Abstract
Rett syndrome (RTT) is one of the most prevalent female mental disorders. De novo mutations in methyl CpG-binding protein 2 (MeCP2) are a major cause of RTT. MeCP2 regulates gene expression as a transcription regulator as well as through long-range chromatin interaction. Because MeCP2 is present on the X chromosome, RTT is manifested in an X-linked dominant manner. Investigation using murine MeCP2 null models and post-mortem human brain tissues has contributed to understanding the molecular and physiological function of MeCP2. In addition, RTT models using human induced pluripotent stem cells derived from RTT patients (RTT-iPSCs) provide novel resources to elucidate the regulatory mechanism of MeCP2. Previously, we obtained clones of female RTT-iPSCs that express either wild-type or mutant MECP2 due to the inactivation of one X chromosome. Reactivation of the X chromosome also allowed us to have RTT-iPSCs that express both wild-type and mutant MECP2. Using these unique pluripotent stem cells, we investigated the regulation of gene expression by MeCP2 in pluripotent stem cells by transcriptome analysis. We found that MeCP2 regulates genes encoding mitochondrial membrane proteins. In addition, loss of function in MeCP2 results in de-repression of genes on the inactive X chromosome. Furthermore, we showed that each mutation in MECP2 affects a partly different set of genes. These studies suggest that fundamental cellular physiology is affected by mutations in MECP2 from early development, and that a therapeutic approach targeting to unique forms of mutant MeCP2 is needed.
INTRODUCTION
Rett syndrome (RTT) is one of the most prevalent female neurodevelopmental disorders. RTT symptoms include seizures, developmental regression, typical hand wringing and motor abnormalities. Most RTT patients have a mutated X-linked gene encoding methyl-CpG-binding protein 2 (MeCP2) (1,2). MeCP2 regulates the expression of neuronal developmental genes such as BDNF and DLX5/6 (3,4). Expression of MeCP2 at high dosage also causes detrimental RTT phenotypes (5,6). The molecular mechanism by which MeCP2 regulates gene expression is under active investigation. Since MeCP2 interacts with repressive histone modification enzymes such as SIN3A and HDAC1, MeCP2 was initially proposed as a transcription repressor (7,8). However, global gene expression analysis in the hypothalamus of Mecp2 null and transgenic mice implicates MeCP2 in transcription activation as well (9). In addition, ChIP analysis of MeCP2 in human neuronal cells showed that MeCP2 binds promoters of active genes, further supporting the notion that MeCP2 acts as a transcription activator (10). MeCP2 is also involved in RNA splicing (11), silent-chromatin looping (4), regulation of ubiquitin ligase (12), cell cycle regulation, apoptosis (13) and repression of LINE-1 retrotransposition (14,15).
In female cells, one of the two X chromosomes is inactivated (XCI) to balance the expression of genes on X chromosome to that in male cells. During early development, XCI occurs when XIST transcripts from one of the X chromosomes coat in cis the chosen X chromosome, which is then repressed by heterochromatic modification through trimethylation of histone H3 lysine 27 (H3K27me3) (16). Human female embryonic stem cells (hESCs) display three distinct patterns of XCI: class I cells have two active X chromosomes and recapitulate XCI during differentiation; class II cells have one inactive X chromosome with detectable XIST and H3K27me3; and class III cells also have one inactive X chromosome, but do not show H3K27me3 and XIST(17). Induced pluripotent stem cell (iPSC) clones that we previously isolated express either wild-type MECP2 (RTT-wt-iPSCs), mutant MECP2 (RTT-mu-iPSCs) or both (RTT-bi-iPSCs) (18). Whereas RTT-wt-iPSCs and RTT-mu-iPSCs show the distribution of class II hESC-like patterns, RTT-bi-iPSCs show that of class I. Recent studies demonstrated that long-term culture, oxygen concentration and feeder cell change X chromosome status (19–21).
Success in generating iPSCs gives an opportunity to develop efficient human disease models (22–24). The patient-derived iPSCs grow indefinitely in culture and can be used for monitoring pathophysiology of the diseases and testing drug responses in vitro (25,26). Previously, we and other groups have derived iPSCs from fibroblasts of RTT patients with retroviruses expressing four reprogramming factors (OCT4, SOX2, KLF4 and MYC) (18,27–29). Neurons differentiated from RTT-iPSCs showed the morphological and functional phenotypes that are known in RTT patients and murine models. In addition to the essential function of MeCP2 in neurons, recent studies showed that abnormal function of MeCP2 in non-neuronal cells, including astrocytes and microglia, is critical in RTT (30,31). These results suggest that MeCP2 plays a critical role in other non-neuronal cells expressing MeCP2, including pluripotent stem cells.
Here, we set out to examine the regulation of pluripotent genes by MeCP2 using human RTT-iPSCs. We performed a transcriptome analysis of normal hESCs, iPSCs and RTT-iPSCs with massively parallel RNA sequencing (RNA-seq). Comparative analysis of global gene expression patterns indicates that mutant iPSCs are separable from normal iPSCs and ESCs. Our data showed that some sets of genes are differentially expressed between mutant RTT-iPSCs and wild-type iPSCs, although the expression of MeCP2 is relatively lower in pluripotent stem cells compared with neurons. In addition to genes involved in neuronal development or function, MECP2 mutant RTT-iPSCs showed the up-regulation of mitochondria-related genes. Furthermore, comparative analysis among RTT patients revealed that distinct genes are affected by different mutations in MeCP2. Together, our results show that MeCP2 regulates physiologically fundamental genes from very early embryonic development, suggesting the importance of investigating the role of MeCP2 in diverse cell types.
RESULTS
Global expression similarity among hESCs, iPSCs and RTT-iPSCs
RTT-iPSCs were obtained from five different patients, who have distinct mutations in different domains of MeCP2: RTT1 (T158M), RTT2 (Q244X), RTT3 (E235fs), RTT4 (R306C) and RTT5 (X487W) (Supplementary Material, Table S1). The X chromosome status in human ESCs, normal iPSCs, and RTT-iPSCs were evaluated by staining cells using a H3K27me3 antibody as in previous studies (18,32) (Supplementary Material, Fig. S1A). Furthermore, we applied MeCP2 transcripts of each iPSC line to Sanger sequencing to determine whether RTT-iPSC clone expresses either wild-type MECP2 (RTT-wt-iPSCs), mutant MECP2 (RTT-mu-iPSCs), or both wild-type and mutant MECP2 (RTT-bi-iPSCs) (Supplementary Material, Fig. S1B). X chromosome status was further assessed by fluorescent in situ hybridization (FISH) and qPCR to determine the presence of XIST coating (16). Consistent with the corroborative mechanism in XCI, XIST foci were found in clones with H3K27me3 foci (Supplementary Material, Fig. S1C). RTT-wt-iPSCs and RTT-mu-iPSCs show higher XIST expression than RTT-bi-iPSCs (Supplementary Material, Fig. S2). In addition to RTT-iPSCs, the X chromosome status of normal male (H1) and female ESCs (H7 and H9), iPSCs derived from normal males (PGP1) and females (PGP9 and Detroit 551) was assessed. The XIST expression level in RTT-bi-iPSCs is similar to that in male.
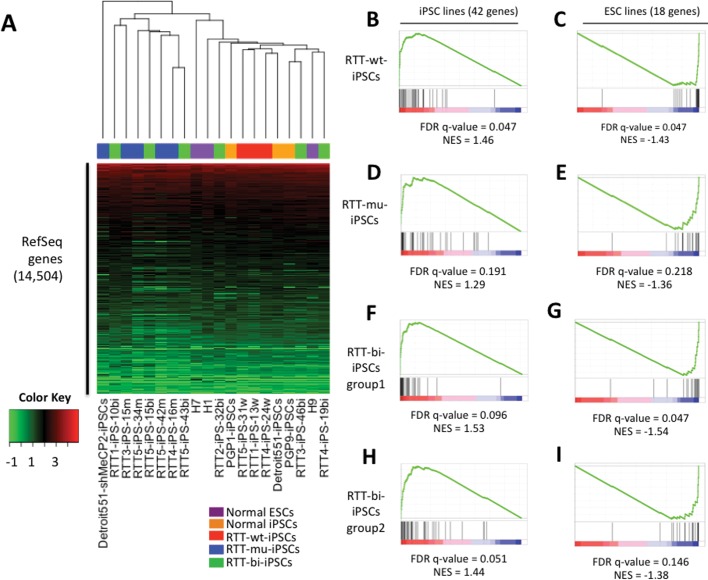
In order to investigate the regulatory role of MeCP2 in pluripotent stem cells, RNA-seq was performed. More than 20 million reads were sequenced in each sample, using the Illumina GAII or Hi-Seq 2000. 83–95% of reads were successfully mapped on the human genome (Supplementary Material, Table S1). Expression of mutant MeCP2 in RTT-mu-iPSCs was also observed in RNA-seq (Supplementary Material, Fig. S3). Hierarchical clustering was then performed for the expression patterns of 14, 504 RefSeq genes. The clustering dendrogram distinguished RTT-mu-iPSCs from RTT-wt-iPSCs and normal iPSCs or ESCs (Fig. 1A). This result suggests that MeCP2 plays a role in global gene regulation in pluripotent stem cells. In contrast, global expression patterns of RTT-bi-iPSCs were not separable from RTT-wt-iPSCs and RTT-mu-iPSCs, because RTT-bi-iPSCs express both wild-type and mutant MeCP2. Whereas three RTT-bi-iPSCs (RTT2-iPS-32bi, RTT3-iPS-46bi and RTT4-iPS-19bi; group1) were closer to RTT-wt-iPSCs and the others (RTT1-iPS-10bi, RTT5-iPS-15bi and RTT5-iPS-43bi; group2) were closer to RTT-mu-iPSCs.
Figure 1.
Global Expression similarity of hESCs, cell line-derived iPSCs and RTT-iPSCs. (A) Supervised hierarchical clustering analysis was performed using 14 504 expressed RefSeq genes. Log2(FPKM) values were represented by colors from green (low) to red (high). (B–I) Enrichment of iPSCs and hESCs-specific genes in RTT-iPSCs. False discovery rate (FDR) and normalized enrichment score (NES) were calculated by Gene Set Enrichment Analysis (GSEA) software at Broad institute.
Human somatic cell reprogramming was shown to leave reprogramming-specific epigenetic and genetic signatures (33). In order to determine whether MeCP2 status has influence on reprogramming-specific gene expression, we performed a direct comparison of hESCs and iPSCs. We identified genes differentially expressed between ESCs and iPSCs. Eighteen genes are highly expressed in ESCs, and 42 genes are highly expressed in iPSCs, respectively (P < 0.05; Supplementary Material, Fig. S4). ESC-specific genes include TMEM132D, FAM19A5 and TCERG1L, which were previously identified as genes that are repressed in iPSCs (33). Interestingly, only in RTT-wt-iPSCs, iPSC-specific genes were significantly over-represented (Fig. 1B, false discovery rate (FDR) q < 0.05), and the ESC-specific genes were significantly depleted (Fig. 1C, q < 0.05). However, no significant enrichment of iPSC- or ESC-specific genes was observed in RTT-mu-iPSCs and group 2 of RTT-bi-iPSCs compared with ESCs (Fig. 1D–I; q > 0.05). Group1 of RTT-bi-iPSCs showed significant deletion of ESC-specific genes (q < 0.05), but did not show significant enrichment of iPSC-specific genes (q > 0.05). These results indicate that RTT-wt-iPSCs are closer to normal iPSCs than RTT-bi-iPSCs and RTT-mu-iPSCs.
Genes regulated by MeCP2 in pluripotent stem cells
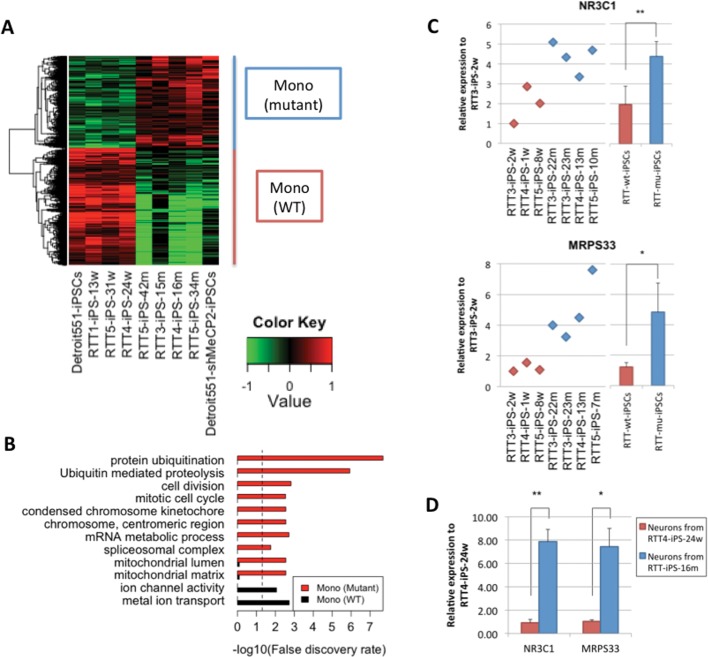
To determine the gene regulation by MeCP2, the gene expression patterns between wild-type and mutant were directly compared. In addition to RTT-mu-iPSCs, we depleted MeCP2 in normal Detroit551-iPSCs by knocking down (KD) MeCP2 with shRNAs against MeCP2 mRNA, and selected clones with MeCP2 depletion (Supplementary Material, Fig. S5). We detected that 2,038 and 1,587 genes show significantly higher expression in RTT-wt-iPSCs and RTT-mu-iPSCs, respectively (Fig. 2A and Supplementary Material, Table S2A and B, P < 0.05). KEGG pathway and Gene Ontology (GO) analysis revealed that genes involved in splicing and ubiquitin-mediated proteolysis are highly enriched in RTT-mu-iPSCs, which were previously shown to be associated with MECP2 mutations (11–13). (Fig. 2B). Furthermore, genes involved in mitochondrial functions were also highly enriched in RTT-mu-iPSCs, including gene encoding mitochondrial transcription factor (NR3C1) and mitochondrial ribosomal protein (MRPS33) (Fig. 2C). These genes are also upregulated in the mutant cells after neuronal differentiation (Fig. 2D). Consistent with our results, previous gene expression analysis in Mecp2 null mouse brain showed the overexpression of mitochondrial genes (34). In addition, human RTT brain showed the enrichment of mitochondrial genes compared with normal brain (Supplementary Material, Fig. S6) (35). The functional implication of abnormal regulation of mitochondrial genes in pluripotent cells and their derivatives in RTT will be an important question in elucidating RTT pathogenesis.
Figure 2.
Differentially-expressed genes between RTT-wt-iPSCs and RTT-mu-iPSCs. (A) Heatmap represents genes, which were highly expressed in RTT-wt-iPSCs and RTT-mu-iPSCs, respectively. Relative expression values to the average expression values across all samples were represented by colors from green to red (log2 scale). (B) Over-representation of GO terms and KEGG pathways is shown by bar plot. Dashed line represents 0.05 FDR. qPCR validation of differential expression of NR3C1 and MRPS33 in (C) RTT-iPSCs and (D) neurons derived from RTT-iPSCs (*P < 0.05 and **P < 0.01).
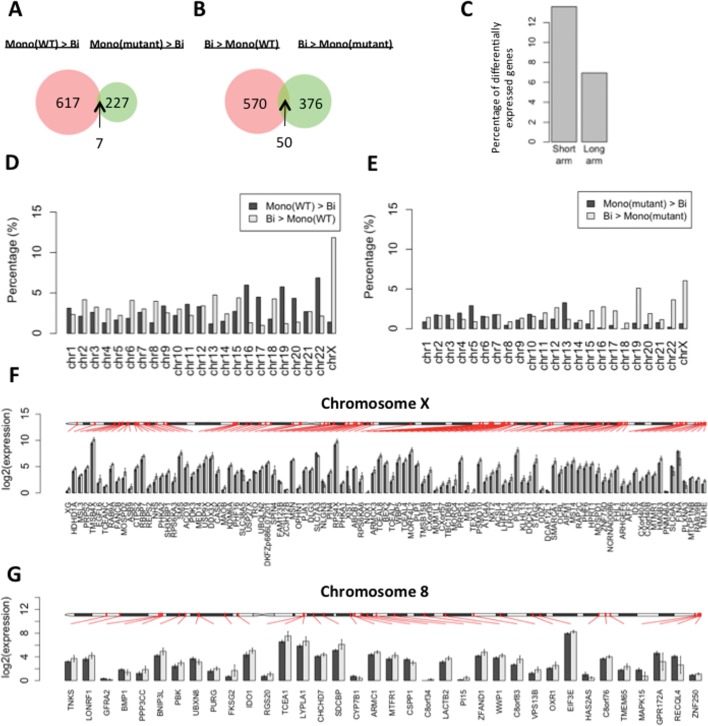
Gene expression was compared in RTT-iPSCs exhibiting biallelic or monoallelic X chromosome expression. We found that only seven genes are highly expressed in monoallelic RTT-wt-iPSCs and RTT-mu-iPSCs compared with RTT-bi-iPSCs (Fig. 3A). One X-linked gene, XIST are included in the monoallelic iPSC-specific genes (Supplementary Material, Table S2C), corresponding to FISH and qPCR experiments. Fifty genes are highly expressed in biallelic RTT-iPSCs compared with monoallelic cells, of which 18 genes are on the X chromosome (Fig. 3B and Supplementary Material, Table S2D). The number of X-linked genes among the biallelic-specific genes is significantly higher than that expected by chance (P = 9.53e–15 by hypergeometric test). The biallelic iPSCs-specific genes did not show any significant overrepresentation of GO terms and KEGG pathways, but include active histone modification enzymes (KDM4DL and MLL3).
Figure 3.
Differentially expressed genes between monoallelic and biallelic RTT-iPSCs. (A and B) Venn diagram represents the number of (A) highly or (B) lowly expressed genes in RTT-wt-iPSCs and RTT-mu-iPSCs compared with RTT-bi-iPSCs (P < 0.05). Thus, the common genes in (A) were defined as monoallelic iPSC-specific and in (B) as biallelic iPSC-specific genes. (C) Comparison of the ratio of differentially expressed X-linked genes between short (<60 600 kbp) and long arms (>60 600 kbp). (D and E) (D) The percentage of differentially expressed genes in each chromosome between RTT-wt-iPSCs and RTT-bi-iPSCs and (E) between RTT-mu-iPSCs and RTT-bi-iPSCs. The ratio was calculated by dividing the absolute amount of differentially expressed genes by the total number of genes per chromosome. (F and G) Chromosomal distribution of differentially-expressed genes in (F) chromosome X and (G) chromosome 8. Black and gray bars represent average of log2(FPKM) value in RTT-wt-iPSCs and RTT-bi-iPSCs, respectively. Error bar represents standard deviation of log2(FPKM).
When compared with RTT-wt-iPSCs, RTT-bi-iPSCs have a larger number of X-linked genes highly expressed than do autosomes (Fig. 3D). While 92 genes on the X chromosome are differentially expressed (Fig. 3F), 29 genes in chromosome 8 that has a similar size as chromosome X showed differential expression (Fig. 3G). The differentially expressed X-linked genes were enriched on the short arm of the X chromosome, which were proposed to be evolutionally added later and to have large number of XCI escaping genes (36) (Fig. 3C). A few number of X-linked genes, including XIST, showed higher expression in RTT-wt-iPSCs than in RTT-bi-iPSCs. Among them, four X-linked genes (PNMA6A, SLC6A8, FLNA and PLXNA3) are proximal to the telomeric region, which is epigenetically variable in iPSCs (33,37).
In comparison between RTT-mu-iPSCs and RTT-bi-iPSCs, most of the X-linked genes are highly expressed in RTT-bi-iPSCs. However, less number of X-linked genes are differentially expressed, compared with those between RTT-wt-iPSCs and RTT-bi-iPSCs (Fig. 3E). This illustrates that MeCP2 contributes to repression of X-linked genes and the MeCP2 mutation in RTT-mu-iPSCs de-represses genes on inactive X chromosome. In autosome, there is no clear difference in the number of differentially expressed genes between monoallelic and biallelic iPSCs (Fig. 3D and E), suggesting the role of MeCP2 in both gene activation and repression in autosomal genes.
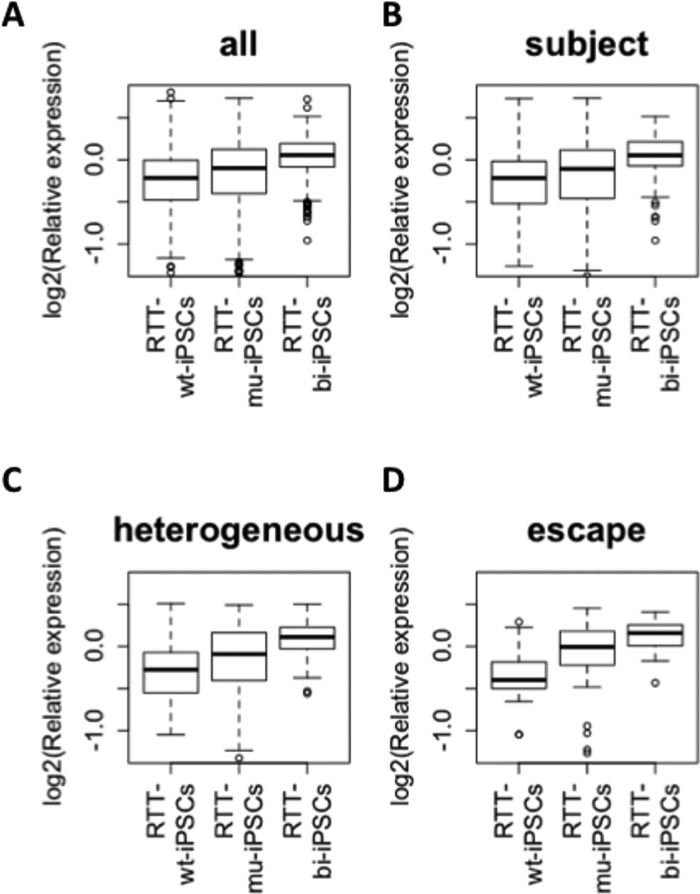
Furthermore, the expression of X-linked genes is significantly different among RTT-wt-iPSCs, RTT-mu-iPSCs and RTT-bi-iPSCs (Fig. 4A). RTT-mu-iPSCs showed higher gene expression values than RTT-wt-iPSCs (q = 4.43e−4), but lower expression than RTT-bi-iPSCs (q = 6.60e–16; Fig. 4A). This result suggests that MeCP2 and XIST are important factors for the regulation of X-linked gene expression. Several X-linked genes were shown to escape from XCI. Carrel et al. (38) extensively classified X-linked genes into three classes: escape, subject and heterogeneous, which represent genes escaping XCI, those strictly subject to XCI, and those showing loose XCI. We used their categories and compared the expression values of X-linked genes. In the subject category, RTT-mu-iPSCs showed a similar expression pattern to total X chromosome genes, and exhibited significantly a higher gene expression value than RTT-wt-iPSCs (q = 2.96e–2), and significantly lower expression than RTT-bi-iPSCs. (q = 6.11e − 9; Fig. 4B). Interestingly, expression values of heterogeneous genes were significantly lower in RTT-mu-iPSCs compared with RTT-bi-iPSCs (q = 1.64e–2), but there is no significant difference in heterogeneous gene expression between RTT-wt-iPSCs and RTT-mu-iPSCs (q = 0.214; Fig. 4C). In contrast, in RTT-mu-iPSCs expression values of escape genes were significantly greater compared with RTT-wt-iPSCs (q = 1.66e–3), but similar to RTT-bi-iPSCs (q = 0.116; Fig. 4D). Previously, the escape genes were defined by biallelic expression regardless of XCI. However, our analysis suggests that expression of escape genes is subjected to XCI in pluripotent stem cells. Interestingly MeCP2 seems to play an active role in repressing the escape genes, but not in the heterogeneous genes. It will be interesting to examine whether MeCP2 has a similar role in differentiated cells and affects genes on X chromosomes, especially escape genes.
Figure 4.
Expression of genes in X chromosome among mono-allelic wild type and mutant, and biallelic RTT-iPSCs. Relative expression level to average of all RTT-iPSCs are shown (log2 scale): (A) average all; (B), ‘heterogeneous’; (C), ‘subject’; (D), ‘escape’ X-linked genes which are defined by Carrel et al. (38).
Regulation of distinct genes in each RTT-iPSC
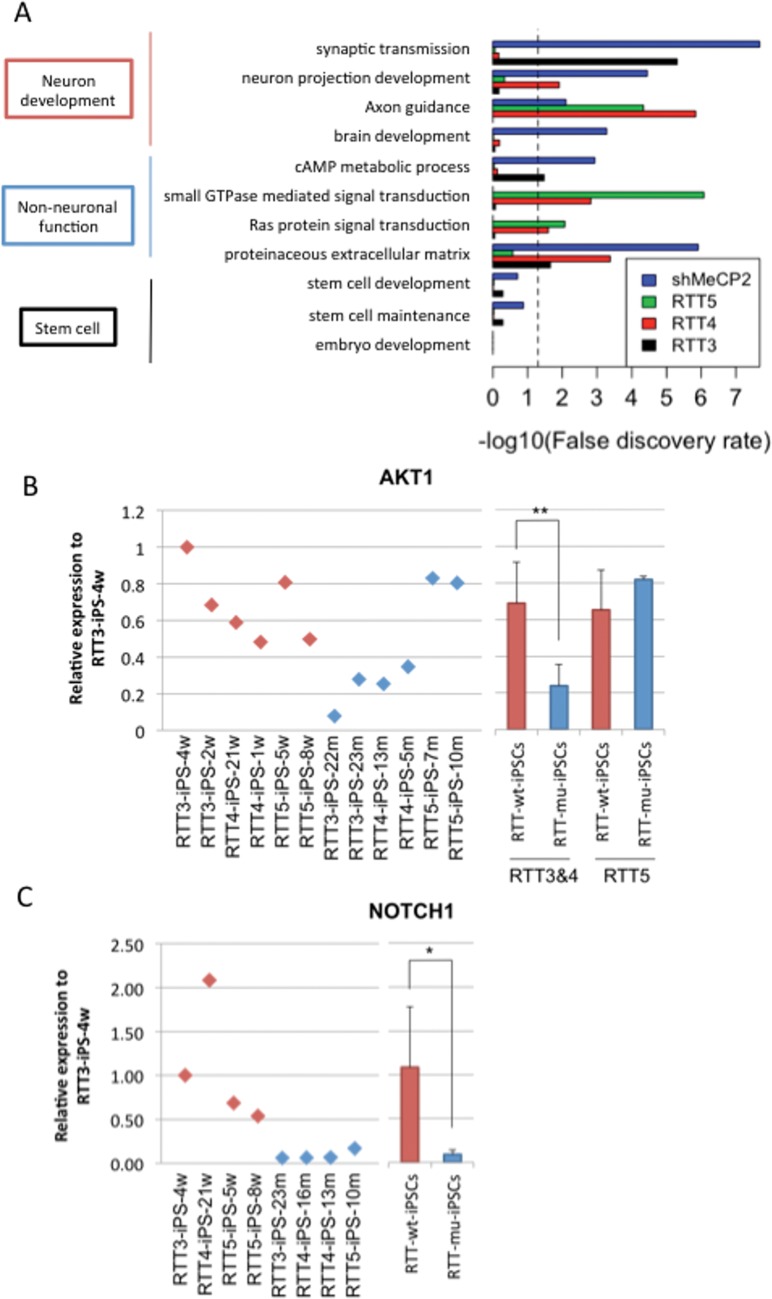
Although different mutations in MeCP2 cause similar RTT symptoms, distinct cognitive and clinical consequences in patients with different mutations suggest that different functions are affected in MeCP2 (39). In order to determine the regulation of gene expression in pluripotent stem cells by different mutations in MECP2, genes with ≥1.5-fold changes between wild-type and mutant RTT-iPSCs were compared. In order to analyze the effect of MeCP2 loss on gene regulation with those from each MECP2 mutation, differential gene expression from MeCP2 KD was included for comparison. Among affected pathways are included those involved in neurodevelopment as well as signaling transduction pathways (Fig. 5A). Pathways, such as neuron projection development and axon guidance, were affected in MeCP2 KD, RTT4 and RTT5 mutant iPSCs, while the pathway in synaptic transmission is affected in MeCP2 KD and RTT3-iPSCs. Among non-neuronal functions, RTT4 and RTT5 mutant iPSCs showed affected pathways in small GTPase-mediated signaling transduction and Ras protein signaling pathway. Proteinaceous extracellular matrix is affected in MeCP2 KD, RTT3 and RTT4. Different types of biological pathways were shown affected by different mutations. We have performed qPCR analysis of AKT1 and NOTCH1, which are required for neuronal development. In RNA-seq, AKT1 showed differential expression in RTT2 and RTT3, whereas NOTCH1 is downregulated in all RTTs. Like RNA-seq data, AKT1 showed lower expression in RTT2 and RTT3 but not in RTT5-iPSC (Fig. 5B). Meanwhile, we confirmed that NOTCH1 is repressed in RTT3, RTT4 and RTT5-iPSCs (Fig. 5C). The pathways involved in stem cell development and differentiation were not affected in any of the mutations of MeCP2 or KD iPSCs (Fig. 5A), suggesting that MeCP2 mutations do not affect stem cell development and differentiation. The functional implication of the different pathways affected by different mutations in MeCP2 will be important in elucidating the neuro- and non-neuronal pathophysiology of RTT.
Figure 5.
Differential gene expression in RTT iPSCs with distinct mutations. (A) Over-represented GO terms and KEGG pathways of downregulated genes in RTT-iPSCs with different mutations and MeCP2 KD iPSCs. Pathways in non-neuronal and neuronal functions were represented. Dashed line represents 0.05 FDR. (B and C) qPCR validation of differential expression of neuronal developmental genes (AKT1 and NOTCH1) using multiple wild-type and mutant RTT-iPSCs. (*P < 0.05 and **P < 0.01)
DISCUSSION
In this study, comparative analyses of gene expression in RTT-iPSCs and normal ESCs/iPSCs were made using RNA-seq. We found several functions of MeCP2 in pluripotent stem cells. First, we found that the MeCP2 pathway is involved in both transcriptional up- and down-regulation in pluripotent stem cells. Previous analysis of the transcriptome in Mecp2 null or transgenic neurons showed that the major function of MeCP2 in neurons is transcription activation (6,9,10). It is conceivable that the regulatory role of MeCP2 is different among different cell types, and further investigation will be needed to elucidate the mechanism of gene regulation by MeCP2. A recent study found that MeCP2 also binds to 5hmC in vitro (40). 5hmC is enriched in active promoters, enhancers and gene bodies of ESC and postnatal brain, and is negatively correlated with MeCP2 dosages (41,42), suggesting that the control of 5hmC level is one of the potential mechanisms of MeCP2-mediated gene activation. Another potential possibility for controlling MeCP2 function is a brain-specific post-translational modification of MeCP2. Phosphorylation of MeCP2 in neurons controls nervous system maturation processes such as dendritic growth and spine maturation (3,43,44). Serine 80 of MeCP2, which is conserved in human, mouse and rat, is known to be specifically phosphorylated in brain and alters chromatin structure to modulate transcription levels of genes for neuronal activity (45). Therefore, cell type-specific post-translational modification in MeCP2 may determine repressive or active functions.
Second, we found that in pluripotent stem cells, MeCP2 modulates the expression of mitochondria-related genes whose expression is also altered in other neuronal diseases. In Parkinson's and Huntington's disease, the deficiency of these genes induces mitochondria-mediated apoptosis that causes excessive death of mature neurons (46,47). Reduction of apoptosis was previously demonstrated by down-regulation of MeCP2 in mesenchymal stem cells (MSCs) (13). Gene expression analysis in a murine RTT model also showed an abnormality of the mitochondrial genes in RTT brain, suggesting that MeCP2 regulates common signaling pathways in pluripotent cells and brain cells (34).
In addition to the common regulatory pathways, each MECP2 mutation in RTT-iPSCs shows different expression in genes related to neuronal development in pluripotent stem cells. Since the depletion of MeCP2 protein by KD leads to the down-regulation of all pathways observed in each patient line, MECP2 KD cells have advantages to understand all potential targets of MeCP2, but are not suitable for studying patient-specific RTT pathogenesis. Our results further support the critical importance of using patient-derived iPSCs to understand the unique phenotypes and drug response in individual RTT patient.
Third, we found that MeCP2 is also important in inactivation of X-linked genes. Our data showed that MeCP2 represses X-linked genes. The repression by MeCP2 is not uniform across all X-linked genes and expression differences between RTT-wt-iPSCs and RTT-mu-iPSCs were smaller in genes, which are prone to XCI. These results suggest that the repressive effect of MeCP2 on some genes present in X chromosome is independent of XIST-mediated XCI.
In summary, our data highlight the MeCP2 regulatory pathway in undifferentiated cells. These data provide novel insights into MeCP2 functions in the undifferentiated stage and implicate the importance of dissecting the function of MeCP2 in diverse cell types for RTT.
MATERIALS AND METHODS
Cell culture
We used RTT-iPSCs generated in (18). Retroviral preparation and infection with retroviruses encoding OCT4, SOX2, KLF4 and c-MYC complementary DNA (cDNA) driven by the LTR promoter of a retroviral vector, pMIG (MSCV-IRES-GFP) were performed as previously described (18,48). Briefly, retrovirus stocks were prepared by transient transfection of 293T cells using X-tremgene9 (Roche), and supernatants were harvested 48 h later. For viral infection, RTT fibroblasts were plated onto a six-well plate followed by retrovirus infection in the presence of protamine sulfate. The plates were incubated at 37°C for 24 h, and then the viral supernatant was replaced with fresh culture medium (DMEM with 10%FBS, non-essential amino acid and P/S). Five days after viral infection, the cells were transferred onto irradiated MEF cells in human ES cell culture media (DMEM/F12 with 20% of knockout serum replacer, 2 mm of glutamine, 0.1 mm of β-mercaptoethanol, 4 µg/ml of FGF-2, 0.1 mm of non-essential amino acid and P/S). After 30 days, iPSC colonies, which showed absence of GFP fluorescence, were picked using a 20 µl pipette under a fluorescence microscope, and expanded on MEFs and on Matrigel (BD biosciences)
Immunostaining and FISH
Cells were grown on Matrigel-coated plates. Samples were washed with phosphate buffered saline (PBS), fixed with 4% PFA/PBS for 15 min at RT, permeabilized with 0.2% Triton X-100/PBS for 15 min at RT and blocked with 3% BSA/PNS for 2 h. Then, the following H3K27me3 antibody (Rabbit mAb #9733 from Cell Signaling Technology) was added overnight at 4°C. The cells were washed with PBS and then incubated with a secondary antibody for 1 h at 4°C.
RNA FISH was carried out as described previously (49). iPSCs were grown on gelatin-coated coverslips and permeabilized by means of sequential transfer into twice ice-cold PBS for 30 s, ice-cold CSK buffer (100 mm NaCl, 300 mm sucrose, 3 mm MgCl2 and 10 mm pH 6.8 PIPES and RNase-inhibitor RNasin) for 30 s, and then CSK buffer containing 0.2% Triton X-100 buffer for 15 min, followed twice with CSK buffer for 30 s each. Cells were then fixed with 4% formaldehyde in PBS for 15 min and dehydrated through sequential changes of 70, 85 95 and 100% EtOH for 3 min each, followed by air drying before hybridization of the XIST probe. For the XIST probe, double-strand shorter DNA probes were generated form a G1A plasmid (50) and labeled with fluorescein-12-dUTP using the Prime-It Fluor Labeling kit (Stratagene). The probe was then hybridized with a prepared iPSC sample at 37 °C overnight in a humidified chamber.
MeCP2 depletion experiment
pLKO.1-TRC-based lentiviral vectors (shRNA against MeCP2, clones TRCN0000021239 and TRCN0000021240 from Sigma) were transfected into 293T cells together with plasmids MDL, REV and VSVL using X-tremgene9 (Roche) according to the manufacturer's instructions. The virus-containing supernatant was collected 48 h after transfection, filtered through 0.45 µm filters (Millipore, Bedford, MA, USA) and concentrated by ultracentrifugation. iPSCs were incubated in the virus supernatant supplemented with protamine sulfate for 24 h, and then the cells were replaced in fresh ES culturing medium added puromycin at a final concentration of 1 µg/ml. iPSC colonies resistant to puromycin were selected for 2 weeks, and were used to confirm the knock-down of MeCP2.
Real time quantitative PCR
Total RNA was isolated using an RNeasy kit (QIAGEN) followed by cDNA synthesis using Superscript Reverse Transcriptase kit (iScript™ cDNA Synthesis Kit, Bio-rad). The knock-down efficiency of MeCP2 and the expression level of XIST, mitochondrial genes and neuronal developmental genes were calculated by a quantitative PCR with iQ™ SYBR® Green Supermix (Bio-rad). The expression level of each gene was normalized to that of beta-ACTIN. The difference in the expression level between the wild type and the mutant was evaluated by one-side T test. The primers for qPCR are listed in Supplementary Material, Table S3.
Western blotting
Cells were lysed directly in RIPA lysis buffer (50 mm pH 7.5 Tris–HCl, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate and protease inhibitor cocktail) and western blot analysis was performed with an antibody recognizing only the C-terminus of MeCP2 (C-17, Santa Cruz). ACTIN was used as a loading control.
RNA-seq library preparation and Illumina sequencing
Libraries were prepared according to Illumina's mRNA-Sequencing Sample Preparation Guide for mRNA-Seq 8 Sample Prep Kit (Part# RS-100-0801). Briefly, poly-A containing mRNA molecules were purified from 10 µg of total RNA using poly-T oligo-attached magnetic beads, then fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments are copied into the first strand cDNA using reverse transcriptase and random primers followed by second strand cDNA synthesis using DNA Polymerase I and RNaseH. These cDNA fragments then go through an end repair process using a combination of T4 DNA polymerase, E. coli DNA Pol I large fragment (Klenow polymerase) and T4 polynucleotide kinase. The blunt, phosphorylated ends were treated with Klenow fragment (3′–5′ exo minus) and dATP to yield a protruding 3- ‘A’ base for ligation of Illumina's adapters, which have a single ‘T’ base overhang at the 3′ end. These products are then purified on 2% E-Gel EX Gel (Invirotgen #G4020-02) and enriched with 15 cycles of PCR to create the final cDNA library. Library fragments of 150–350 bp (insert plus adaptor and PCR primer sequences) were isolated from 2% E-Gel EX Gel (Invirotgen #G4020-02). The purified DNA was captured on an Illumina flow cell for cluster generation. Libraries were sequenced on the Genome Analyzer following the manufacturer's protocols.
Data processing of RNA-seq
Human genomic sequences and coordinates of RefSeq genes (version hg19) were downloaded from UCSC database. All RNA-seq reads were aligned to the reference genome by Tophat (v1.1.4) with default parameter (51). Cufflinks (v0.9.3) were then applied to assemble the mapped reads and to calculate gene expression values (Fragments Per Kilobase of exon model per Million mapped fragments: FPKM), which is normalized by the number of mapped reads and gene size, by using RefSeq genes as reference annotation (52). We used genes, which showed average FPKM ≥ 0.3, for further analyses. Differentially expressed genes between two phenotypes (ESCs versus cell line-derived iPSCs, RTT-wt-iPSCs versus RTT-mu-iPSCs, RTT-wt-iPSCs versus RTT-bi-iPSCs and RTT-mu-iPSCs versus RTT-bi-iPSCs) were identified by the Wilcoxon rank-sum test. The number of X-linked genes in biallelic-specific genes compared with that by chance was statistically evaluated by a hypergeometric test with phyper function in R.
Enrichment analysis of ESC- and iPSC-specific genes
The expression of ESC -and iPSC-specific genes in RTT-iPSCs was analyzed by GSEA v2.0.10 software at the Broad Institute. Relative expression of RTT-iPSCs to hESCs was rank-ordered by signal-to-noise ratios. The enrichment was evaluated with 1,000 permutations of phenotype and weighted enrichment statistic
Gene annotation analysis
GO and KEGG pathway analysis were performed by GOstats in Bioconductor package (http://www.bioconductor.org/packages/release/bioc/html/GOstats.html). The P-value of overrepresentation was adjusted by Benjamini and Hochberg (BH) multiple-test correction with a FDR cutoff of 0.05.
XCI-escaped gene analysis
Categories of X-linked genes (‘escape’, ‘heterozygous’ and ‘subject’) were obtained from the previous report (38). All primers were mapped to hg19 Refseq cDNA sequences with exact match. Genes, for which both forward and reverse primers are assembled to unique RefSeq X-linked gene, were retained. Finally, 44 escape, 50 heterogeneous and 257 subject genes were obtained by the above criterion. Significant differences in gene expression values among all RTT-iPSCs were evaluated by the Wilcoxon rank-sum test. Then, the q-value was calculated by the BH method using p.adjust function in R.
SUPPLEMENTARY MATERIAL
FUNDING
I.-H. P. was partly supported by NIH (GM0099130-01), CSCRF (12-SCB-YALE-11), Charles Hood Foundation, KRIBB/KRCF research initiative program (NAP-09-3) and by CTSA Grant UL1 RR025750 from the National Center for Advancing Translational Science (NCATS), a component of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. X.P. and S.M.W. were supported by NIH (GM0099130-01).
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the Park laboratory members for helpful comments and discussion. Computation time was provided by Yale University Biomedical High Performance Computing Center.
Conflict of Interest statement: none declared.
REFERENCES
- 1.Gadalla K.K., Bailey M.E., Cobb S.R. MeCP2 and Rett syndrome: reversibility and potential avenues for therapy. Biochem. J. 2011;439:1–14. doi: 10.1042/BJ20110648. [DOI] [PubMed] [Google Scholar]
- 2.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 3.Chen W.G., Chang Q., Lin Y., Meissner A., West A.E., Griffith E.C., Jaenisch R., Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 4.Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 5.Chao H.T., Zoghbi H.Y., Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samaco R.C., Mandel-Brehm C., McGraw C.M., Shaw C.A., McGill B.E., Zoghbi H.Y. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat. Genet. 2012;44:206–211. doi: 10.1038/ng.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 8.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 9.Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasui D.H., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., Lasalle J.M. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl Acad. Sci. USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young J.I., Hong E.P., Castle J.C., Crespo-Barreto J., Bowman A.B., Rose M.F., Kang D., Richman R., Johnson J.M., Berget S., et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl Acad. Sci. USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samaco R.C., Hogart A., LaSalle J.M. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squillaro T., Alessio N., Cipollaro M., Renieri A., Giordano A., Galderisi U. Partial silencing of methyl cytosine protein binding 2 (MECP2) in mesenchymal stem cells induces senescence with an increase in damaged DNA. FASEB J. 2010;24:1593–1603. doi: 10.1096/fj.09-143057. [DOI] [PubMed] [Google Scholar]
- 14.Yu F., Zingler N., Schumann G., Strätling W.H. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muotri A.R., Marchetto M.C., Coufal N.G., Oefner R., Yeo G., Nakashima K., Gage F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 17.Silva S.S., Rowntree R.K., Mekhoubad S., Lee J.T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K.Y., Hysolli E., Park I.H. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc. Natl Acad. Sci. USA. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomoda K., Takahashi K., Leung K., Okada A., Narita M., Yamada N.A., Eilertson K.E., Tsang P., Baba S., White M.P., et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lengner C.J., Gimelbrant A.A., Erwin J.A., Cheng A.W., Guenther M.G., Welstead G.G., Alagappan R., Frampton G.M., Xu P., Muffat J., et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Saha K., Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung Y.W., Hysolli E., Kim K.Y., Tanaka Y., Park I.H. Human induced pluripotent stem cells and neurodegenerative disease: prospects for novel therapies. Curr. Opin. Neurol. 2012;25:125–130. doi: 10.1097/WCO.0b013e3283518226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung A.Y., Horvath L.M., Grafodatskaya D., Pasceri P., Weksberg R., Hotta A., Carrel L., Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum. Mol. Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomp O., Dreesen O., Leong D.F., Meller-Pomp O., Tan T.T., Zhou F., Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maezawa I., Swanberg S., Harvey D., LaSalle J.M., Jin L.W. Rett syndrome astrocytes are abnormal and spread MeCP2 deficiency through gap junctions. J. Neurosci. 2009;29:5051–5061. doi: 10.1523/JNEUROSCI.0324-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derecki N.C., Cronk J.C., Lu Z., Xu E., Abbott S.B., Guyenet P.G., Kipnis J. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 33.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S., et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriaucionis S., Paterson A., Curtis J., Guy J., Macleod N., Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol. Cell Biol. 2006;26:5033–5042. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng V., Matagne V., Banine F., Frerking M., Ohliger P., Budden S., Pevsner J., Dissen G.A., Sherman L.S., Ojeda S.R. FXYD1 Is an MeCP2 target gene overexpressed in the brains of rett syndrome patients and Mecp2-null mice. Hum. Mol. Genet. 2007;16:640–650. doi: 10.1093/hmg/ddm007. [DOI] [PubMed] [Google Scholar]
- 36.Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang T., Wu H., Li Y., Szulwach K.E., Lin L., Li X., Chen I.P., Goldlust I.S., Chamberlain S.J., Dodd A., et al. Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat. Cell Biol. 2013;15:700–711. doi: 10.1038/ncb2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 39.Neul J.L., Fang P., Barrish J., Lane J., Caeg E.B., Smith E.O., Zoghbi H., Percy A., Glaze D.G. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellén M., Ayata P., Dewell S., Kriaucionis S., Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szulwach K.E., Li X., Li Y., Song C.X., Han J.W., Kim S., Namburi S., Hermetz K., Kim J.J., Rudd M.K., et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I., et al. 5-hmC-mediated Epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z., Hong E.J., Cohen S., Zhao W.N., Ho H.Y., Schmidt L., Chen W.G., Lin Y., Savner E., Griffith E.C., et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cukier H.N., Perez A.M., Collins A.L., Zhou Z., Zoghbi H.Y., Botas J. Genetic modifiers of MeCP2 function in Drosophila. PLoS Genet. 2008;4:e1000179. doi: 10.1371/journal.pgen.1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao J., Hu K., Chang Q., Wu H., Sherman N.E., Martinowich K., Klose R.J., Schanen C., Jaenisch R., Wang W., et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl Acad. Sci. USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Exner N., Lutz A.K., Haass C., Winklhofer K.F. Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31:3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quintanilla R.A., Johnson G.V. Role of mitochondrial dysfunction in the pathogenesis of Huntington's disease. Brain Res. Bull. 2009;80:242–247. doi: 10.1016/j.brainresbull.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park I.H., Lerou P.H., Zhao R., Huo H., Daley G.Q. Generation of human-induced pluripotent stem cells. Nat. Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 49.Tchieu J., Kuoy E., Chin M.H., Trinh H., Patterson M., Sherman S.P., Aimiuwu O., Lindgren A., Hakimian S., Zack J.A., et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C., Pachter L., Salzberg S.L. Tophat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.