a moderately reduced eye size and roughened surface (Fig. 1A and B), and this phenotype has been used successfully in our prior studies to identify second-site genetic interactors (14,17). RNAi lines were co-expressed with Dicer2 to potentiate gene silencing (24,25). Of the five genes evaluated, we found that genetic manipulation of cindr, the fly ortholog of human CD2AP (26), robustly enhanced Tau toxicity. Co-expression of cindr.RNAi with Tau caused further reduction in eye size and increased surface architectural disruptions (Fig. 1, Supplementary Material, Fig. S1). At the low expression levels in which interaction with Tau was documented, cindr. RNAi was not associated with any retinal toxicity when expressed independently of Tau (Supplementary Material, Fig. S2). Thus, our results suggest that cindr protects against Tau retinal toxicity, identifying a potential mechanism for the association of human CD2AP with AD susceptibility.
Figure 1.
RNAi-mediated disruption of AD candidate genes enhances Tau toxicity in Drosophila. Compared with control animals (A, GMR-Gal4, UAS-Dcr2/+), expression of human Tau generates a reduced eye size and moderate roughened appearance (B, UAS-TauV337M/+; GMR-Gal4, UAS-Dcr2/+). RNAi directed against several candidate genes enhanced Tau toxicity, exacerbating the rough eye phenotype: oxt (C, UAS-TauV337M/+; GMR-Gal4, UAS-Dcr2/UAS-oxt.RNAi); cindr (D, UAS-TauV337M/+; GMR-Gal4,UAS-Dcr2/+; UAS-cindr.RNAi3.73+81/+); Fit1 (E, UAS-TauV337M/+; GMR-Gal4,UAS-Dcr2/UAS-Fit1.IR.v46495); scb (F, UAS-TauV337M/+; GMR-Gal4, UAS-Dcr2/UAS-scb.IR.JF02696); Lar (G, UAS-TauV337M/UAS-Lar.IR.v36270; GMR-Gal4, UAS-Dcr2/+); SmB (H, UAS-TauV337M/+; GMR-Gal4, UAS-Dcr2/UAS-SmB.IR.HM05097); aret (I, UAS-TauV337M/+; GMR-Gal4,UAS-Dcr2/+; UAS-aret.IR.v41567/+) and CG6498 (J, UAS-TauV337M/+; GMR-Gal4,UAS-Dcr2/UAS-CG6498.IR.v35100). All modifier effects were scored using a semi-quantitative rating scale and found to be significantly different (P < 0.001) from controls, using pairwise independent sample t-tests (Supplementary Material, Fig. S1). The following RNAi lines showed consistent modifier effects, providing further independent confirmation: UAS-cindr.IR.JF02695, UAS-Fit1.IR.v46494, UAS-Lar.IR.HMS00822, UAS-SmB.IR.v110713, UAS-aret.IR.v41568, UAS-CG6498.IR.JF02778 and UAS-CG6948.IR.GL00220. RNAi lines were not associated with any significant toxicity when expressed independently of Tau (Supplementary Material, Fig. S2). RNAi lines used in the screen were obtained from publicly available collections (24,25) or were requested from other Drosophila laboratories (26,44).
Based on available GWAS summary data from the Alzheimer's Disease Genetics Consortium (ADGC) (3) and the Cohorts for Hearts and Aging in Genomic Epidemiology (CHARGE) (19), 147 additional independent loci (66 ADGC and 81 CHARGE) were considered for functional screening based on suggestive evidence of disease association (5 × 10−8 < P < 10−4) (Table 1 and Supplementary Material, Table S2). For determination of candidate genes, we defined a genomic window for each independently associated SNP, based on regional linkage disequilibrium patterns (r2 > 0.5) defined in the International HapMap Project (27). This yielded an initial list of 119 candidate genes from 70 distinct loci, of which 63 genes (53%) were strongly conserved in the Drosophila genome, and 84 homologous fly genes were identified (Supplementary Material, Table S3) (22). The genes promoted to our functional screen represent 44 independently associated loci from AD GWAS. A tabulated flowchart of our validation screen is presented in Table 1, and a full list of SNPs, candidate genes, including those with and without Drosophila orthologs is presented in Supplementary Material, Tables S2–S4. Following the strategy we deployed for the established AD loci (mentioned earlier), we identified and obtained 233 distinct RNAi transgenic lines (∼3 per candidate gene) to enable gene disruption at each targeted locus, and an additional 60 fly lines known or predicted to activate gene expression were also available for the majority of candidate genes (28). Supplementary Material, Table S5 details all of the Drosophila reagents tested in our screen.
Table 1.
Flowchart for screening of suggestive AD loci
| ADGC | CHARGE | Total | |
|---|---|---|---|
| Independently associated loci | 66 | 81 | 149 |
| Loci containing genes | 30 | 40 | 70 |
| Gene candidates | 51 | 68 | 119 |
| Conserved genes (loci) | 24 (18) | 39 (26) | 63 (44) |
| Fly orthologs | 35 | 49 | 84 |
| Tau modifiers | 6 | 3 | 9 |
Linkage-disequilibrium (LD)-based pruning was used to determine independently associated loci with suggestive evidence (5 × 10−8 < P < 1 × 10−4) for association with AD based on published GWAS from the ADGC (3) and CHARGE (19). A genomic window based on regional LD patterns (r2 > 0.5) was used to identify regional gene candidates. Not all loci had gene candidates based on these criteria. The DIOPT (22) was used to identify evolutionarily conserved genes and corresponding fly orthologs, which were promoted to the functional screen to identify Tau modifiers.
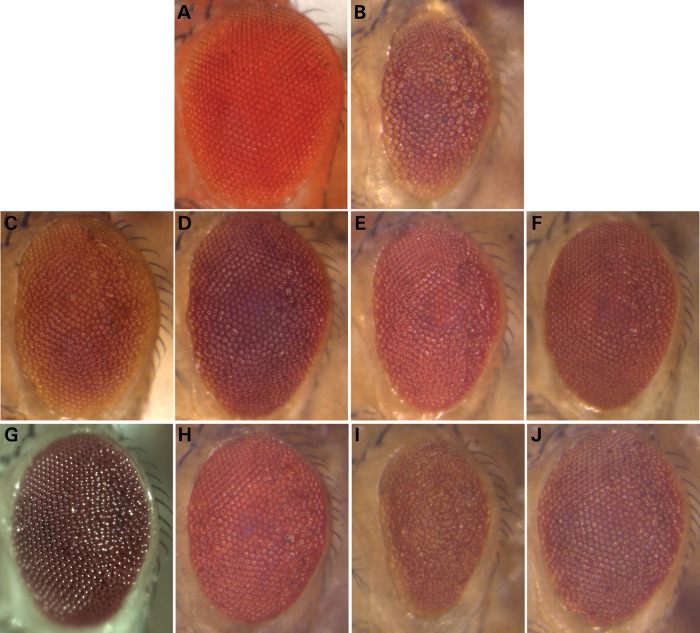
As with the established AD susceptibility loci, we systematically crossed all available lines to the TauV337M screening stock, identifying lines that enhanced or suppressed the rough eye phenotype. Eight additional genes were identified as robust Tau interactors (Table 2). In all cases, RNAi-mediated gene disruption enhanced Tau toxicity (Fig. 1), and we confirmed that RNAi lines did not disrupt eye morphology when tested independent of Tau in control crosses to GMR-GAL4; UAS-Dcr2 (Supplementary Material, Fig. S2). Reciprocally, we found that activating expression of 7 genes suppressed the Tau rough eye (Fig. 2). All reported genetic modifiers showed statistically significant differences from control Tau transgenic animals when scored using a semi-quantitative scale (P < 0.001, Supplementary Material, Fig. S1).
Table 2.
AD loci showing functional interactions with Tau in Drosophila
| SNP | CHR | P-value | Human gene | Fly ortholog(s) |
|---|---|---|---|---|
| ADGC | ||||
| rs9349407 | 6 | 8.6 × 10−9 | CD2AP | cindr |
| rs7175782 | 15 | 2.6 × 10−5 | SNRPN | SmB |
| rs2136530 | 9 | 3.4 × 10−5 | PTPRD | Lar |
| rs6498673 | 16 | 3.9 × 10−5 | XYLT1 | oxt |
| rs17125924 | 14 | 5.2 × 10−5 | FERMT2 | Fit1, Fit2 |
| rs7206295 | 16 | 7.1 × 10−5 | ITGAM | scb |
| CHARGE | ||||
| rs2242081 | 11 | 2.3 × 10−5 | CELF1 | aret |
| rs7722928 | 5 | 2.9 × 10−5 | MAST4 | CG6498 |
| rs267526 | 3 | 9.6 × 10−5 | ITGA9 | scb |
Figure 2.
Activating AD candidate genes modifies Tau toxicity in Drosophila. Compared with control animals (A, GMR-Gal4/+), expression of human Tau generates a reduced eye size and moderate roughened appearance (B, UAS-TauV337M/+; GMR-Gal4/+). Lines predicted to activate expression of candidate genes were suppressors of the Tau rough eye: Lar (C,UAS-TauV337M/+; GMR-Gal4/UAS-Lar); Fit2 (D, UAS-TauV337M/+; GMR-Gal4/+; Fit2EY08530); Fit1 (E, UAS-TauV337M/+; GMR-Gal4/UAS-Fit1); oxt (F, UAS-TauV337M/+; GMR-Gal4/+; oxtEPG4946/+); SmB (G, UAS-TauV337M/+; GMR-Gal4/+; UAS-SmB/+) and aret (H, UAS-TauV337M/+; GMR-Gal4/+; UAS-aret/+). Two different lines predicted to activate expression of scb either enhanced (I, UAS-TauV337M/+; GMR-Gal4/UAS-scbVolL) or suppressed (J, UAS-TauV337M/+; GMR-Gal4/scbEY10270) Tau toxicity, and consistent results were seen with the independent lines scbEY02806and UAS-scbVolL(III), respectively. All modifier effects were scored using a semi-quantitative rating scale and found to be significantly different (P < 0.001) from controls, using pairwise independent sample t-tests (Supplementary Material, Fig. S1). Activating lines used in the screen were obtained from publicly available collections (28) or were requested from other Drosophila laboratories (45–48); the UAS-Fit1 transgenic line was generated de novo (see Materials and Methods).
Notably, 2 of the 8 suggestively associated genes highlighted by our screen, FERMT2 and CELF1, have now been independently validated as AD susceptibility loci based on the latest GWAS meta-analysis with substantially enhanced sample size (6). Two Drosophila genes, Fit1 and Fit2, are strongly homologous with human FERMT2, and both genes were promoted to our screen based on suggestive disease association of rs17125924 (P = 5.2 × 10−5) (3). RNAi against either Fit1 or Fit2 enhanced the Tau rough eye (Fig. 1, Supplementary Material, Fig. S1), whereas lines predicted to activate expression of these genes suppressed Tau (Fig. 2). Similarly, loss- or gain-of-function in aret, the fly ortholog of CELF1, enhanced and suppressed Tau retinal toxicity, respectively. CELF1 was evaluated along with 7 other conserved genes out of 10 candidates from the genomic locus identified by the rs2242081 association with AD (P = 2.3 × 10−5) (19). aret was the only homologous fly gene nominated by this locus that interacted with Tau toxicity, illustrating the utility of our strategy to annotate the most likely causal genes within susceptibility loci.
Of the established AD susceptibility loci, both CD2AP and FERMT2 have been implicated in cell adhesion (29,30), and as discussed earlier, both loci possess fly orthologs that modify Tau toxicity. Specifically, FERMT2 encodes a member of the kindlin protein family, which transduces signals from integrin adhesion receptors (31), and CD2AP similarly functions with integrins to maintain glomerular adhesion and structural integrity in the mammalian kidney (32). Intriguingly, among the comprehensive list of suggestively associated loci were several human genes encoding integrin receptor alpha-subunits, including ITGAM, ITGA8 and ITGA9 (Supplementary Material, Table S2). Consequently, we evaluated homologous Drosophila integrins in our screen, including scab, inflated, alphaPS4 and alphaPS5 (Supplementary Material, Table S3). RNAi against scab, encoding a fly integrin alpha subunit homologous to both ITGAM and ITGA9, enhanced the Tau rough eye (Fig. 1, Supplementary Material, Fig. S1). Further, lines that activate expression of scab either suppressed or enhanced Tau (Fig. 2), likely due to dominant-negative interactions at higher expression levels, as previously reported for Drosophila integrins (33). We also tested lines disrupting inflated (if), encoding another Drosophila integrin alpha subunit homologous to human ITGA8. Although RNAi against if provided some support for an enhancing interaction with Tau (data not shown), we were unable to confirm with a second, independent reagent (either another RNAi line or activating insertion), precluding definitive functional validation of this gene based on our screening assay. Several other genes showed interactions with Tau, as detailed in Table 1 and Figures 1 and 2, including additional potential regulators of cell adhesion pathways.
DISCUSSION
By integrating results from AD GWAS with a Drosophila functional screen, we identify nine genes with potential roles in AD pathogenesis, supported by association with disease susceptibility in humans and genetic interactions with Tau in vivo. Following the success of GWAS to identify risk loci for complex genetic disorders such as AD, the critical next step will be to confirm the responsible genes and begin to understand the relevant molecular mechanisms. Our screening strategy is well suited to achieve these goals. In the case of three loci with established links to AD—CD2AP, FERMT2 and CELF1—we show that genetic manipulation of fly gene homologs modulates Tau neurotoxicity. These results help to (1) identify the gene responsible for a locus association and (2) establish a potential mechanism based on the functional screening paradigm (in our case, Tau toxicity). In an independent study, BIN1 was similarly implicated to alter Tau-mediated neuronal injury (18). At present, the refinement and fine mapping of GWAS signals to determine the causal gene remains a challenge (34). The haplotypes identified by associated polymorphisms often contain multiple genes, including many that a priori appear to be equally good candidates. For example, in the case of the CELF1 locus considered in our analysis, the most strongly associated SNP tag identified an LD-based genomic interval overlapping 10 gene candidates (Supplementary Material, Table S2), of which 8 were sufficiently conserved in the Drosophila genome for promotion to our screen. However, only aret modified the Tau rough eye, validating CELF1 as the most likely gene responsible for the locus association with AD susceptibility. While our results suggest that many AD susceptibility genes may influence Tau-mediated neuronal injury, future studies will be required to establish the detailed mechanisms. For example, it will be important to determine, using fly models or other experimental systems, whether these genes directly or indirectly lead to Tau hyperphosphorylation, misfolding or aggregation, which are all established correlates of increased toxicity and disease progression (2).
Importantly, neither the FERMT2 nor CELF1 loci were established, based on the most stringent genome-wide significance criteria (P < 5 × 10−8), when we initiated this study. Rather, these candidates were among a comprehensive list of 119 genes from 70 loci with suggestive evidence of association (5 × 10−8 < P < 10−4) based on earlier studies (3,19). The independent validation of these two loci in the recently reported GWAS meta-analysis (6) powerfully demonstrates the validity of the Drosophila screening approach to enhance human genetic studies. As we screened 84 fly gene homologs and identified only 9 Tau modifiers, it is highly unlikely that we would have identified homologs of FERMT2 and CELF1 (fit1, fit2 and aret) simply by chance (P < 0.001). In fact, the overall ‘hit’ rate of Tau modifiers in our study (∼10%) is ∼10-fold higher than that in similar screening efforts of unselected Drosophila genes (14), suggesting that our candidate gene list was indeed enriched for modulators of Tau-mediated neurodegeneration. It is now recognized that susceptibility for complex genetic disorders, including neuropsychiatric diseases like AD, is likely influenced by a large number of genetic variants, including perhaps hundreds of distinct genomic loci (20,21). With the completion of the recent international meta-analysis including 74 000 subjects (6), we are likely approaching the upper limit of sample size and statistical power achievable using current GWAS designs. Therefore, innovative approaches are needed if we are to identify additional loci among those falling short of the genome-wide significance threshold. While such variants may individually have relatively weak effects on disease risk, a comprehensive list may facilitate identification of important cellular pathways in AD pathogenesis. Our results suggest that model organism screening might be one successful strategy to accomplish this goal. Thus, like CELF1 and FERMT2, we suggest that six other genes—SNRPN, PTPRD, XYLT1, ITGAM, ITGA9 and MAST4—with similar suggestive disease associations and shown here to possess fly orthologs that also interact with Tau, become excellent candidates for further study in both human subjects and model systems relevant to AD.
Several genetic loci identified by AD GWAS, including CD2AP and FERMT2, have been similarly implicated with roles in cellular adhesion, and more specifically function coordinately with integrin receptors (31,32). Besides the Drosophila orthologs of these loci (cindr, Fit1 and Fit2), we additionally find that the fly integrin receptor gene scab interacts with Tau toxicity, which was evaluated based on suggestive associations of the human ITGA9 and ITGAM loci with AD. Other genes identified by our functional screen, PTPRD and XYLT1, also have established roles in cellular adhesion (35,36). Notably, a recent analysis of co-expression networks in the brain transcriptome highlighted functional modules representing integrin adhesion, cell adhesion and the extracellular matrix among those most significantly dysregulated in AD (37). Understanding the mechanism by which neuronal and/or synaptic adhesion may modulate Tau toxicity and AD susceptibility will require more detailed investigation; however, we hypothesize that dynamic changes in the actin cytoskeleton might be important, as suggested by prior work (38,39). Importantly, CD2AP is an actin-associated protein (40), and integrin signaling—mediated by FERMT2—can re-organize the actin cytoskeleton (31).
Ultimately, multiple strategies will be required for the comprehensive functional evaluation of candidate disease susceptibility loci, and no single approach will be without limitations. While Drosophila offers powerful and rapid genetics, supporting a medium-to-high-throughput screening strategy, not all AD susceptibility genes are conserved between human and flies (Supplementary Material, Table S5). Lack of conservation precluded evaluation of genes involved in lipid metabolism (APOE and CLU) and immunity (CR1, CD33 and MS4A), for example. Further, while the literature supports a central role for Tau leading to neuronal death in AD (2), many risk loci may impact alternative disease mechanisms. The Tau transgenic model utilized in our screen does not address APP cleavage or the clearance of amyloid-ß, which are key determinants of AD pathogenesis, although Tau is likely an important mediator of amyloid-ß toxicity (1,2). Thus, it may be important in future work to iteratively screen AD susceptibility genes with several complementary assays. It is notable that all of the genes highlighted by our screen were found to be loss-of-function enhancers of Tau toxicity, consistent with a potential protective role in AD. In prior studies using a similar screening strategy (14,17), we have additionally identified loss-of-function suppressors, although such genes appear to be less common overall. It is possible that this reflects a greater sensitivity of our screening assay for enhancers or rather could be a consequence of the underlying genetic determinants of Tau-induced neuronal injury. Future functional studies in flies or other systems may additionally help identify susceptibility genes, which function as endogenous promoters of AD pathogenesis. Finally, our strategy required several assumptions to identify candidate genes from associated polymorphisms. While linkage-disequilibrium criteria provide a conservative estimate for the genomic window containing a potential causal variant, regulatory variants could potentially act over much longer ranges to impact candidate genes outside this interval. Accounting for such distant effects, however, would have required screening a much greater number of gene candidates. Given that regulatory variants indeed appear to be enriched among AD-associated variants (41), one potential refinement to our strategy would be to prioritize additional genes in which brain expression is linked to associated polymorphisms.
In conclusion, we demonstrate how a model organism screening strategy can identify AD susceptibility variants and enhance human GWAS results by providing clues to molecular mechanisms. Our results suggest that many AD risk genes are determinants of Tau-mediated neuronal injury and further highlight cell adhesion as one important pathway for further study. When coupled with emerging human genomic data, simple animal models, such as the Drosophila system used here, show great promise to accelerate the functional genetic dissection of complex diseases.
MATERIALS AND METHODS
Identification of candidate genes and orthologs
Significant (P < 5 × 10−8) and suggestive (5 × 10−8 < P < 1 × 10−4) SNP associations with AD were based on published GWAS (3,19). Linkage-disequilibrium (LD)-based clumping (r2 > 0.1), implemented within PLINK (42), was used to define the top-ranked and independently associated SNPs. Next, a genomic window was defined around each associated SNP based on regional LD patterns (0.5 < r2 < 1) from HapMap (27). Any gene from the human reference genome (build hg18) overlapping this defined interval was designated a candidate causal gene and considered further for functional analyses. For identification of fly gene orthologs, we used the Drosophila Integrated Ortholog Prediction Tool (DIOPT) (22), available as a web-based tool (http://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl). DIOPT facilitates rapid query of fly orthologs based on 10 distinct bioinformatic algorithms. We applied a stringent standard requiring a ‘DIOPT score’ of 2 or higher for promotion of a gene to our screen, meaning that at least two distinct algorithms agreed on the ortholog pairing. For the 67 human candidate genes with identifiable fly orthologs, the mean DIOPT score was 5.0 (SD = 2.2). Thirteen genes had more than one identifiable fly ortholog meeting our pre-specified criteria, and in these cases, all corresponding fly genes were considered for screening. Supplementary Material, Tables S4 and S5 detail the identification of Drosophila orthologs based on human genes, including DIOPT scores and the evidence of homology.
Drosophila genetics
Fly gene orthologs were queried within FlyBase to identify available genetic reagents for manipulation of candidate gene function (23). For each candidate susceptibility gene, we obtained RNA-interference (RNAi) stocks and lines to activate gene expression. Transgenic RNAi lines exist for virtually all Drosophila genes, and we obtained all available lines to evaluate target genes (24,25). Additional lines consisted of either previously characterized, published reagents or those available from collections of transposon alleles predicted to activate gene expression based on their insertion sites (28). Reagents were obtained from the Bloomington Drosophila Stock Center, the Harvard/Exelixis collection, the Vienna Drosophila RNAi Center, the Harvard Transgenic RNAi project (TRiP), or requested from laboratories within the fly research community. The UAS-Eph transformant was re-established by injecting pUAST-Eph plasmid generously provided by Dr R. Dearborn and Dr S. Kunes (43). The UAS-Fit1.GFP flies were established by amplifying the Fit1 coding sequence and inserting into a pUAS vector for P-element transformation. The coding sequence for green fluorescent protein flanked by 3 serines was inserted within a poorly conserved loop in domain F1, after residue 228. A genomic rescue construct with GFP in this position rescued Fit1 mutations. All Drosophila reagents used in this study are detailed in Supplementary Material, Table S5, and full genotypes are also included in the figure legends with the relevant references.
The UAS-TauV337M line used for evaluation of candidate genes has been previously described (12,14,17). All functional validation tests were the product of a single generation genetic cross and assessed the ability of lines to enhance or suppress the moderate rough eye phenotype of UAS-TauV337M/+; GMR:gal4/+ animals. All RNAi reagents were evaluated with co-expression of Dicer2, using animals of the genotype, UAS-TauV337M/+; UAS-Dcr2, GMR-Gal4/+. All reported crosses were carried out at 25°C. Reported modifier effects (Table 1) are based on lines that showed strong and consistent effects to enhance or suppress the TauV337M rough eye phenotype in repeat experiments. Modifier effects were photographed and quantified in female animals, but all modifiers showed consistent effects in both sexes. All lines found to enhance Tau toxicity were evaluated with GMR-Gal4, UAS-Dcr2 in isolation, and none demonstrated significant toxicity in the absence of Tau (Supplementary Material, Fig. S2). To minimize the chance that observed modifier interactions were caused by off-target effects of RNAi transgenic stocks, we required consistent activity of at least two independent RNAi lines, or independent confirmation of modifier effects based on activating gain-of-function lines.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (K08AG034290, C06RR029965), the Burroughs Wellcome Fund (Career Award for Medical Scientists to J.M.S.), the Ellison Medical Foundation (to M.B.F.) and the Wellcome Trust (Grant 086451 to N.H.B.). The TRiP at Harvard Medical School, which provided fly stocks, is supported by R01GM084947.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr U. Pandey, Dr R. Bodmer, Dr C. Micchelli, Dr P. Macdonald, Dr V. Panin, Dr L.S. Sashidhara, Dr R. Benton, Dr S. Hayashi, Dr H. Bellen, Dr A. Diantonio, Dr S. Nishihara, Dr R. Johnson, Dr S. Kunes, Dr R. Dearborn and Dr R. Cagan for generously providing Drosophila reagents. We also thank the Bloomington Drosophila stock center, the Vienna Drosophila RNAi Center and the TRiP at Harvard Medical School for providing fly stocks. We are grateful to Dr S. Seshadri and the CHARGE consortium for sharing results of top-ranked, discovery-stage SNP associations from the published GWAS (4).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Querfurth H.W., LaFerla F.M. Alzheimer's disease. N. Engl. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 2.Spillantini M.G., Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 3.Naj A.C., Jun G., Beecham G.W., Wang L.-S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M., Bis J.C., Smith A.V., Carrasquillo M.M., Lambert J.C., et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.-C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert J.-C., Ibrahim-Verbaas C., Harold D., Naj A.C., Sims R., Bellenguez C., Jun G., DeStefano A.L., Bis J.C., Beecham G.W., et al. Meta-analysis in More Than 74,000 Individuals Identifies 11 new Susceptibility Loci for Alzheimer's Disease. Boston, MA: Platform Presentation at the Alzheimer's Association International Conference; 2013. [Google Scholar]
- 7.Shulman J.M., Chen K., Keenan B.T., Chibnik L.B., Fleisher A.S., Thiyyagura P., Roontiva A., McCabe C., Patsopoulos N.A., Corneveaux J., et al. Genetic susceptibility for Alzheimer's disease neuritic plaque pathology. JAMA Neurol. 2013;70:1150–1157. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chibnik L.B., Shulman J.M., Leurgans S.E., Schneider J.A., Wilson R.S., Tran D., Aubin C., Buchman A.S., Heward C.B., Myers A.J., et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann. Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griciuc A., Serrano-Pozo A., Parrado A.R., Lesinski A.N., Asselin C.N., Mullin K., Hooli B., Choi S.H., Hyman B.T., Tanzi R.E. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradshaw E.M., Chibnik L.B., Keenan B.T., Ottoboni L., Raj T., Tang A., Rosenkrantz L.L., Imboywa S., Lee M., Von Korff A., et al. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruchaga C., Kauwe J.S.K., Harari O., Jin S.C., Cai Y., Karch C.M., Benitez B.A., Jeng A.T., Skorupa T., Carrell D., et al. GWAS of cerebrospinal fluid Tau levels identifies risk variants for Alzheimer's disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wittmann C.W., Wszolek M.F., Shulman J.M., Salvaterra P.M., Lewis J., Hutton M., Feany M.B. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- 13.Moloney A., Sattelle D.B., Lomas D.A., Crowther D.C. Alzheimer's disease: insights from Drosophila melanogaster models. Trends Biochem. Sci. 2009;35:228–235. doi: 10.1016/j.tibs.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shulman J.M., Feany M.B. Genetic modifiers of tauopathy in Drosophila. Genetics. 2003;165:1233–1242. doi: 10.1093/genetics/165.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blard O., Feuillette S., Bou J., Chaumette B., Frébourg T., Campion D., Lecourtois M. Cytoskeleton proteins are modulators of mutant tau-induced neurodegeneration in Drosophila. Hum. Mol. Genet. 2007;16:555–566. doi: 10.1093/hmg/ddm011. [DOI] [PubMed] [Google Scholar]
- 16.Ambegaokar S.S., Jackson G.R. Functional genomic screen and network analysis reveal novel modifiers of tauopathy dissociated from tau phosphorylation. Hum. Mol. Genet. 2011;20:4947–4977. doi: 10.1093/hmg/ddr432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shulman J.M., Chipendo P., Chibnik L.B., Aubin C., Tran D., Keenan B.T., Kramer P.L., Schneider J.A., Bennett D.A., Feany M.B., et al. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am. J. Hum. Genet. 2011;88:232–238. doi: 10.1016/j.ajhg.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapuis J., Hansmannel F., Gistelinck M., Mounier A., Van Cauwenberghe C., Kolen K.V., Geller F., Sottejeau Y., Harold D., Dourlen P., et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. [epub ahead of print, February 12, 2013] Mol. Psychiatry. doi: 10.1038/mp.2013.1. doi:10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M., Bis J.C., Smith A.V., Carassquillo M.M., Lambert J.C., et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'donovan M.C., Sullivan P.F., Sklar P. International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McQuilton P., St Pierre S.E., Thurmond J. FlyBase Consortium. Flybase 101–the basics of navigating flyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 25.Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Tao R., Handler D., Karpowicz P., et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Meth. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R.I., Seppa M.J., Cagan R.L. The Drosophila CD2AP/CIN85 orthologue cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J. Cell Biol. 2008;180:1191–1204. doi: 10.1083/jcb.200706108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frazer K., Ballinger D., Cox D., Hinds D., Stuve L., Gibbs R., Belmont J., Boudreau A., Hardenbol P., Leal S., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellen H.J., Levis R.W., He Y., Carlson J.W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K.L., et al. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai-Cheong J.E., Parsons M., McGrath J.A. The role of kindlins in cell biology and relevance to human disease. Int. J. Biochem. Cell Biol. 2010;42:595–603. doi: 10.1016/j.biocel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Wolf G., Stahl R.A.K. CD2-associated Protein and glomerular disease. Lancet. 2003;362:1746–1748. doi: 10.1016/S0140-6736(03)14856-8. [DOI] [PubMed] [Google Scholar]
- 31.Moser M., Legate K.R., Zent R., Fässler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 32.Chiang C.-K., Inagi R. Glomerular diseases: genetic causes and future therapeutics. Nat. Rev. Nephrology. 2010;6:539–554. doi: 10.1038/nrneph.2010.103. [DOI] [PubMed] [Google Scholar]
- 33.Brabant M.C., Fristrom D., Bunch T.A., Brower D.L. Distinct spatial and temporal functions for PS integrins during Drosophila wing morphogenesis. Development. 1996;122:3307–3317. doi: 10.1242/dev.122.10.3307. [DOI] [PubMed] [Google Scholar]
- 34.Ioannidis J.P.A., Thomas G., Daly M.J. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 2009;10:318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chagnon M.J., Uetani N., Tremblay M.L. Functional significance of the LAR receptor protein tyrosine phosphatase family in development and diseases. Biochem. Cell Biol. 2004;82:664–675. doi: 10.1139/o04-120. [DOI] [PubMed] [Google Scholar]
- 36.Sarrazin S., Lamanna W.C., Esko J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004952. doi:10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B., Gaiteri C., Bodea L.-G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulga T.A., Elson-Schwab I., Khurana V., Steinhilb M.L., Spires T.L., Hyman B.T., Feany M.B. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol. 2007;9:139–148. doi: 10.1038/ncb1528. [DOI] [PubMed] [Google Scholar]
- 39.DuBoff B., Götz J., Feany M.B. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75:618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehtonen S., Zhao F., Lehtonen E. CD2-associated protein directly interacts with the actin cytoskeleton. Am. J. Physiol. Renal Physiol. 2002;283:F734–F743. doi: 10.1152/ajprenal.00312.2001. [DOI] [PubMed] [Google Scholar]
- 41.Allen M., Zou F., Chai H.S., Younkin C.S., Crook J., Pankratz V.S., Carrasquillo M.M., Rowley C.N., Nair A.A., Middha S., et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–228. doi: 10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M., Bender D., Maller J., Sklar P., de Bakker P., Daly M., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dearborn R., He Q., Kunes S., Dai Y. Eph receptor tyrosine kinase-mediated formation of a topographic map in the Drosophila visual system. J. Neurosci. 2002;22:1338–1349. doi: 10.1523/JNEUROSCI.22-04-01338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueyama M., Takemae H., Ohmae Y., Yoshida H., Toyoda H., Ueda R., Nishihara S. Functional analysis of proteoglycan galactosyltransferase II RNA interference mutant flies. J. Biol. Chem. 2008;283:6076–6084. doi: 10.1074/jbc.M709189200. [DOI] [PubMed] [Google Scholar]
- 45.Wada A., Kato K., Uwo M.F., Yonemura S., Hayashi S. Specialized extraembryonic cells connect embryonic and extraembryonic epidermis in response to Dpp during dorsal closure in Drosophila. Dev. Biol. 2007;301:340–349. doi: 10.1016/j.ydbio.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 46.Gonsalvez G.B., Rajendra T.K., Wen Y., Praveen K., Matera A.G. Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development. 2010;137:2341–2351. doi: 10.1242/dev.042721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snee M.J., Harrison D., Yan N., Macdonald P.M. A late phase of oskar accumulation is crucial for posterior patterning of the Drosophila embryo, and is blocked by ectopic expression of bruno. Differentiation. 2007;75:246–255. doi: 10.1111/j.1432-0436.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 48.Krueger N.X., Van Vactor D.D., Wan H.I., Gelbart W.M., Goodman C.S., Saito H. The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell. 1996;84:12. doi: 10.1016/s0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.