Abstract
Mutations of mitochondrial DNA are linked to many human diseases. Despite the identification of a large number of variants in the mitochondrially encoded rRNA (mt-rRNA) genes, the evidence supporting their pathogenicity is, at best, circumstantial. Establishing the pathogenicity of these variations is of major diagnostic importance. Here, we aim to estimate the disruptive effect of mt-rRNA variations on the function of the mitochondrial ribosome. In the absence of direct biochemical methods to study the effect of mt-rRNA variations, we relied on the universal conservation of the rRNA fold to infer their disruptive potential. Our method, named heterologous inferential analysis or HIA, combines conservational information with functional and structural data obtained from heterologous ribosomal sources. Thus, HIA's predictive power is superior to the traditional reliance on simple conservation indexes. By using HIA, we have been able to evaluate the disruptive potential for a subset of uncharacterized 12S mt-rRNA variations. Our analysis revealed the existence of variations in the rRNA component of the human mitoribosome with different degrees of disruptive power. In cases where sufficient information regarding the genetic and pathological manifestation of the mitochondrial phenotype is available, HIA data can be used to predict the pathogenicity of mt-rRNA mutations. In other cases, HIA analysis will allow the prioritization of variants for additional investigation. Eventually, HIA-inspired analysis of potentially pathogenic mt-rRNA variations, in the context of a scoring system specifically designed for these variants, could lead to a powerful diagnostic tool.
INTRODUCTION
Currently, more than 550 mitochondrial DNA (mtDNA) mutations and sequence variants have been associated with human disease (1). Since it has been estimated that the minimal prevalence of clinically manifesting mtDNA disease in adults is ∼1 in 10 000 (2), establishing whether mtDNA mutations are pathogenic is of major importance for genetic counseling and patient support. Whilst important, the assignment of pathogenicity to mtDNA mutations is a difficult task. This is in no small part due to high levels of mtDNA variation in human populations and the extremely heterogeneous clinical presentations of many known mtDNA mutations. Due to these and other difficulties, the evidence for pathogenicity has been rather weak in some cases, especially in the context of rRNA variation. In fact, some authors have suggested that a number of these associations might be erroneous (3–7).
DiMauro and Schon (8) were the first to describe criteria for the identification of mutations in mtDNA linked to disease. Since then, our understanding of the elements associated with pathogenicity has substantially changed. For example, the first element cited by DiMauro and Schon for the identification of pathogenic mutations was that the variant must be heteroplasmic (i.e. the mutated and wild-type variants co-existing in the cells of the affected tissues). More recently, it has become appreciated that certain pathogenic variants may exist as homoplasmic mutations with variable degrees of penetrance (9). These changes in our appreciation of the elements required for pathogenicity, together with the fact that the data available in databases has grown enormously, makes it imperative to find a new framework for the analysis of mitochondrially encoded rRNA (mt-rRNA) variations. The emergence of scoring systems has provided a much needed tool for the evaluation of certain categories of mtDNA mutations and should provide guidance to future research on mt-rRNA variation (6,7,10–12).
With all probability, an important subset of pathogenic mtDNA variations map to the mt-rRNA genes; however, they remain the most difficult genes in which to confirm the pathogenicity of an mtDNA variant. Since the mitochondrial ribosome (mitoribosome) translates all 13 proteins encoded by the mitochondrial genome, mutations leading to ribosome dysfunction should result in a respiratory chain defect and, therefore, cause mitochondrial disease. This prediction was confirmed by the identification of alterations in the nuclear and mitochondrial genome that affect the mitochondrial translational machinery. For example, a small number of highly deleterious mutations have been identified in the nuclear genes encoding protein components of mitoribosomes, mitochondrial translation factors and elements of the apparatus involved in the maturation of mitoribosomes (13–20). The severity of these mutations clearly indicates that interfering with mitochondrial protein synthesis can lead to highly deleterious phenotypes. Not surprisingly, deleterious mutations have also been identified in the rRNA component of the mitoribosome. Two such mutations, 908A>G (m.1555A>G) and 847C>U (m.1494C>T), are well-known causes of hearing loss (21). In addition, there is a large list of other mt-rRNA mutations whose pathogenic role is yet to be convincingly proven.
Thus, despite the identification of base substitutions in the mt-rRNA of patients, the evidence supporting the pathogenicity of mt-rRNA variations is, at best, circumstantial, except for the two cases mentioned above. The reasons for this are many. First, ascertaining the magnitude of the actual biochemical defect caused by such variations is often complicated by incomplete penetrance, as well as by the possible existence of nuclear modifiers. Secondly, the existence of numerous human haplogroups further complicates the correct assignment of mutations, as the penetrance of some variants might change in different genetic contexts (22). Thirdly, the intractability of the mitochondrial translation apparatus and the lack of methods for the manipulation of the mitochondrial genome make it impossible to test the effect of these mutations directly. Finally, all of this is complicated by the lack of agreement among authors on the criteria that should be used to evaluate pathogenicity in the context of mt-rRNA mutations (5,23). For all these reasons, it is imperative that a robust system is devised that allows the assessment of the biochemical defect caused by mt-rRNA mutations.
Scoring systems have been developed and validated for the analysis of the potential pathogenicity of both mt-tRNA and mitochondrially encoded complex I subunits (6,7,10–12). The existence of a substantial number of known mutations and gold-standard experimental procedures, including biochemical and transmitochondrial-cybrid evidence, were important in developing these scoring systems. Unfortunately, these elements are largely lacking in the case of mt-rRNA variations. To fill this void, we propose taking advantage of the universal conservation of the rRNA structure (24) in order to assess the disruptive potential of mt-rRNA variations. Our approach, which we have named heterologous inferential analysis (HIA) predicts the structural and/or functional role of a particular mt-rRNA residue by extrapolating conclusions drawn from evidence pertaining to its structurally equivalent position in a heterologous ribosome. HIA is used in this paper to analyze a subset of extremely rare variations mapping to the small ribosomal subunit (SSU) of the mitoribosome, which are suspected to be associated with a respiratory chain defect. Our goal is to establish whether these mt-rRNA variations have a potentially disruptive effect on the function of the mitoribosome. While HIA alone cannot, in most cases, be used to establish pathogenicity beyond doubt, we believe that it constitutes an important first step toward building a scoring system with clinical relevance that can be used to predict the pathogenic potential of mt-rRNA variations. At the same time, HIA's predictions will allow the prioritization of variants for additional functional investigation. Additionally, for certain mt-rRNA mutations for which sufficient information regarding the genetic and pathological manifestation of the mutations are available, HIA data can be used to predict their pathogenicity.
RESULTS
Our search through the literature for mt-12S rRNA variations possibly associated with disruption of mitochondrial function, as defined in the section Materials and Methods, resulted in the identification of 161 different variations, corresponding to 152 different sites, almost 16% of the molecule (not shown). Evaluation of evidence used to identify mtDNA mutations in other mtDNA genes indicates that the absence of the variant in controls is important evidence of pathogenicity. In the absence of proper experimental controls in most of the cases studied here, we turned to the GenBank and Phylotree databases to screen for extremely rare variations, i.e. variations with zero appearances in these databases. While we know this criteria would excluded the known pathogenic variants, 1555A>G with 29 appearances and 1494C>T with three appearances, it was hoped that such a criterion would identify a group of variants most likely to yield pathogenic or potentially pathogenic mutations.
Correlation between phylogenetic conservation and the distribution of variations in mt-12S rRNA
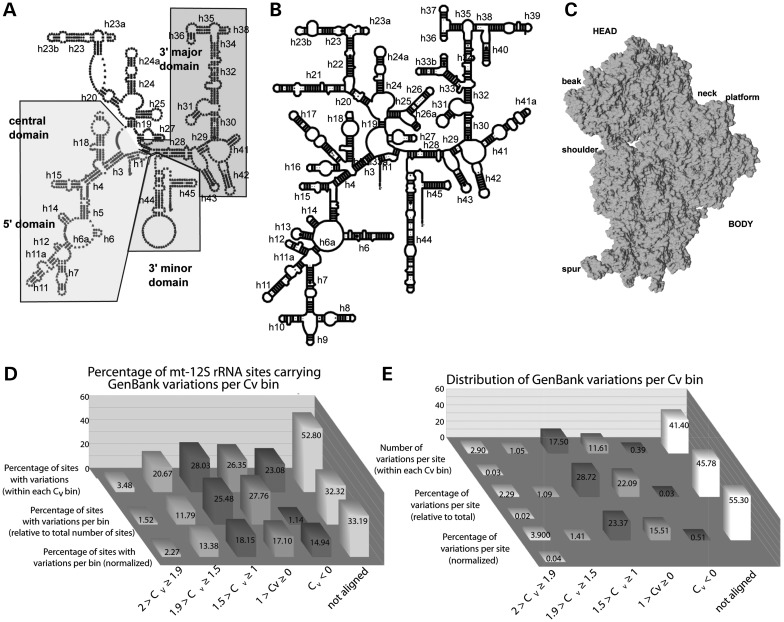
The accuracy of phylogeny-driven algorithms for the prediction of rRNA structure has been amply demonstrated (24). Therefore, we reasoned that the universal conservation of the rRNA fold must be central to any inferential attempt to ascertain the potentially disruptive effect of mutations in mt-rRNA. Figure 1A shows the secondary structure map of the human 12S rRNA with all domains and helices numbered according to the nomenclature used for the bacterial 16S rRNA, shown in Figure 1B (25). Figure 1C shows the three dimensional structure of the Escherichia coli SSU with landmarks indicated (accession code 2I2P). Despite the loss of many peripheral RNA segments in the human 12S rRNA, the overall fold is clearly conserved (Fig. 1A and B), thus allowing the identification of equivalent residues in both structures. Only for positions lying in areas of little structural homology does the identification of heterologous structural equivalents become impossible.
Figure 1.
(A and B) SSU secondary-structure maps. The secondary-structure map of the human mt-12S rRNA is shown in (A). Helices numbered according to the E. coli secondary structure model, shown in B. (C) Three-dimensional structure of the E. coli SSU with landmarks indicated. (D) Percentage of mt-12S rRNA positions carrying GenBank variations within each Cv bin. Cv bins were defined as described at the Comparative RNA Web (CRW) Site (25,26). Back row: per-bin percentage of sites with GenBank variations, relative to the total number of residues in each Cv bin. Middle row: per-bin percentage of sites with GenBank variations, relative to the total number of mt-12S rRNA sites with variations present in GenBank. Front row: normalized, per-bin percentage of sites with GenBank variations, relative to the normalized total number of mt-12S rRNA sites with such variations (normalization is to the Cv bin with the highest number of sites, i.e. 1 > Cv ≥ 0; the normalized values used for these calculations are shown in Table 1). X axis: Cv bins. Y axis: percentage value. (E) Distribution of mt-12S rRNA GenBank variations per Cv bin. The distribution is shown as the number of GenBank variations per site within each Cv bin (two back rows, see main text for calculation details); as the per-bin percentage of variations, relative to the total number of mt-12S rRNA variations present in GenBank (two middle rows); and as the per-bin, normalized percentage of variations, relative to the normalized total number of mt-12S rRNA variations with such variations (normalization as in D). Odd rows: as adjacent even row but corrected after discarding the outlier residue 180A (m.827A) (see main text. Only shown for most conserved bin). X axis: Cv bins. Y axis: number of variations per site (two back rows), percentage value (four front rows).
The aforementioned idea that rare variations might be good candidates for pathogenic mutations would require a non-random distribution of variations in the MT-RNR1gene, i.e. that an inverse correlation exists between phylogenetic conservation and the number of variations per residue. However, our analysis of near 18 000 complete sequences of the human mt-genome currently deposited in GenBank showed a high prevalence of mt-12S rRNA sites carrying variations, i.e. 27.6% of all sites (Table 1). This prompted us to consider whether such a correlation between phylogenetic conservation and the distribution of variations per mt-12S rRNA residue actually exists. Conservation values Cvs for SSU rRNA residues have been calculated by using 8513 SSU rRNA sequences from all three phylogenetic domains, Eubacteria, Archaea and Eucarya (including nuclear, mitochondria and chloroplast sequences) (25). According to the authors, 100% conservation would yield a Cv of 2, 90% conservation would result in Cvs around 1.5, 60–80% conservation in Cvs near 1.0, 40–50% conservation in values approaching 0.5 and total lack of conservation in values below 0 (26). Since Cvs constitute measurements of universal rRNA conservation and, thus, should have relevance in all ribosomal systems, including organellar ones, one would expect that the number of variations at every position should correlate with their Cv. Accordingly, positions with higher Cvs, i.e. more highly conserved, would display a lower number of variations than positions with lower Cvs. To test this hypothesis we aligned the 954 residues of mt-12S rRNA and the 1542 residues of E. coli 16S rRNA by using the secondary-structure maps as a reference (not shown). As a source of mt-12S rRNA variations we used the GenBank database. After extracting all the variation data for every mt-12S rRNA position, we compared these data to the calculated Cvs for the same position.
Table 1.
Distribution of GenBank variations within conservation bins
| Conservation value (Cv) | Number of sites per category | Number of sites carrying variations per category | Normalized number of sites carrying variations per category | Number of variations per category | Normalized number of variations per category |
|---|---|---|---|---|---|
| 2 > CV ≥ 1.9 | 115 | 4 (3)a | 9.72 | 333 (3)a | 809.13 (7.29)a |
| 1.9 > Cv ≥ 1.5 | 150 | 31 | 57.25 | 158.00 | 291.77 |
| 1.5 > Cv ≥ 1 | 239 | 67 | 77.65 | 4182.00 | 4846.92 |
| 1 > Cv ≥ 0 | 277 | 73 | 73.00 | 3216.00 | 3216.00 |
| Cv < 0 | 13 | 3 | 63.92 | 5.00 | 106.54 |
| Not aligned | 161 | 85 | 146.24 | 6666.00 | 11 468.83 |
| Totals | 955 | 263 | 427.78 | 14 560.00 | 20 739.20 |
Corrected values after subtraction of values corresponding to outlier residues shown in brackets. Normalization is to the bin with the highest number of sites carrying variations (1 > Cv ≥ 0). In brackets are the values obtained for the most conserved bin (2 > Cv ≥ 1.9) after neglecting the 330 appearances of the outlier residue 180A (m.827A).
aValues calculated for the 2 > CV ≥ 1.9 category after neglecting the 330 appearances at position 827.
When the percentage of mt-12S rRNA positions carrying GenBank variations in every Cv bin is plotted, a clear picture emerges. Only 4 of 115 virtually universally conserved residues carried variations, corresponding to 3.48% of all such sites (Table 1 and Fig. 1D, back row). The percentage of sites carrying variations increases sharply as we move into less conserved categories. By definition, the category of residues with Cvs < 0 is not well represented within mt-12S rRNA (with only 13 aligned residues in this bin). Thus, this bin must be considered as a non-significant element of the analysis. Instead, 161 non-conserved mt-12S rRNA residues were placed in an additional category, named ‘not aligned’ and the associated percentage of variations was calculated. Strikingly, almost 53% of the residues in this category carried variations, an observation that is in clear agreement with our hypothesis of a non-random distribution of variations in human mtDNA. Also shown in Figure 1D is the percentage of sites with variations within each bin, relative to the total number of such sites in the whole mt-12S rRNA (middle row). Additionally, to account for the different bin sizes, we normalized the number of sites with variations in every bin to the bin with the highest number of such sites, namely the 1 > Cv ≥ 0 bin (Table 1). These data were used to calculate a normalized percentage of sites with variations per bin (front row of Fig. 1D). Again, the normalized data clearly follows a trend that is consistent with our hypothesis, i.e. the existence of an inverse correlation exists between phylogenetic conservation and the number of variations per residue. The trend is particularly strong for the bins with highest (2 > Cv ≥ 1.9 and 1.9 > Cv ≥ 1.5) and lowest (‘not aligned’) Cvs values.
We also investigated the correlation between the total number of variations recorded in GenBank (all possible variations per site were taken into account for this purpose) and Cv values. This was done by dividing the total number of variations recorded at all sites within a Cv bin by the number of sites in the bin (Fig. 1E, rows 5 and 6). The two most conserved categories clearly display the lowest numbers of variations per site, i.e. 2.90 variations/site for the most conserved bin (2>Cv ≥ 1.9) and 1.05 variations/site for the next bin (1.9 > Cv ≥ 1.5) (Fig. 1E, 6th row). Notably, a single residue from the nearly invariant bin (2 > Cv ≥ 1.9), namely position A827, accounts for 330 out of the 333 variations in the bin (Table 1). When this outlier is excluded from the calculations, the number of variations per site for the nearly invariant bin becomes 0.03 (Fig. 1E, 5th row from the front), in agreement with the extremely high conservation of the residues in this bin. Similarly to Figure 1D, the number of variations per site increases sharply as we move into the less conserved bins, reaching a maximum at the ‘not-aligned’ bin (41.40 variations/site). When the total number of variations per bin is plotted as a percentage of the total number of mt-12S rRNA variations recorded in GenBank, the two most conserved bins account for only 3.38% of the total number of variations (1.11% if the aforementioned outlier is excluded). On the contrary, the ‘not-aligned’ bin accounts for >45% of the variations (Fig. 1E, 3rd and 4th rows). A similar trend is observed after normalization of the percentages to account for bin size (Fig. 1E, 1st and 2nd rows). Again, the data shown in Figure 1E are in agreement with our hypothesis.
In summary, our comparison of phylogenetically derived data on SSU rRNA conservation and the human mtDNA records currently available in GenBank clearly shows that the distribution of variations in mt-12S rRNA is non-random. This distribution shows a strong inverse correlation with the phylogenetically derived conservation value, Cv. In addition to confirming the strength of comparative methods for the study of rRNA structure, these results validate the use of the GenBank and Phylotree databases as criteria for the identification of extremely rare variations. Doing this, one would expect that an important proportion of such variations could map to highly conserved residues with large disruptive potentials and therefore, have important roles in the function of the mitoribosome.
For all the reasons evidenced above, a group of very rare mt-12S rRNA variants were examined in this study. Specifically, we have focused on 52 variations, including 46 mutations with zero appearances in published controls and in the MITOMAP, GenBank and Phylotree databases, plus the m.896A>G variant, included due to their relevance to the presented results (Table 2 and Supplementary Material, Table S1). We have also analyzed the following five variants: the ‘proven’ pathogenic mutations 847C>U (m.1494C>T) and 908A>G (m.1555A>G), included as HIA positive controls (Table 2); the two common population changes with an abundance over 5% of the GenBank population, 791A>G (m.1438A>G) and 62G>A (m.709G>A), included as negative HIA controls ( Supplementary Material) and the aforementioned, highly conserved outlier 180A>G (m.827A>G), included to test whether a rationale to its strange behavior could be discovered (Supplementary Material).
Table 2.
Classification of mt-SSU rRNA mutations according to their predicted disruptive potential on ribosomal function
| Disruptive power | E. coli equivalent | Conservation of position according to CRWa | CiUniv | Ci1ry | Ci2ry | CiTot | |
|---|---|---|---|---|---|---|---|
| Unlikely | |||||||
| 249A>G (m.896A>G) | G524 | g | 3 | 4 | 5 | 19 | |
| Average | NA | NA | NA | 19 | |||
| SD | NA | NA | NA | NA | |||
| Undetermined | |||||||
| 67A>U (m.714A>T) | G112 | g | 3 | 3 | 5 | 17 | |
| 90C>U (m.737C>T) | Unknown | ? | NI | NI | NI | NI | |
| 103G>C(m.750G>C) | Unknown | ? | NI | NI | NI | NI | |
| 125A>G (m.772A>G) | C271b | x | 0 | 2 | 2 | 6 | |
| 167A>G(m.814A>G) | G351 | o | 2 | 4 | 4 | 16 | |
| 282A>U (m.929A>T) | G557 | x | 0 | 0 | 3 | 3 | |
| 296G>A (m.943G>A) | G577 | g | 3 | 3 | 5 | 17 | |
| 308A>G (m.955A>G) | Unknown | ? | NI | NI | NI | NI | |
| 313C>A(m.960C>A) | Unknown | ? | NI | NI | NI | NI | |
| 314U>A(m.961T>A) | Unknown | ? | NI | NI | NI | NI | |
| 314InsC(5)(m.961InsC(5)) | Unknown | ? | NI | NI | NI | NI | |
| 314delU(m.961delT) | Unknown | ? | NI | NI | NI | NI | |
| 361A>U (m.1008A>T) | G711 | w | 1 | 2 | 4 | 10 | |
| 364C>A(m.1011C>A) | G714 | g | 3 | 2 | 4 | 14 | |
| 474A>G (m.1121A>G) | Unknown | ? | NI | NI | NI | NI | |
| 485U>C (m.1132T>C) | C879 | w | 1 | 4 | 4.5 | 14.5 | |
| 878C>G(m.1525C>G) | Unknown | ? | NI | NI | NI | NI | |
| 888U>C(m.1535T>C) | Unknown | ? | NI | NI | NI | NI | |
| Average | 1.71 | 2.63 | 3.63 | 8.69 | Average | ||
| SD | 0.98 | 0.90 | 0.90 | 0.92 | SD | ||
| NEE | |||||||
| 40G>A(m.687G>A) | G46 | w | 1 | 3 | 5 | 13 | |
| 257C>U(m.904C>T) | A532 | o | 2 | 4 | 5 | 17 | |
| 265U>A(m.912T>A) | G540 | w | 1 | 1 | 4 | 8 | |
| 277A>U (m.924A>T) | U552 | w | 1 | 1 | 4 | 8 | |
| 620U>A (m.1267T>A) | U1062 | u | 3 | 3 | 4.5 | 16.5 | |
| 642G>C (m.1289G>C) | U1083 | o | 2 | 2 | 5 | 13 | |
| 698G>A (m.1345G>A) | G1215 | o | 2 | 3 | 5 | 15 | |
| 701G>A (m.1348G>A) | G1218 | w | 1 | 2 | 5 | 11 | |
| 727A>G (m.1374A>G) | C1245 | w | 1 | 1 | 2.5 | 6.5 | |
| 733G>A (m.1380G>A) | G1294 | w | 1 | 1 | 4.5 | 8.5 | |
| 742G>A (m.1389G>A) | G1304 | g | 3 | 2 | 5 | 15 | |
| 897A>U (m.1544A>T) | C1479 | x | 0 | 3 | 4 | 10 | |
| 929G>A (m.1576G>A) | G1511 | o | 2 | 4 | 5 | 17 | |
| 939G>A(m.1586G>A) | C1521 | w | 1 | 4 | 5 | 15 | |
| Average | 1.38 | 1.79 | 4.34 | 11.13 | Average | ||
| SD | 0.78 | 1.19 | 0.74 | 3.83 | SD | ||
| Likely | |||||||
| 450G>A (m.1097G>A) | G809 | o | 2 | 4 | 5 | 17 | |
| 507A>C (m.1154A>C) | A901 | a | 3 | 3 | 5 | 17 | |
| 676G>A (m.1323G>A) | G1193 | o | 2 | 4 | 4.5 | 16.5 | |
| 680G>A (m.1327G>A) | A1197 | w | 1 | 3 | 5 | 13 | |
| 852U>C (m.1499T>C) | G1415 | w | 1 | 1 | 4.5 | 8.5 | |
| Average | 1.50 | 2.31 | 4.79 | 13.41 | Average | ||
| SD | 0.84 | 1.22 | 0.27 | 3.70 | SD | ||
| Expectedly | |||||||
| 232U>C(m.879T>C) | C507 | c | 3 | 3 | 5 | 17 | |
| 255G>A (m.902G>A) | G530 | G | 4 | 5 | 5 | 23 | |
| 522G>A (m.1169G>A) | U916 | o | 2 | 5 | 5 | 19 | |
| 533U>G (m.T1180T>G) | G927 | w | 1 | 4 | 5 | 15 | |
| 579C>G (m.1226C>G) | C972 | C | 4 | 5 | 5 | 23 | |
| 580G>A (m.1227G>A) | G973 | g | 3 | 4 | 5 | 19 | |
| 910A>C (m.A1557A>C) | A1492 | A | 4 | 4 | 4 | 20 | |
| 915G>A (m.1562G>A) | G1497 | G | 4 | 6 | 5 | 25 | |
| 919C>U (m.1566C>T) | C1501 | c | 3 | 5 | 5 | 21 | |
| Average | 2.57 | 4.39 | 4.86 | 19.77 | Average | ||
| SD | 1.05 | 0.88 | 0.33 | 3.15 | SD | ||
| Proven | |||||||
| 847C>U (m.1494C>T) | A1410 | w | 1 | 4 | 4 | 14 | |
| 908A>G (m.1555A>G) | U1490 | w | 1 | 3 | 4 | 12 | |
| Average | 1.89 | 2.99 | 1.89 | 11.90 | Average | ||
| SD | 1.31 | 1.45 | 1.50 | 6.71 | SD | ||
NI, no information; NA, not applicable. The table includes all mitochondrial mutations meeting the criteria described in the section Materials and Methods. Only the mutations within the ‘likely’ and ‘expectedly’ categories have been analyzed in detail in this paper. Cis were calculated as described in the section Materials and Methods. Whenever possible, Cis were averaged in a per-category fashion and a standard deviation was calculated.
aCannone et al. (25) nomenclature for primary standard conservation across all three phylogenetic kingdoms is included. A, C, G, U, N, nucleotide is conserved in 98–100% of the sequences in the alignment; a, c, g, u, n, nucleotides is conserved in 90–100% of the sequences in the alignment; o, nucleotide is conserved in 80–90% of the sequences in the alignment; w, nucleotide is conserved in <80% of the sequences in the alignment; x, position is conserved in <95% of the sequences in the alignment.
bTentative assignment.
Conservation index (Ci) analysis
Many authors have relied heavily on very simple Cis for assessing the potential pathogenicity of mt-rRNA mutations. We have calculated three different types of Cis: two based on primary structure conservation, CiUniv and Ci1ry; one based on secondary structure conservation, Ci2ry; and a composite Ci which combined the information obtained from the other three, CiTot. The results are displayed in Table 2. A quick glance of the table clearly shows the failure of Cis to predict pathogenicity. For example, the two known pathogenic mutations 847C>U (m.1494C>T) and 908A>G (m.1555A>G) display rather low Cis. The implications of this are discussed below.
HIA analysis
The results of HIA analysis are shown in Table 2. Out of the 49 mutations analyzed (the negative controls and outlier were not included in the table), we found 1 whose disruptive potential on the function of mitoribosomes is ‘unlikely disruptive’, 18 which cannot be classified due to local structural differences between the mitochondrial and the heterologous ribosomes (‘undetermined’), 14 which are regarded as ‘not enough evidence (NEE)’, 5 which are ‘likely disruptive’ and 9 which are ‘expectedly disruptive’ (see Materials and Methods). Finally, the pathogenic mutations 847C>U (m.1494C>T) and 908A>G (m.1555A>G) were classified within the ‘proven’ category.
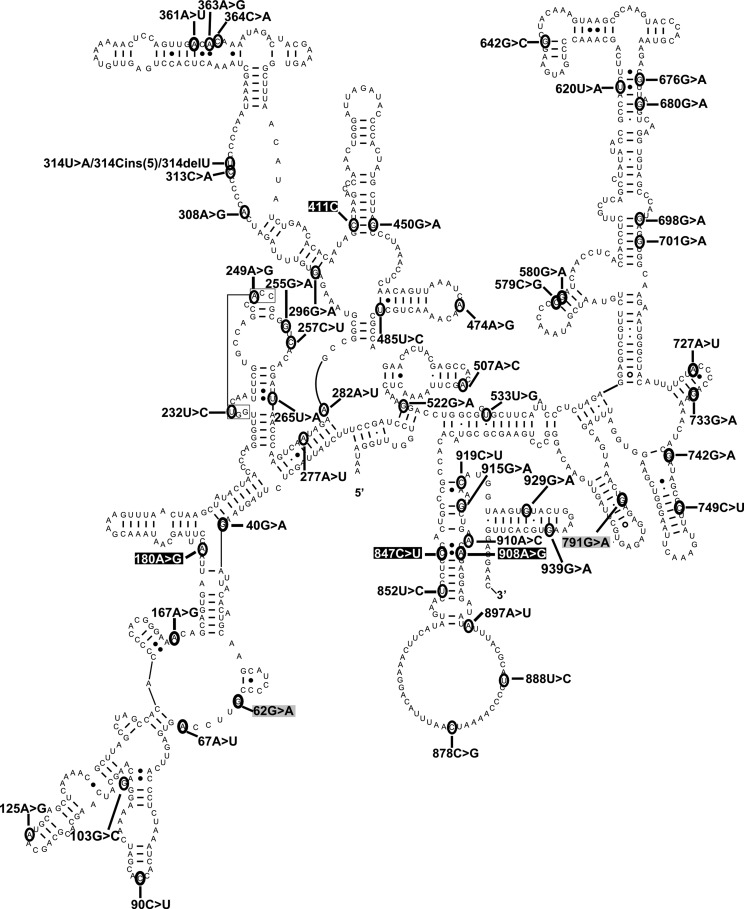
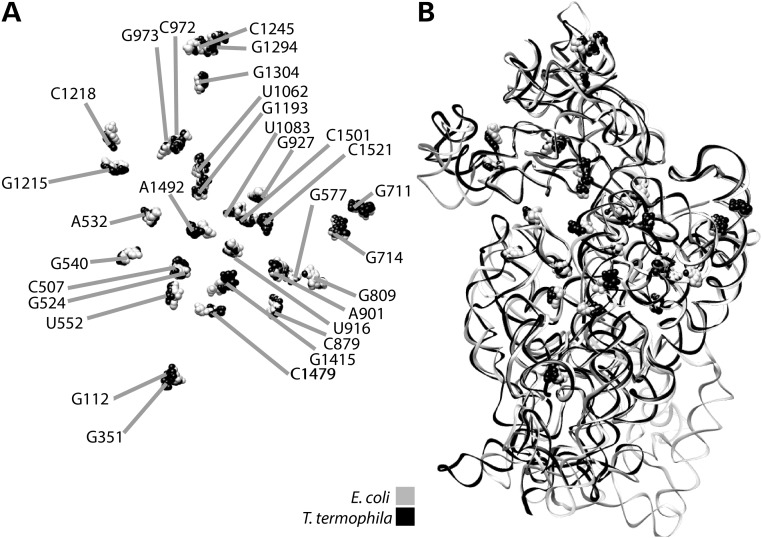
Figure 2 shows the location of the mutations with the highest disruptive potential, namely the ‘likely’ and ‘expectedly disruptive’ categories, on the secondary-structure map of human mt-12S rRNA. The location of the equivalent residues in E. coli is shown in Figure 3. While the predictive power of phylogenetically derived secondary structure maps has been amply confirmed experimentally by high-resolution structures of ribosomes from all three domains of life (24,27–29), we sought to confirm the accuracy of our assignment of heterologous residues by independently performing such an assignment in two SSU rRNAs from distantly related organisms. This was followed by their visualization in superposed high-resolution structures. To this end, we used phylogenetically derived secondary-structure maps and high-resolution crystal structures of SSUs from Tetrahymena thermophila (Eucarya) and E. coli (Eubacteria), both organisms more distant phylogenetically than eubacteria and mitochondria. The results of this comparison are displayed superposed onto the models of SSU rRNAs from these two organisms (Fig. 4 and Supplementary Material, Movie S1). Our results show a perfect structural match for all the studied mitochondrial positions as long as structurally equivalent residues can be located in phylogenetically derived, secondary structure maps of the heterologous ribosomes. The structural agreement between the predicted positions in both SSUs demonstrates that equivalence at the secondary structure level translates into accurate superposition in 3D. Thus, these data demonstrate the validity of comparative analysis to accurately predict the location of such structural equivalents in heterologous 3D ribosomal structures. Structural placement of mitochondrial mutations in heterologous ribosome structures provides the basis for HIA.
Figure 2.
Secondary structure map of the human 12S mt-rRNA showing sites of mutation. The secondary-structure map of human mt-12S rRNA was obtained from the CRW site (25). Sites of mutation analyzed in the main text are shown in black font. Other important sites described in the text (including HIA positive controls) are shown in white font over the black background. HIA negative controls are shown in black font over the gray background.
Figure 3.
Bacterial equivalents of mitochondrial mutations. Bacterial equivalents of analyzed mutations are shown in black font on the secondary-structure map of E. coli 16S rRNA (25). Other important sites described in the text (including HIA positive controls) are shown in white font over the black background. HIA negative controls are shown in black font over the gray background.
Figure 4.
Placement of heterologous equivalents of mitochondrial mutations on the high-resolution structures of T. thermophila and E. coli. (A) Ball representation of superposed heterologous residues. T. thermophila, grey; E. coli, black. E. coli numbering indicated. (B) Same residues onto ribbon models of the T. thermophila and E. coli SSUs. RCSB accession codes 2I2P and 2XZM.
Below, we briefly describe the results of the HIA analysis for the mutations arranged within the ‘likely’ and ‘expectedly disruptive’ categories.
Bridge B2c
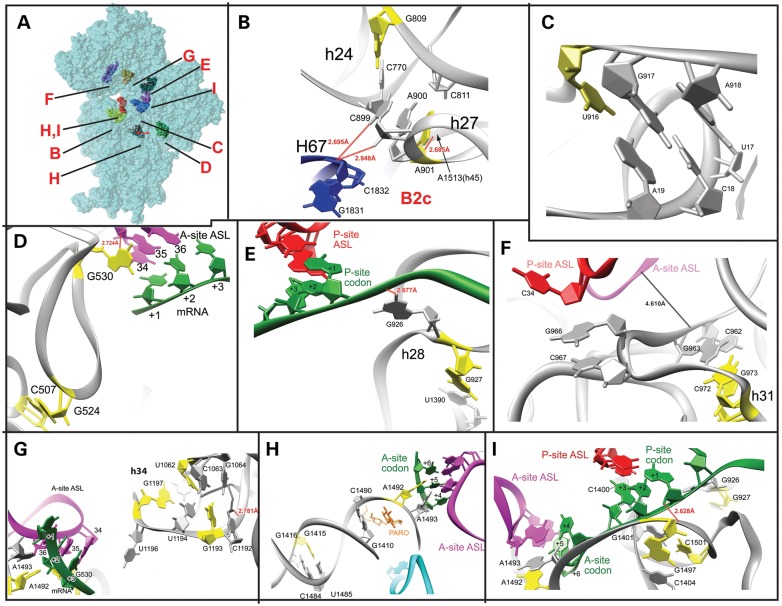
Ribosomal bridge mt-B2c has the same chemical composition in human mitochondria and in bacteria (30). Bridge b-B2c is formed in E. coli by the packing of residues 770:771 of b-h24, 899:901 of b-h27 and 1512:1513 of b-h45, against the LSU residues 1831:1833 of b-H67 (Fig. 3) (29,31,32). The mutation 450G>A (m.1097G>A), which was identified in a COX-deficient colonic crypt, maps to this region of 12S rRNA (33) (Figs 2 and 3). The equivalent position in E. coli, G809, is base paired to b-B2c residue C770 (Figs 3 and 5B). The evidence for the possible involvement of the 450G (m.1097G) in the incorrect functioning of bridge mt-B2c appears quite likely, since the disruption of the G809:C770 base pair by mutagenesis has been shown to be detrimental to ribosomal function (34,35). In addition, mutations disrupting Watson:Crick geometry at the adjacent G769:C810 base pair were highly deleterious (36,37). In the specific case of the 450G>A (m.1097G>A) mutation, the resulting C·A mismatch between 411C and 450G could adopt wobble geometry and thus, impose a rather small structural distortion at the base of m-h24. Indeed, such a configuration has been observed in ∼3.1% of mitochondrial SSU rRNAs (25). Further evidence is necessary to establish whether such geometry would be tolerated in this important region of the human mitoribosome.
Figure 5.
Placement of mitochondrial mutations in the structures of bacterial SSUs. (A) Surface representation of the E. coli SSU showing the location of the residues shown in B–I. (B) 530 pseudoknot. RCSB accession code 2J00. (C) Central pseudoknot. RCSB accession code 2I2P. (D) Bridge b-B2c. RCSB accession codes 2I2P and 2I2T. (E) 927 region. RCSB accession code 2J02. (F) 972 region. RCSB accession code 2J02. (G) Mutations in b-h34 and A-site codon–anticodon interaction. RCSB accession code 2J00. (H) b-h44. The aminoglycoside antibiotic paromomycin is shown in orange. RCSB accession code 2J00. (I) Decoding region of b-h44. RCSB accession code 2J00. (B–I) Bacterial equivalents of mitochondrial mutations shown in yellow. Other residues mentioned in the main text are shown in gray. 16S rRNA shown in gray. P-site ASL in red, A-site ASL in purple, mRNA in green, ribosomal proteins in black and 23S rRNA in blue. Inter-atomic distances are indicated, but note that ribbon representations do not necessarily reflect accurate interatomic distances involving backbone atoms.
One more variation in the neighborhood of mt-B2c, an A>C transition at position 507 (m.1154A>C), was identified in the 12S rRNA of a patient with hearing loss (38). The E. coli equivalent to this residue is A901, located in the highly conserved tetraloop that caps b-h27 (Fig. 5B). In the E. coli ribosome A901 is involved in tertiary contacts with the ribose of residue 811C at the base of b-h24, and with the backbone of residue A1513 in b-h45 (39), underscoring its important role in the stabilization of the complicated structure of the region. The importance of base identity in the tetraloop, as well as that of the correct packing of b-h27 and b-h24 for ribosomal function has been demonstrated in mutagenesis studies (40). In particular, variations at A901 resulted in less than half of the normal ribosome activity and moderate misreading effects (40).
In summary, the functioning of bridge mt-B2c could be compromised by two mutations found in human 12S rRNA, 450G>A (m.1097G>A) and 507A>C (m.1154A>C) (33,38). Mutagenesis studies performed in bacteria around the heterologous equivalent of 450G (m.1097G) indicated an important role of this region in ribosomal function. However, it is still possible that the helical geometry introduced by the G>A mutation in the organellar ribosome could be accommodated without a major deleterious effect. As for 507A>C (m.1154A>C), the paternal inheritance of the deafness pattern rules out a primary causative effect for the mutation (38). However, the disruptive potential of the base change is quite high, in agreement with the possibility that 507A>C (m.1154A>C) might contribute to the hearing defect. As a result, we are prompted to assign both mutations to the ‘likely disruptive’ category.
Central pseudoknot
A G to A mutation at position 522 (m.1169G>A) in the central pseudoknot of the SSU has been found in a patient with renal carcinoma (41). Figure 5C shows position U916, the E. coli equivalent to 522G (m.1169G) that is base paired to residue A19 within the central pseudoknot (Figs 3 and 5C). This universally conserved structure, whose formation is believed to constitute a crucial step during the maturation of the SSU (42), has been thoroughly studied by mutagenesis in bacteria. The results were clear. Essentially, base complementarity and not primary sequence were required to maintain ribosomal function (43–45). Thus, it appears reasonable to assume that the 522G>A (m.1169G>A) mutation should result in a defective SSU and so we have assigned this mutation to the ‘expectedly disruptive’ category.
The 530 loop
The 530 loop is one of the most important functional centers of the ribosome and, as such, its primary sequence is highly conserved phylogenetically across all three kingdoms of life (Figure 5). A homoplasmic G to A mutation was identified at position 255 (m.902) in a COX-deficient sample of urothelial cell patches (46). The bacterial equivalent is key residue G530 from which the loop receives its name. G530 has been shown to participate in monitoring the fidelity of decoding by checking the codon-anti-codon interaction at the A site (Fig. 5D and G) (47,48). Since alterations at G530 have been shown to result in dominant lethal phenotypes (49), we must include the 255G>A (m.902G>A) mutation in the ‘expectedly disruptive’ category.
Two more mutations map to this important structural element of the human mitoribosome. The extremely rare variation 232U > C (m.879T > C) was identified in a COX-deficient colonic crypt and in a cancer patient (33,50). The equivalent position in E. coli, 507C is base paired to 524G within a pseudoknot motif that provides structural stability to the 530 loop (Figs 2 and 3) (51). While the primary sequence of the pseudoknot is highly conserved, the identity of the bases constituting the 507:524 base pair is different in bacteria and human mitochondria. The bacterial pseudoknot was originally dissected by site-directed mutagenesis (51). It was shown that single base changes on either side of this structural element resulted in severe growth defects that could be restored by second-site, compensatory mutations (51). Interestingly, an A>G base change at the human equivalent of G524, namely 249A (m.896A) appears in 20 GenBank sequences and in a MITOMAP unpublished sequence (http://mitomap.org/bin/view.pl/MITOMAP/Submissions/20100804015). These observations suggest that certain mutations within the pseudoknot structure are tolerated in the organellar ribosome. A benign phenotype for 249A>G (m.896A>G) would not be in disagreement with HIA data and could be explained structurally. This is because G:U/U:G base-pair schemes within the pseudoknot are well tolerated in bacteria, including the very same scheme that would be induced by the 249A>G (m.896A>G) mutation (51,52).
The disruption of the bacterial pseudoknot, involving residues 507 and 524, likely impairs the function of the G530 in A-site decoding (47,48). In addition, disruption of the pseudoknot could also affect the maturation of SSU rRNA, as shown in E. coli (53). Taken together, these data allow the assignment of 232U>C (m.879T > C) within the ‘expectedly disruptive’ category, whereas 249A>G (m.896A>G) must be considered an ‘unlikely disruptive’ polymorphism.
Helix 28
The 533U>G (m.1180T>G) mutation was identified in one pediatric subject with non-syndromic hearing loss (54). The E. coli equivalent is G927 that is base paired to U1390 in helix h28 (Figs 2 and 3). Next to this site is the universally conserved G926 (conservation between 98 and 100%) (25), which is hydrogen bonded to the phosphate linking the P- and E-site codons on the mRNA (55) (Fig. 5E). In agreement with the important role of this region in translation, mutagenesis studies performed in yeast have shown that several base changes at the equivalent positions to 926, 927 or 928 confer lethal phenotypes (56). Similarly, functional studies in bacteria demonstrated that all three mutations at G926 drastically reduce the activity of the mutant ribosomes (57), albeit with only moderate effects on cell growth (58). Finally, b-h28 has been recently proposed to serve as the spring of the ratcheting process undergone by the ribosome during translocation (31). In summary, the available evidence obtained from different model organisms, points to an important involvement of h-28 residues in ribosomal function. As a result, the 533U>G (m.1180T > G) transversion is a good candidate for an ‘expectedly disruptive’ mutation.
Helix 31
A C to G transversion has been identified at residue 579C (m.1226C) in a pediatric subject with non-syndromic hearing loss (54). Interestingly, a G>A transition at the adjacent 580G (m.1227G) was reported to be associated with schizophrenia in a single patient by Ueno et al. (59). While the 580G>A (m.1227G>A) mutation was properly placed on a phylogenetically derived, secondary-structure map of the mammalian mt-12S rRNA by the authors (59), no further inquiries regarding the structural and/or functional role of the residue were pursued. The E. coli equivalents to 579C (m.1226C) and 580G (m.1227G) are the universally conserved residue C972 and the highly conserved G973 (Figs 2 and 3). These residues map to the vicinity of the P- and A-site anticodon stem loops (ASLs) in the decoding site (Fig. 5F). Residue C972 is base paired to G963, which is also universally conserved and places one of its phosphate oxygens <5 Å away from the backbone of the A-site ASL (Fig. 5F) (25,60). This base pair and the one formed by G973 and C962 appear to be important for the stabilization of the complicated structure of the loop atop helix b-h31 (Fig. 5F). Not surprisingly, mutations in this region have been found to disrupt ribosome activity in E. coli. For example, base changes at C972, G963 and A964 produced moderately deleterious, but dominant growth defects, whereas mutations at G973 strongly impaired cell growth (61). Finally, the physical proximity of the C972:G963 to the A-site ASL explains the observed effects of mutations in the region on the fidelity of A-site decoding (62). In summary, both the 579C > G (m.1226C > G) and the 580G>A (m.1227G>A) mutations are expected to be highly disruptive to the function of the mitoribosome.
Helix 34
The mutations 620U>A (m.1267T>A) (63) and 676G>A (m.1323G>A) (64), mapping to mt-h34, have been identified in three cancer patients (see Supplementary Material, Table S1). This area of the ribosome has been implicated in the movement of the SSU head during translocation and in the binding of the translocation inhibitor spectinomycin (31,65,66). Indeed, the residue equivalent to 620U>A (m.1267T>A) in the chloroplasts of Nicotiana tabacum and of Chlamydomonas is a spectinomycin resistance (specR) site (65). In E. coli, the equivalent residues to 620U>A (m.1267T > A) and 676G>A (m.1323G>A) are 1062U and 1193G, which base pair to U1194 and C1063, respectively. Adjacent to the C1063:G1193 base pair are residues G1064 and C1192 (Fig. 5G), which are part of the binding site for the antibiotic spectinomycin (55,66), an antibiotic proposed to restrict head movement by increasing the rigidity of the region around these residues (55,66). Despite their identity and location on the secondary structure map, G1064 and C1192 do not form a Watson–Crick base pair in the bacterial ribosome. Indeed, the human mt-SSU rRNA contains a U·C mismatch at the positions equivalent to G1064 and C1192 (compare Figs 2 and 3). Mutagenesis analysis of G1064 and C1192 has shown that mutations at both sites are well tolerated and confer resistance to spectinomycin (66,67). In contrast, mutations at G1193, also conferring spectinomycin resistance in E. coli, do so with a clear fitness cost (68). While no mutagenesis data exist for either U1062 or its non-canonical base -pairing partner U1194, it is striking that the identity of both residues, forming a U·U mismatch, is conserved in human mitochondria. Analysis of the quaternary structure of the region shows that the N-terminal region of protein b-S3 makes direct contacts with the base of U1062 and with the RNA backbone at C1192 (Fig. 5G). The significance of this observation cannot be extrapolated to the organellar case, however, as a protein homologous to b-S3 is not present in the mitoribosome.
In summary, while some mutations in this region can be well tolerated in bacteria, others result in deleterious phenotypes. Specifically, the heterologous evidence appears quite solid for a potentially disruptive role of 676G>A (m.1323G>A). On the other hand, a prediction of the disruptive potential of 620U>A (m.1267T>A) cannot be based solely on the available heterologous data.
Another mt-h34 mutation, namely 680G>A (m.1327G>A), has been identified as homoplasmic in atrophic prostate epithelium (69). Its bacterial equivalent is position 1197, which in Thermus thermophilus is also a G (A in E. coli). While mutagenesis studies at position 1197 have not been carried out, the following information provides evidence for an important role in translation. First, position A1196 of E. coli has been cross linked to residues +5, +8 and +9 of the mRNA and shown to be protected by the binding of elongation factor G (70–72). Indeed, position 1196 is modeled in close proximity of the A-site codon–anticodon interaction in high-resolution structures of the bacterial ribosome (Fig. 5) (60). All these data taken together prompt us to consider 680G>A (m.1327G>A) as a ‘likely disruptive’ mutation.
Helix 44
A U to C transition at position 852 (m.1499T>C) has been identified in a breast cancer patient (73). The equivalent position in E. coli would be G1415 within b-h44 (Figs 2 and 3). G1415 is base paired to U1485, which is part of inter-subunit bridge b-B3, a structure that appears to be conserved in the mitoribosome (Fig. 5H) (30,32). We have recently targeted the adjacent position C1484 of E. coli SSU's rRNA for site-directed mutagenesis to find that alterations of the helical stability of this region can cause highly deleterious phenotypes (74). Similarly, mutations of the b-B3 partner region on the bacterial LSU also resulted in defective ribosomes (75), in agreement with the finding that bridge b-B3 is the pivot point for inter-subunit ratcheting (31). Thus, all the available evidence points at an important role of residue 852U (m.1499T) in the mitoribosome and argue for a likely disruptive role for the U to C transition at this position.
A highly interesting variation was recently listed in the MITOMAP unpublished variant database. The variation 910A>C (m.1557A>C) was identified in a Russian individual suffering from deafness (http://www.mitomap.org/bin/view.pl/MITOMAP/Submissions/20070709002). Its bacterial equivalent, the universally conserved A1492 is, together with A1493, one of the most important residues of the ribosome. The functional role of these adenosines is to monitor the geometry of the codon–anticodon interaction at the A-site (Fig. 5H and I) (47,48). In bacteria, mutations at A1492 or A1493 result in dominant lethal phenotypes (57,76,77). Clearly, 910A>C (m.1557A>C) constitutes an extremely well-qualified candidate for a highly disruptive mutation.
The 915G>A (m.1562G>A) mutation was identified in a COX-deficient colonocyte in a study investigating clonally expanded mtDNA mutations associated with aging (78). The bacterial equivalent to this residue is the universally conserved G1497, adjacent to the P-site codon on the mRNA and is base paired to the also universally conserved C1404 (Fig. 5H and I). The phosphate between G1497 and U1498 is within h-bonding distance of the 2′OH of position +1 of the mRNA, i.e. the first base of the P-site codon (60). Not surprisingly, mutagenesis studies showed that disrupting the C1404:G1497 base pair resulted in serious impairment of tRNA binding at both P and A sites (79). Due to all these observations regarding the bacterial equivalent of 915G>A (m.1562G>A), the organellar mutation is clearly expected to result in a deleterious mitochondrial phenotype.
A 919C>U (m.1566C>T) transition has been identified in a cancer patient (41). The bacterial equivalent is the highly conserved 1501C, which is base paired to 1401G in the vicinity of the codon–anticodon interaction at the P site (Figs 2, 3 and 5I). Indeed, the adjacent 1400C is stacked under position 34 of P-site tRNA (55). In agreement with their location, mutagenesis studies have shown that altering the 1401G:1501C base pair completely abolishes ribosomal function (80,81). Hence, the heterologous evidence clearly places this mutation in the ‘expectedly disruptive’ category.
DISCUSSION
Currently, all available structural information pertaining to the mammalian mitoribosome is contained within phylogenetically derived secondary structure maps of its rRNA components and in a single low-resolution reconstruction of the bovine organellar ribosome (25,82). Here, we have made the first systematic demonstration that this information can be combined with data obtained from heterologous sources to make predictions on the disruptive potential of mt-rRNA mutations. We believe that this exercise constitutes a valid first approximation toward understanding the role of these mutations in human disease. Indeed, we propose that these methods should become one of the prime functional tests for the examination of novel or rare mt-rRNA variants seen in the mtDNA of patients.
The predictive power of comparative analysis for the generation of structural predictions on the folding of large RNAs has been confirmed by the high-resolution structures of the ribosome (24,28). For this reason, it has been surprising to find that, except for one report (59), no attempt has been made to use phylogenetically derived secondary-structure maps for the interpretation of the available mutational data. This has, in some cases, resulted in quite inaccurate structural predictions that remain uncontested in the literature. For example, Mutai et al. (83) have used the program CENTROIDFOLD (84) to predict the disruptive potential of the 257C>U (m.904C>T) mutation. According to the authors, the C–U base change would disrupt the canonical G–C base pair between 242G and 257C (83). However, such a canonical base pair is not predicted by phylogenetic methods (Fig. 2) and is not substantiated by high-resolution data from heterologous sources, as shown in Supplementary Material, Figure S4.
HIA and Ci analysis
In addition to HIA, we have also attempted to assign a Ci to all mutations, as described in the section Materials and Methods. This has been a common practice among researchers attempting to make sense of mutational data pertaining to mt-rRNAs. One of the main novelties of our Ci analysis is to include primary and secondary structure information from the region surrounding the tested residue (Ci1ry and Ci2ry), as opposed to most reported Cis that merely reflect the conservation of said residue (54,83,85–87). Regarding conservation, we have measured this parameter across the width of phylogenetic tree (CiUniv). In contrast, most authors have restricted their conservation analysis to a narrow range of phylogenetic space (54,83,85–87). While this practice works for other mitochondrial genes, we believe that the potential importance of the high number of exceptionally well-conserved residues of rRNA would be underestimated by Cis based on a narrow proportion of the total rRNA variation found on the tree of life. While our results show that Cis have some potential to predict the assignment of an important subset of mutations according to their HIA-predicted disruptive power, especially those within the ‘expectedly’ category, Cis cannot be relied upon for the assignment of pathogenicity to most mutations. One striking example is that of the ‘unlikely disruptive’ 249A>G (m.896A>G) mutation, for which all four Cis are well over the average value obtained for the ‘expectedly disruptive’ category. Similarly, some mutations within the ‘undetermined’ category display Ci scores that would place them within the ‘likely’ or even the ‘expectedly’ category, e.g. 67A>U(m.714A>T), 167A>G(m.814A>G) and 296G>A (m.943G>A). However, the best proof of the very limited predictive power of Cis in the context of mt-rRNA mutations is obtained when the ‘proven’ pathogenic mutations 847C>U (m.1494C>T) and 908A>G (m.1555A>G) are taken into account. The low Cis assigned to these mutations clearly demonstrate the limitations regarding their use as a way to obtain insightful information on the pathogenicity of mt-rRNA mutations.
In the case of universally conserved residues, both CiUniv and HIA predictions agree. It should be noted, however, that harmless or mild mutations at universally conserved rRNA residues do exist (58). As a result, the agreement between CiUniv and HIA predictions would be lost for such mutations. In summary, while the predictive potential of Cis remains an open issue, it appears clear that Ci analysis should always be applied in conjunction with HIA. This leads to a situation similar to that found while reporting mt-tRNA variants (6,7,10). Accordingly, a reliable assessment of pathogenicity of an mt-rRNA variant might require collaborative efforts between research centers to bring together the required analytical skills (9,88).
Disruptive power and pathogenicity
How much damage to ribosomal function is necessary for pathogenicity, and how do other factors modulate the amount of damage caused by mt-rRNA variants? Here we make the assumption that ribosome disruption leads to defective organellar protein synthesis, which translates into mitochondrial dysfunction, and finally into clinical symptoms. This highly simplistic assumption, substantiated by an enormous amount of evidence both from the point of view of the ribosome and that of the mitochondrion, will serve as a first approximation toward the analysis of the pathogenic potential of mt-rRNA mutations. The only two mt-rRNA mutations whose pathogenicity has been established with certainty are 908A>G (m.1555A>G) and 847C>U (m.1494C>T). The bacterial equivalents of these residues, namely positions 1490 and 1410, are part of the binding site for aminoglycoside antibiotics (Fig. 5H) (55). Despite their clear pathogenicity, when 908A>G (m.1555A>G) and 847C>U (m.1494C>T) mutations were recreated in bacterial systems, the cell growth was not completely arrested and the translation rate was not markedly decreased (89). Instead, a clear reduction in accuracy was observed in these studies (89). These results indicate that pathogenicity of mt-rRNA mutations can be achieved with ribosomes that can fully support translation, albeit with decreased accuracy. The fact that external modifiers, such as aminoglycoside antibiotics or nuclear markers, are needed to achieve the full pathogenic potential of 908A>G (m.1555A>G) and 847C>U (m.1494C>T) (21,90), suggests that, much like in the bacterial case, the mitochondrial mutations also result in functional ribosomes.
One would expect that mutations resulting in translation defects more severe than those conferred by 908A>G (m.1555A>G) and 847C>U (m.1494C>T) will not need the presence of additional factors to result in mitochondrial failure. Some examples of mutations expected to result in complete disruption of ribosomal function are contained within the ‘expectedly disruptive’ category. In principle, it seems reasonable to assume that ribosomes carrying severely disruptive mutations could co-exist in heteroplasmy with their wild-type counterparts. A similar co-existence of wild-type and mutant ribosomes has been frequently recreated in bacteria, where the dominance/recessiveness of highly deleterious mutations can be easily modulated by shifting the level of the mutant particles (91). The maintenance of bacterial growth is then dependent on the existence of enough wild-type ribosomes to support a sufficient level of protein synthesis. Extrapolating from the bacterial case, the heteroplasmy threshold above which the levels of disruptive mt-rRNA mutations would become incompatible with normal mitochondrial function would be expected to depend on two main factors. First, the severity of the deleterious effect caused by the base changes on the function of the mutated ribosomal particles (61,91,92). Secondly, the presence of phenotypic modifiers either in cis or in trans with the rRNA mutation. In the absence of direct biochemical data, a recent study in which the load of the 908A>G (m.1555A>G) mutation in a Chinese family was monitored becomes highly illustrative. The study showed that higher levels of heteroplasmy consistently resulted in more severe symptoms (93), thus mirroring the bacterial situation. In this study, heteroplasmy levels <20% were found in asymptomatic individuals, levels ∼50% in slightly affected individuals and >70% in severely affected patients (93).
Based on the large body of direct structural, mutational and functional data gathered in heterologous ribosomes, the mutations with the highest disruptive power described in this paper are 255G>A (m.902G>A) and 910A>C (m.1557A>C) (46) (http://www.mitomap.org/bin/view.pl/MITOMAP/Submissions/20070709002) which, in mitochondria, affect the key A-site residues in the mitochondria, equivalent to the bacterial G530 and 1492A, respectively (49,57,76). These nucleotides are amongst the most important ribosomal residues from the functional point of view and, therefore, it comes as no surprise that mutations at both G530 and 1492A have been found to cause dominant lethal phenotypes in bacteria (49,57,76). While in the case of the mitochondrial equivalent to 1492A, namely 910A>C (m.1557A>C) (http://www.mitomap.org/bin/view.pl/MITOMAP/Submissions/20070709002), we lack data regarding heteroplasmy levels, the bacterial data strongly suggest that this mutation can only be present at low levels if the mitochondria are to retain their function (49,77). However, what constitutes the actual threshold for a defect caused by this mutation is currently unknown. As for the homoplasmic 255G>A (m.902G > A) (46), the fact that the all point mutations at its bacterial equivalent resulted in dominant lethal phenotypes, together with the central role of this residue in decoding, is a strong indication that all ribosomes in the mutant mitochondria are incapable of performing protein synthesis. As a result, this mutation meets all pathogenicity criteria proposed for a homoplasmic mutation: absence from healthy controls, remarkable phylogenetic conservation and unequivocal evidence of a functional defect of the mutant ribosome (9). While the evidence for a functional defect has been obtained in heterologous systems, the available data are so strong that pathogenicity can hardly be contested.
Two more ‘expectedly disruptive mutations’ 533U>G (m.1180T>G) and 579C>G (m.1226C>G) have also been found in homoplasmy (54). The disruption of the yeast equivalent to the mitochondrial U533:G827, formed between positions 927 and 1390 (bacterial numbering), gave rise to recessive lethal phenotypes. While such phenotypes are not as drastic as the dominant lethality caused by mutations at the bacterial G530 and A1492, the presence of 533U>G (m.1180T>G) in homoplasmy indicates that the affected mitochondria should be highly impaired in their ability to carry out protein synthesis, thus leading to mitochondrial dysfunction. The case of 579C>G (m.1226C>G) is different. Despite the fact that the CiTot for this position is the same as that of 255G>A (m.902G>A) and higher than that of 910A>C (m.A1557A>C), it has been shown that disrupting the heterologous equivalent of the mitochondrial 570G:579C base pair, namely b-963G:972C, results in moderate dominant defects. Thus, the 579C>G (m.1226C>G) mutation is expected to result in at least suboptimal protein synthesis. Whether this can lead to enough organelle dysfunction to cause clinical phenotype remains unclear. The case of 579C>G (m.1226C>G) constitutes yet another example of the fact that Cis cannot be used to predict the disruptive potential of rRNA mutations even in the heterologous systems used as references in these studies. At the same time, this mutation underscores the enhanced discriminatory power gained by using HIA.
In summary, whilst a clear case for pathogenicity can be made for the 255G>A (m.902G>A) and the 533U>G (m.1180T>G) mutations, the 579C>G (m.1226C>G) mutation cannot be considered pathogenic by HIA alone.
Role of HIA within a scoring system for mt-rRNA mutations
The existence of direct experimental data showing a mitochondrial defect caused by a mtDNA mutation is an essential pre-requisite of the current scoring systems to evaluate the pathogenicity of mt-tRNA and complex I gene mutations (6,7,9–11). In the absence of direct biochemical data, HIA provides an educated estimation of the disruptive potential of an important subset of mt-rRNA mutations. This situation is reminiscent of the pre-crystallography days (1990s) in which the study of ribosome structure had to rely on evidence obtained from low-resolution studies. Notably, the quality of HIA predictions can be substantially improved as additional heterologous data emerge. As such, this research provides new impetus for studies aimed at testing the effect of equivalent mutations in heterologous systems and constitutes an invitation to ‘ribosomologists’ to contribute to the improvement of HIA predictions, thus leading to higher diagnostic accuracy in mitochondrial clinics. An obvious first aim of such a collaborative effort between clinical and basic science should be to move as many of the ‘NEE’ mutations to the ‘likely disruptive’ or, perhaps, to the ‘expectedly disruptive’ category, by means of site-directed mutagenesis and biochemical studies in heterologous systems.
The importance of mutagenesis studies in heterologous systems as a way to ascertain the effects of mt-rRNA alteration studies is best exemplified by the case of the 908A>G (m.1555A>G) and 847C>U (m.1494C>T) mutations. When the mutations were introduced in bacteria, the observed phenotypes were in perfect agreement with the mitochondrial data (89,94). This, together with structural evidence (95), provided a molecular explanation to the pathological symptoms related to the mitochondrial mutations, namely that misreading of the genetic code in mitochondria can lead to a defective organelle (89). Therefore, HIA would clearly result in an ‘expectedly disruptive’ score for these mutations.
We have also shown that in cases where sufficient information exists regarding the genetic and pathological manifestation of the mutation, HIA alone can be used to predict the pathogenicity of certain mt-rRNA mutations. In these cases, the heterologous data are strong enough to replace direct biochemical evidence. In other cases, the disruptive potential predictions will be incorporated into a full-fledged scoring system for mt-rRNA that will allow an evaluation of their pathogenic potential. In addition to the usual elements, the mt-rRNA scoring system will include HIA predictions and CI scores. As mentioned above, the level of heteroplasmy, together with the degree of disruption of ribosomal function, will constitute major elements of the scoring system. Here again, future HIA-motivated studies performed in heterologous systems can provide extremely useful information. For example, quantitative studies with hybrid populations of ribosomes in bacteria (91) could be used to assess the heteroplasmy threshold at which mt-rRNA mutations become detrimental to normal protein synthesis.
Final conclusions
Here, we have presented what we believe will be a central element of a scoring system for mt-rRNA mutations. HIA analysis appears to be a powerful tool that could help overcome the lack of biochemical and/or trans-mitochondrial cybrid data, which are considered to be crucial elements of the scoring systems for mt-tRNA and complex I genes (6,7,10,11). It should be noted that a simple ‘no-appearance-in-controls’ requisite, as established ever since the canonical criteria of DiMauro and Schon were established, will result in the discarding of well-known pathogenic mutations such as 908A>G (m.1555A>G) and 847C>U (m.1494C>T), underscoring the variability in penetrance of this group of mutations. One important challenge for the future must be to account for any effect of haplogroup background, as there is increasing evidence that this parameter is important in both the presentation and penetrance of mtDNA disease (22). Future analysis of mt-rRNA mutations will require that authors report their data fully including all the variants from the rCRS (particularly variants mapping to rRNA and tRNA genes), haplogroup definition, heteroplasmy levels, patient/control group sizes and data on the family/cohort structure. Similarly, mitochondrial genome sequences submitted to GenBank should clearly state whether the subject is considered a patient or a control.
While HIA predictions alone would not be sufficient for a definitive assignment of pathogenicity, except for some key residues with critical functional roles such as 255G>A (m.902G>A) and 533U>G (m.1180T>G), the method does provide a robust indicator of disruptiveness for a large group of mutations lacking direct evidence regarding their role in mitochondrial dysfunction. At the same time, HIA naturally allows the prioritization of variants for additional investigation.
MATERIALS AND METHODS
Nomenclature
Since the focus of this paper is the mitoribosome, all mt-rRNA base changes will be referred to by their gene, rather than their genomic position. For clarity, however, the genomic position will also be provided in brackets. Bacterial rRNA residues will be italicized throughout the text. Mitochondrial specific notation will be used to denote organellar base changes, e.g. m.1111A>G. Likewise, bacterial nomenclature will be used to denote bacterial base changes, i.e. A1111G. To distinguish structural elements in the bacterial and mitochondrial ribosomes their name will be preceded by b- and mt-, respectively. Small subunit helices are denoted by ‘h’ and large subunit helices are denoted by ‘H’.
Sources of mutational data
The following sources of mutational data were used in this study: (a) scientific literature, (b) MITOMAP list of unpublished variants (1), (c) GenBank, (d) Phylotree (96) and (e) three unpublished variations (see below).
The criterion required for mt-rRNA mutations to be included in this study was their identification in human subjects, with either a suspected mitochondrial disease or somatic mtDNA mutations that had undergone clonal expansions in cancer patients, or COX-deficient cells located in aged tissues (97). In the case of studies involving tumorigenic tissue, the existence of non-cancerous control tissue from the same patient was a requirement. Similarly, the existence of COX-proficient surrounding tissue was a requirement in the case of COX-deficient samples of aged individuals. This allowed the variant to be identified as a somatic mutation that had undergone clonal expansion.
mtDNA sequencing
Mutation 878C>G (m.1525C>G) was detected as follows. Total DNA was extracted from patient tissues using standard DNA extraction methodologies. The entire mitochondrial genome was amplified using 36 overlapping primer pairs as previously described (33). PCR products were cycle sequenced using ABI BigDye chemistry by following standard manufacturer's protocols and analyzed on an ABI3130xl genetic analyzer (Applied Biosystems). Mutations 125A>G (m.772A>G) and 361A>U (m.1008A>T) were identified as follows. The entire mtDNA of the patients was evaluated using a standard mitochip analysis (GeneChip® Human Mitochondrial Resequencing Array 2.0, Affymetrix, Inc.). Percentage heteroplasmy was quantified using the Pyrosequencing technology (Biotage AB, Uppsala, Sweden) on the PSQ96 MA platform according to the protocol of the manufacturer.
Analysis of GenBank mitochondrial genome data
As of February 2013, there existed 17 850 human mtDNA sequences in GenBank [query = ‘Homo[Organism] + AND + gene_in_mitochondrion[PROP] + AND + 14 000:19000[SLEN] + NOT + pseudogene[All Fields]’ (98)]. All sequences were downloaded with Entrez Programming Utilities and our own PERL scripts (99). In addition, we also downloaded 210 sequences from http://www.phylotree.org/ that were not included in the GenBank collection as of May 2012. The data were downloaded in FASTA format and sequentially aligned to the Cambridge Reference (NC_012920.1) (100) by means of the MAFFT package (101), which provided a quick algorithm for massive sequence alignment. The generated alignments were then analyzed by means of a PERL script that recorded the total number of appearances for every variation. For variations with 15 or less appearances in the combined data set, including the data from both the GenBank and Phylotree databases, the possibility that the individual sequences could be linked to mitochondrial disease was investigated, by inspecting the related publication sources. In all cases, it was observed that no new patient sequences, other than the ones previously found in the literature, were present within GenBank.
Assignment of the disruptive power potential by HIA
Phylogenetically derived, secondary-structure rRNA maps carry an enormous amount of conservation information that can be used to infer the relative positions of equivalent residues from organisms located at opposite ends of the universal phylogenetic tree. HIA uses this information as follows. First, comparison of phylogenetically derived secondary structures of the human mt-12S rRNA and the E. coli 16S rRNA (normally used as the standard numbering system for rRNA residues) (25) allowed the location of structurally equivalent residues. Secondly, once the position of a structurally equivalent residue was identified, its location in three dimensions (3D) was ascertained by visualization in high-resolution models of heterologous ribosomes (normally atomic models of the E. coli or T. thermophilus ribosomes). Subsequent analysis of the region surrounding the heterologous residue within the high-resolution structures provided a first approximation of the structural role of the mutated mitochondrial residue. Thirdly, an exhaustive search for additional structural and functional information regarding the heterologous residues was performed. Finally, the existence of a low-resolution reconstruction of the mitoribosome provided important clues regarding the conservation of gross features of the organellar particle.
The disruptive power potential was estimated for the equivalent heterologous positions by the following criteria: N = ‘certainly not disruptive’, supportive direct mutagenesis data in favor of this assignment exists for the tested residue or its base-pairing partner, U = ‘unlikely disruptive’, no direct mutagenesis data exist but enough indirect data exist in favor of this conclusion; NEE = ‘not enough evidence’, no direct or indirect evidence argues against a potential disruptive power; L = ‘likely disruptive’, no direct mutagenesis data exist but enough indirect data exist in favor of this conclusion and C = ‘certainly disruptive’, supportive direct mutagenesis data exist for the tested residue or its base -pairing partner. A mtDNA mutation would be considered to have equally disruptive power to its heterologous counterpart as long as the available structural data for the former is not in disagreement with the heterologous evidence, e.g. if the higher order structural elements taken into consideration are conserved in both the mitochondrial and heterologous ribosomes. Two additional categories were used to classify the mitochondrial mutations: und = ‘undetermined’, no heterologous data exist to evaluate the disruptive potential or the existing structural differences are too large to allow the extrapolation of conclusions made in the heterologous case and E = ‘expectedly disruptive’, which would contain the mitochondrial equivalents to residues assigned to the ‘certainly disruptive’ category in the heterologous system.
Conservation index analysis
We have devised three simple conservation indexes (Cis). CiUniv was designed to monitor the universal conservation of the tested residue in SSU rRNAs encompassing the whole width of the universal phylogenetic tree, i.e. archaeal, bacterial, eukaryotic nuclear, mitochondria and chloroplast SSU rRNAs. It was calculated by assigning a value from 0 to 5 to the conservation categories defined by Cannone et al. (25) (see Table 2). Ci1ry measures the degree of primary sequence conservation around the tested residue. It was calculated by assigning 1 point per matched residue within a five-residue window centered on the tested nucleotide. In addition, an extra point was assigned whenever the position of the heterologous residue had been predicted by primary sequence alignment between the human and E. coli SSU rRNA (102). Ci2ry is similar to Ci1ry but measures the degree of secondary structure around the tested residue. To calculate this index, we assumed structural equivalence between G:U/U:G and Watson–Crick base-pairing schemes, as assumed during the construction of phylogenetically derived secondary-structure maps (25). Again, 1 point was assigned per matched residue within the five-residue window, whenever the same secondary structure was predicted in phylogenetically derived secondary-structure maps of the human and E. coli SSU rRNA (25). A half-a-point penalty would be in place whenever a phylogenetically predicted mismatch was replaced by a canonical or G:U/U:G base pair, or vice versa. Finally, an arbitrary composite Ci was calculated as follows: CiTot = (2 × CiUniv) + (2 × Ci1ry) + Ci2ry. The weights of CiUniv and Ci1ry were doubled to account for their higher predictive power (see Results).
Structural analysis
The following crystal structures were obtained from rcsb (www.rcsb.org): 2I2P and 2I2T (39), 2J02 and 2J03 (60), 1VSA and 2OW8 (103), 2Y0U (104), 2XZM (27) and 3R8N (31). Structural analysis was performed with UCSF Chimera (105) and Phenix (106).
Figure 4 and Supplementary Material, Movie S1 were created as follows. The position of all mutations that had equivalents in E. coli 16S rRNA was located on the secondary-structure map of T. thermophila SSU (27). The Tetrahymena positions were then displayed on a 3D model of the SSU T. thermophila 18S rRNA that had been previously superposed onto the 3D model of the E. coli 16S rRNA with Chimera UCSF (27,106).
SUPPLEMENTARY MATERIAL
FUNDING
A.V.S. was supported by the Marie Curie Actions programme of the European Commission (Grant PIIF-GA-2010-274660) and the Spanish Ministerio de Economía y Competitividad (AGL2012-39274-C02-02). R.W.T., R.N.L. and Z.M.A.C.L. were supported by the Wellcome Trust (Strategic Award 096919/Z/11/Z). L.C.G. was supported by the Newcastle University Centre for Brain Ageing and Vitality supported by BBSRC, EPSRC, ESRC and MRC as part of the cross council Lifelong Health and Wellbeing Initiative (G0700718). J.L.E. was supported by an Academic fellowship from the Research Council UK. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust (Strategic Award 096919/Z/11/Z).
Supplementary Material
ACKNOWLEDGEMENTS
A.V.-S. would like to acknowledge the following: Fidedigna Vila for balance and coherence and Michael O'Connor and Steven Gregory for their comments on the manuscript. SBW and RJTR would like to thank the Roels Smeets and Edwin van Kaauwen from the Nijmegen Centre for Mitochondrial Disorders (NCMD) for technical assistance. This paper is dedicated to the memory of Dr Carl R. Woese.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaefer A.M., McFarland R., Blakely E.L., He L., Whittaker R.G., Taylor R.W., Chinnery P.F., Turnbull D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 3.Bandelt H.J., Salas A., Taylor R.W., Yao Y.G. Exaggerated status of ‘novel’ and ‘pathogenic’ mtDNA sequence variants due to inadequate database searches. Hum. Mutat. 2009;30:191–196. doi: 10.1002/humu.20846. [DOI] [PubMed] [Google Scholar]
- 4.Bandelt H.J., Salas A., Bravi C.M. What is a ‘novel’ mtDNA mutation —and does ‘novelty’ really matter? J. Hum. Genet. 2006;51:1073–1082. doi: 10.1007/s10038-006-0066-5. [DOI] [PubMed] [Google Scholar]
- 5.Salas A., Yao Y.G., Macaulay V., Vega A., Carracedo A., Bandelt H.J. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005;2:1158–1166. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarham J.W., Al-Dosary M., Blakely E.L., Alston C.L., Taylor R.W., Elson J.L., McFarland R. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum. Mutat. 2011;32:1319–1325. doi: 10.1002/humu.21575. [DOI] [PubMed] [Google Scholar]
- 7.Yarham J.W., McFarland R., Taylor R.W., Elson J.L. A proposed consensus panel of organisms for determining evolutionary conservation of mt-tRNA point mutations. Mitochondrion. 2012;12:533–538. doi: 10.1016/j.mito.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMauro S., Schon E.A. Mitochondrial DNA mutations in human disease. Am. J. Med. Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 9.Tuppen H.A., Fattori F., Carrozzo R., Zeviani M., DiMauro S., Seneca S., Martindale J.E., Olpin S.E., Treacy E.P., McFarland R., et al. Further pitfalls in the diagnosis of mtDNA mutations: homoplasmic mt-tRNA mutations. J. Med. Genet. 2008;45:55–61. doi: 10.1136/jmg.2007.051185. [DOI] [PubMed] [Google Scholar]
- 10.McFarland R., Elson J.L., Taylor R.W., Howell N., Turnbull D.M. Assigning pathogenicity to mitochondrial tRNA mutations: when ‘definitely maybe’ is not good enough. Trends Genet. 2004;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell A.L., Elson J.L., Howell N., Taylor R.W., Turnbull D.M. Sequence variation in mitochondrial complex I genes: mutation or polymorphism? J. Med. Genet. 2006;43:175–179. doi: 10.1136/jmg.2005.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong L.J. Pathogenic mitochondrial DNA mutations in protein-coding genes. Muscle Nerve. 2007;36:279–293. doi: 10.1002/mus.20807. [DOI] [PubMed] [Google Scholar]
- 13.Miller C., Saada A., Shaul N., Shabtai N., Ben-Shalom E., Shaag A., Hershkovitz E., Elpeleg O. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 14.Smits P., Saada A., Wortmann S.B., Heister A.J., Brink M., Pfundt R., Miller C., Haas D., Hantschmann R., Rodenburg R.J., et al. Mutation in mitochondrial ribosomal protein MRPS22 leads to cornelia de lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 2011;19:394–399. doi: 10.1038/ejhg.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serre V., Rozanska A., Beinat M., Chretien D., Boddaert N., Munnich A., Rotig A., Chrzanowska-Lightowlers Z.M. Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochim. Biophys. Acta. 2013;1832:1304–1312. doi: 10.1016/j.bbadis.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galmiche L., Serre V., Beinat M., Assouline Z., Lebre A.S., Chretien D., Nietschke P., Benes V., Boddaert N., Sidi D., et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 2011;32:1225–1231. doi: 10.1002/humu.21562. [DOI] [PubMed] [Google Scholar]
- 17.Akama K., Christian B.E., Jones C.N., Ueda T., Takeuchi N., Spremulli L.L. Analysis of the functional consequences of lethal mutations in mitochondrial translational elongation factors. Biochim. Biophys. Acta. 2010;1802:692–698. doi: 10.1016/j.bbadis.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camara Y., Asin-Cayuela J., Park C.B., Metodiev M.D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Dennerlein S., Rozanska A., Wydro M., Chrzanowska-Lightowlers Z.M., Lightowlers R.N. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 2010;430:551–558. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolanczyk M., Pech M., Zemojtel T., Yamamoto H., Mikula I., Calvaruso M.A., van den Brand M., Richter R., Fischer B., Ritz A., et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell. 2011;22:1–11. doi: 10.1091/mbc.E10-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bindu L.H., Reddy P.P. Genetics of aminoglycoside-induced and prelingual non-syndromic mitochondrial hearing impairment: A review. Int. J. Audiol. 2008;47:702–707. doi: 10.1080/14992020802215862. [DOI] [PubMed] [Google Scholar]
- 22.van der Walt E.M., Smuts I., Taylor R.W., Elson J.L., Turnbull D.M., Louw R., van der Westhuizen F.H. Characterization of mtDNA variation in a cohort of South African paediatric patients with mitochondrial disease. Eur. J. Hum. Genet. 2012;20:650–656. doi: 10.1038/ejhg.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y.G., Salas A., Bravi C.M., Bandelt H.J. A reappraisal of complete mtDNA variation in East Asian families with hearing impairment. Hum. Genet. 2006;119:505–515. doi: 10.1007/s00439-006-0154-9. [DOI] [PubMed] [Google Scholar]
- 24.Gutell R.R., Lee J.C., Cannone J.J. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 25.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortoleva-Donnelly L., Szewczak A.A., Gutell R.R., Strobel S.A. The chemical basis of adenosine conservation throughout the tetrahymena ribozyme. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabl J., Leibundgut M., Ataide S.F., Haag A., Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 28.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 29.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 30.Sharma M.R., Koc E.C., Datta P.P., Booth T.M., Spremulli L.L., Agrawal R.K. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 31.Dunkle J.A., Wang L., Feldman M.B., Pulk A., Chen V.B., Kapral G.J., Noeske J., Richardson J.S., Blanchard S.C., Cate J.H. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 33.Taylor R.W., Barron M.J., Borthwick G.M., Gospel A., Chinnery P.F., Samuels D.C., Taylor G.A., Plusa S.M., Needham S.J., Greaves L.C., et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H.M., Yeom J.H., Ha H.J., Kim J.M., Lee K. Functional analysis of the residues C770 and G771 of E. coli 16S rRNA implicated in forming the intersubunit bridge B2c of the ribosome. J. Microbiol. Biotechnol. 2007;17:1204–1207. [PubMed] [Google Scholar]
- 35.Rackham O., Wang K., Chin J.W. Functional epitopes at the ribosome subunit interface. Nat. Chem. Biol. 2006;2:254–258. doi: 10.1038/nchembio783. [DOI] [PubMed] [Google Scholar]
- 36.Belanger F., Gagnon M.G., Steinberg S.V., Cunningham P.R., Brakier-Gingras L. Study of the functional interaction of the 900 tetraloop of 16S ribosomal RNA with helix 24 within the bacterial ribosome. J. Mol. Biol. 2004;338:683–693. doi: 10.1016/j.jmb.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S., Joseph S. Nonbridging phosphate oxygens in 16S rRNA important for 30S subunit assembly and association with the 50S ribosomal subunit. RNA. 2005;11:657–667. doi: 10.1261/rna.7224305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konings A., Van Camp G., Goethals A., Van Eyken E., Vandevelde A., Ben Azza J., Peeters N., Wuyts W., Smeets H., Van Laer L. Mutation analysis of mitochondrial DNA 12SrRNA and tRNASer(UCN) genes in non-syndromic hearing loss patients. Mitochondrion. 2008;8:377–382. doi: 10.1016/j.mito.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Berk V., Zhang W., Pai R.D., Doudna Cate J.H. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belanger F., Leger M., Saraiya A.A., Cunningham P.R., Brakier-Gingras L. Functional studies of the 900 tetraloop capping helix 27 of 16S ribosomal RNA. J. Mol. Biol. 2002;320:979–989. doi: 10.1016/s0022-2836(02)00550-8. [DOI] [PubMed] [Google Scholar]
- 41.Meierhofer D., Mayr J.A., Fink K., Schmeller N., Kofler B., Sperl W. Mitochondrial DNA mutations in renal cell carcinomas revealed no general impact on energy metabolism. Br. J. Cancer. 2006;94:268–274. doi: 10.1038/sj.bjc.6602929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy-Chaudhuri B., Kirthi N., Culver G.M. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc. Natl Acad. Sci. USA. 2010;107:4567–4572. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poot R.A., van den Worm S.H., Pleij C.W., van Duin J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998;26:549–553. doi: 10.1093/nar/26.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poot R.A., Pleij C.W., van Duin J. The central pseudoknot in 16S ribosomal RNA is needed for ribosome stability but is not essential for 30S initiation complex formation. Nucleic Acids Res. 1996;24:3670–3676. doi: 10.1093/nar/24.19.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brink M.F., Verbeet M.P., de Boer H.A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993;12:3987–3996. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaisa N.T., Graham T.A., McDonald S.A., Canadillas-Lopez S., Poulsom R., Heidenreich A., Jakse G., Tadrous P.J., Knuechel R., Wright N.A. The human urothelium consists of multiple clonal units, each maintained by a stem cell. J. Pathol. 2011;225:163–171. doi: 10.1002/path.2945. [DOI] [PubMed] [Google Scholar]
- 47.Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 48.Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 49.Powers T., Noller H.F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc. Natl Acad. Sci. USA. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kloss-Brandstatter A., Schafer G., Erhart G., Huttenhofer A., Coassin S., Seifarth C., Summerer M., Bektic J., Klocker H., Kronenberg F. Somatic mutations throughout the entire mitochondrial genome are associated with elevated PSA levels in prostate cancer patients. Am. J. Hum. Genet. 2010;87:802–812. doi: 10.1016/j.ajhg.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Powers T., Noller H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Springer B., Kidan Y.G., Prammananan T., Ellrott K., Bottger E.C., Sander P. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 2001;45:2877–2884. doi: 10.1128/AAC.45.10.2877-2884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belotserkovsky J.M., Dabbs E.R., Isaksson L.A. Mutations in 16S rRNA that suppress cold-sensitive initiation factor 1 affect ribosomal subunit association. FEBS J. 2011;278:3508–3517. doi: 10.1111/j.1742-4658.2011.08272.x. [DOI] [PubMed] [Google Scholar]
- 54.Li R., Greinwald J.H., Jr, Yang L., Choo D.I., Wenstrup R.J., Guan M.X. Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J. Med. Genet. 2004;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 56.Dong J., Nanda J.S., Rahman H., Pruitt M.R., Shin B.S., Wong C.M., Lorsch J.R., Hinnebusch A.G. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(met) and AUG selection. Genes Dev. 2008;22:2242–2255. doi: 10.1101/gad.1696608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdi N.M., Fredrick K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vila-Sanjurjo A., Squires C.L., Dahlberg A.E. Isolation of kasugamycin resistant mutants in the 16 S ribosomal RNA of Escherichia coli. J. Mol. Biol. 1999;293:1–8. doi: 10.1006/jmbi.1999.3160. [DOI] [PubMed] [Google Scholar]
- 59.Ueno H., Nishigaki Y., Kong Q.P., Fuku N., Kojima S., Iwata N., Ozaki N., Tanaka M. Analysis of mitochondrial DNA variants in Japanese patients with schizophrenia. Mitochondrion. 2009;9:385–393. doi: 10.1016/j.mito.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Selmer M., Dunham C.M., Murphy F.V., IV, Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 61.Yassin A., Fredrick K., Mankin A.S. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc. Natl Acad. Sci. USA. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kubarenko A., Sergiev P., Wintermeyer W., Dontsova O., Rodnina M.V. Involvement of helix 34 of 16 S rRNA in decoding and translocation on the ribosome. J. Biol. Chem. 2006;281:35235–35244. doi: 10.1074/jbc.M608060200. [DOI] [PubMed] [Google Scholar]
- 63.Gochhait S., Bhatt A., Sharma S., Singh Y.P., Gupta P., Bamezai R.N. Concomitant presence of mutations in mitochondrial genome and p53 in cancer development—a study in north Indian sporadic breast and esophageal cancer patients. Int. J. Cancer. 2008;123:2580–2586. doi: 10.1002/ijc.23817. [DOI] [PubMed] [Google Scholar]
- 64.Zhou S., Kachhap S., Sun W., Wu G., Chuang A., Poeta L., Grumbine L., Mithani S.K., Chatterjee A., Koch W., et al. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc. Natl Acad. Sci. USA. 2007;104:7540–7545. doi: 10.1073/pnas.0610818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fromm H., Edelman M., Aviv D., Galun E. The molecular basis for rRNA-dependent spectinomycin resistance in nicotiana chloroplasts. EMBO J. 1987;6:3233–3237. doi: 10.1002/j.1460-2075.1987.tb02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brink M.F., Brink G., Verbeet M.P., de Boer H.A. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res. 1994;22:325–331. doi: 10.1093/nar/22.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makosky P.C., Dahlberg A.E. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie. 1987;69:885–889. doi: 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- 68.Binet R., Maurelli A.T. Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in chlamydia psittaci 6BC. Antimicrob. Agents Chemother. 2005;49:4455–4464. doi: 10.1128/AAC.49.11.4455-4464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaisa N.T., Graham T.A., McDonald S.A., Poulsom R., Heidenreich A., Jakse G., Knuechel R., Wright N.A. Clonal architecture of human prostatic epithelium in benign and malignant conditions. J. Pathol. 2011;225:172–180. doi: 10.1002/path.2959. [DOI] [PubMed] [Google Scholar]
- 70.Sergiev P.V., Lavrik I.N., Wlasoff V.A., Dokudovskaya S.S., Dontsova O.A., Bogdanov A.A., Brimacombe R. The path of mRNA through the bacterial ribosome: A site-directed crosslinking study using new photoreactive derivatives of guanosine and uridine. RNA. 1997;3:464–475. [PMC free article] [PubMed] [Google Scholar]
- 71.Bucklin D.J., van Waes M.A., Bullard J.M., Hill W.E. Cleavage of 16S rRNA within the ribosome by mRNA modified in the A-site codon with phenanthroline-cu(II) Biochemistry. 1997;36:7951–7957. doi: 10.1021/bi9624954. [DOI] [PubMed] [Google Scholar]
- 72.Matassova A.B., Rodnina M.V., Wintermeyer W. Elongation factor G-induced structural change in helix 34 of 16S rRNA related to translocation on the ribosome. RNA. 2001;7:1879–1885. [PMC free article] [PubMed] [Google Scholar]
- 73.Tseng L.M., Yin P.H., Yang C.W., Tsai Y.F., Hsu C.Y., Chi C.W., Lee H.C. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer. 2011;50:800–811. doi: 10.1002/gcc.20901. [DOI] [PubMed] [Google Scholar]
- 74.Sun Q., Vila-Sanjurjo A., O'Connor M. Mutations in the intersubunit bridge regions of 16S rRNA affect decoding and subunit-subunit interactions on the 70S ribosome. Nucleic Acids Res. 2011;39:3321–3330. doi: 10.1093/nar/gkq1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liiv A., O'Connor M. Mutations in the intersubunit bridge regions of 23 S rRNA. J. Biol. Chem. 2006;281:29850–29862. doi: 10.1074/jbc.M603013200. [DOI] [PubMed] [Google Scholar]
- 76.Cochella L., Brunelle J.L., Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat. Struct. Mol. Biol. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- 77.Yoshizawa S., Fourmy D., Puglisi J.D. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- 78.Greaves L.C., Elson J.L., Nooteboom M., Grady J.P., Taylor G.A., Taylor R.W., Mathers J.C., Kirkwood T.B., Turnbull D.M. Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet. 2012;8:e1003082. doi: 10.1371/journal.pgen.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cunningham P.R., Nurse K., Weitzmann C.J., Ofengand J. Functional effects of base changes which further define the decoding center of Escherichia coli 16S ribosomal RNA: mutation of C1404, G1405, C1496, G1497, and U1498. Biochemistry. 1993;32:7172–7180. doi: 10.1021/bi00079a014. [DOI] [PubMed] [Google Scholar]
- 80.Cunningham P.R., Nurse K., Weitzmann C.J., Negre D., Ofengand J. G1401: a keystone nucleotide at the decoding site of Escherichia coli 30S ribosomes. Biochemistry. 1992;31:7629–7637. doi: 10.1021/bi00148a026. [DOI] [PubMed] [Google Scholar]
- 81.Cunningham P.R., Nurse K., Bakin A., Weitzmann C.J., Pflumm M., Ofengand J. Interaction between the two conserved single-stranded regions at the decoding site of small subunit ribosomal RNA is essential for ribosome function. Biochemistry. 1992;31:12012–12022. doi: 10.1021/bi00163a008. [DOI] [PubMed] [Google Scholar]
- 82.Mears J.A., Sharma M.R., Gutell R.R., McCook A.S., Richardson P.E., Caulfield T.R., Agrawal R.K., Harvey S.C. A structural model for the large subunit of the mammalian mitochondrial ribosome. J. Mol. Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mutai H., Kouike H., Teruya E., Takahashi-Kodomari I., Kakishima H., Taiji H., Usami S., Okuyama T., Matsunaga T. Systematic analysis of mitochondrial genes associated with hearing loss in the Japanese population: DHPLC reveals a new candidate mutation. BMC Med. Genet. 2011;12:135. doi: 10.1186/1471-2350-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamada M., Sato K., Kiryu H., Mituyama T., Asai K. Centroidalign: fast and accurate aligner for structured RNAs by maximizing expected sum-of-pairs score. Bioinformatics. 2009;25:3236–3243. doi: 10.1093/bioinformatics/btp580. [DOI] [PubMed] [Google Scholar]
- 85.Lu J., Li Z., Zhu Y., Yang A., Li R., Zheng J., Cai Q., Peng G., Zheng W., Tang X., et al. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion. 2010;10:380–390. doi: 10.1016/j.mito.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen Z., Zheng J., Chen B., Peng G., Zhang T., Gong S., Zhu Y., Zhang C., Li R., Yang L., et al. Frequency and spectrum of mitochondrial 12S rRNA variants in 440 Han Chinese hearing impaired pediatric subjects from two otology clinics. J. Transl. Med. 2011;9:4. doi: 10.1186/1479-5876-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu Y., Li Q., Chen Z., Kun Y., Liu L., Liu X., Yuan H., Zhai S., Han D., Dai P. Mitochondrial haplotype and phenotype of 13 Chinese families may suggest multi-original evolution of mitochondrial C1494T mutation. Mitochondrion. 2009;9:418–428. doi: 10.1016/j.mito.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 88.Taylor R.W., Giordano C., Davidson M.M., d'Amati G., Bain H., Hayes C.M., Leonard H., Barron M.J., Casali C., Santorelli F.M., et al. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2003;41:1786–1796. doi: 10.1016/s0735-1097(03)00300-0. [DOI] [PubMed] [Google Scholar]
- 89.Hobbie S.N., Bruell C.M., Akshay S., Kalapala S.K., Shcherbakov D., Bottger E.C. Mitochondrial deafness alleles confer misreading of the genetic code. Proc. Natl Acad. Sci. USA. 2008;105:3244–3249. doi: 10.1073/pnas.0707265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guan M.X., Yan Q., Li X., Bykhovskaya Y., Gallo-Teran J., Hajek P., Umeda N., Zhao H., Garrido G., Mengesha E., et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am. J. Hum. Genet. 2006;79:291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vila-Sanjurjo A., Dahlberg A.E. Mutational analysis of the conserved bases C1402 and A1500 in the center of the decoding domain of Escherichia coli 16 S rRNA reveals an important tertiary interaction. J. Mol. Biol. 2001;308:457–463. doi: 10.1006/jmbi.2001.4576. [DOI] [PubMed] [Google Scholar]
- 92.Rodriguez-Correa D., Dahlberg A.E. Genetic evidence against the 16S ribosomal RNA helix 27 conformational switch model. RNA. 2004;10:28–33. doi: 10.1261/rna.5172104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen S.S., Liu C., Xu Z.Y., Hu Y.H., Gao G.F., Wang S.Y. Heteroplasmy levels of mtDNA1555A>G mutation is positively associated with diverse phenotypes and mutation transmission in a Chinese family. Biochem. Biophys. Res. Commun. 2012;420:907–912. doi: 10.1016/j.bbrc.2012.03.100. [DOI] [PubMed] [Google Scholar]
- 94.Hobbie S.N., Akshay S., Kalapala S.K., Bruell C.M., Shcherbakov D., Bottger E.C. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc. Natl Acad. Sci. USA. 2008;105:20888–20893. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kondo J., Westhof E. The bacterial and mitochondrial ribosomal A-site molecular switches possess different conformational substates. Nucleic Acids Res. 2008;36:2654–2666. doi: 10.1093/nar/gkn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 97.Chinnery P.F., Samuels D.C., Elson J., Turnbull D.M. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: Is there a common mechanism? Lancet. 2002;360:1323–1325. doi: 10.1016/S0140-6736(02)11310-9. [DOI] [PubMed] [Google Scholar]
- 98.Pereira L., Freitas F., Fernandes V., Pereira J.B., Costa M.D., Costa S., Maximo V., Macaulay V., Rocha R., Samuels D.C. The diversity present in 5140 human mitochondrial genomes. Am. J. Hum. Genet. 2009;84:628–640. doi: 10.1016/j.ajhg.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sayers E. Entrez Programming. Bethesda, MD: National Center for Biotechnology Information (US); 2008. E-utilities quick start. http://www.ncbi.nlm.nih.gov/books/NBK25501/ Utilities Help [Internet] [Google Scholar]
- 100.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 101.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang X., Miller W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991;12:337–357. [Google Scholar]
- 103.Korostelev A., Trakhanov S., Laurberg M., Noller H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 104.Schmeing T.M., Voorhees R.M., Kelley A.C., Ramakrishnan V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. 2011;18:432–436. doi: 10.1038/nsmb.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 106.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.