Abstract
Fragile X-associated tremor/ataxia syndrome (FXTAS) is a late-onset neurodegenerative disorder in which patients carry premutation alleles of 55–200 CGG repeats in the FMR1 gene. To date, whether alterations in epigenetic regulation modulate FXTAS has gone unexplored. 5-Hydroxymethylcytosine (5hmC) converted from 5-methylcytosine (5mC) by the ten-eleven translocation (TET) family of proteins has been found recently to play key roles in neuronal functions. Here, we undertook genome-wide profiling of cerebellar 5hmC in a FXTAS mouse model (rCGG mice) and found that rCGG mice at 16 weeks showed overall reduced 5hmC levels genome-wide compared with age-matched wild-type littermates. However, we also observed gain-of-5hmC regions in repetitive elements, as well as in cerebellum-specific enhancers, but not in general enhancers. Genomic annotation and motif prediction of wild-type- and rCGG-specific differential 5-hydroxymethylated regions (DhMRs) revealed their high correlation with genes and transcription factors that are important in neuronal developmental and functional pathways. DhMR-associated genes partially overlapped with genes that were differentially associated with ribosomes in CGG mice identified by bacTRAP ribosomal profiling. Taken together, our data strongly indicate a functional role for 5hmC-mediated epigenetic modulation in the etiology of FXTAS, possibly through the regulation of transcription.
INTRODUCTION
The fragile X mental retardation-1 (FMR1) gene is located on the X chromosome and is highly conserved among species (1). The FMR1 gene product, fragile X mental retardation protein (FMRP), is a well-characterized RNA-binding protein essential for normal cognitive development (2,3). There is a CGG trinucleotide repeat in the 5′ untranslated region (UTR) that ranges from 5 to 54 triplets in the general population (4,5). Abnormal expansion of CGG to over 200 repeats (the full mutation) can be hypermethylated, resulting in silencing of FMR1 transcription, resulting in fragile X syndrome (FXS), with symptoms including severe learning deficits or intellectual disability (6–9). Recently, premutation (PM) expansion alleles (55–200 CGG repeats) have been linked to an inherited neurodegenerative disorder termed fragile X-associated tremor/ataxia syndrome (FXTAS) (10). Individuals bearing the PM usually do not exhibit obvious signs in their early life, but they are at greater risk of developing severe symptoms, such as tremor and ataxia, as well as progressive cognitive decline from memory deficits to dementia when they reach the fifth decades of life (5,11). Brain autopsy of symptomatic PM carriers indicates degeneration of the cerebellum, including Purkinje cell death, Bergman gliosis and significant cerebral and cerebellar white matter disease (12).
Although FXS and FXTAS both involve expanded CGG repeats at the 5′UTR of the FMR1, the molecular mechanisms of pathogenesis are distinct (5). In contrast to the complete transcriptional silencing of FMR1 in FXS, FMR1 mRNA levels are increased from alleles carrying the PM expansion (13), leading to the proposal of an RNA toxic gain of function as the primary disease mechanism (10,14). Subsequent studies have revealed that CGG repeats can bind and sequester a number of RNA-binding proteins, including Purα, hnRNP A2B1 and Sam68 in both Drosophila (fly) and mammalian cells (15–18). Several mouse models have been established to study FXTAS, with their phenotypes partially overlapping (19–21). Hashem et al. (21) demonstrated that CGG repeats expressed specifically in Purkinje neurons, outside the context of the FMR1 gene in mice, are sufficient to induce FXTAS phenotypes, supporting an RNA gain-of-function model first described in Drosophila experiments. Although impressive progress has been made, the detailed molecular mechanisms underlying FXTAS require further study. Indeed, one important matter has gone unexplored: how alterations in epigenetic regulation, including cytosine modifications, might impact the transcription state in PM carriers.
Cytosine covalent modifications influence transcriptional states and ultimately cellular identity. DNA methylation (5mC), the best-characterized cytosine modification, usually occurs in genomic contexts where there is a high frequency of GC dinucleotides, termed CpG islands (CGI), and is often associated with a gene repressive environment (22–24). Another cytosine modification, 5-hydroxymethylcytosine (5hmC), has recently been proposed to play a significant biological role in vertebrates (25–28). Ten-eleven translocation 1 (TET1), a 2-oxoglutarate (2OG)- and Fe (II)-dependent enzyme, catalyzes conversion of 5mC to 5hmC (29). Although 5hmC was initially thought to be only an intermediate of active DNA demethylation products (30–33), it was since found to be very stable in mammalian genome and may play specific roles in regulating gene expression (34–38). Remarkably, the overall 5hmC level varies between tissues, with approximately 10 times more found in brain tissues like Purkinje neurons (34–38) than in other tissues (39–42). This differential distribution points to the possible functional importance of 5hmC in normal neuronal development and activity, and aberrant 5hmC patterns may lead to severe neurological and neurodegenerative disease. In fact, our earlier study showed that 5hmC levels are significantly higher in both the cerebellum and hippocampus of adult mice at 6 weeks and 1 year of age than in postnatal day 7 mice, suggesting an age-dependent acquisition of 5hmC and a role for it in the regulation of brain-specific gene expression. Importantly, our data from a Rett syndrome mouse model with methyl-CpG binding protein 2 (MeCP2) knockout revealed increased 5hmC levels, strongly supporting the importance of 5hmC homeostasis in normal neuronal activities (35). A very recent report has also shown a global loss of 5hmC in Huntington's disease (HD) mouse brain, linking 5hmC to neurodegenerative disease (43).
Because CGG expression in Purkinje neurons can cause the FXTAS phenotypes and the 5hmC level is high in Purkinje cells, we chose to investigate the role of 5hmC in an rCGG mouse model. Interestingly, recent advances suggest that PM carriers can develop ‘pre-symptoms’ before the onset of FXTAS, indicating the impact of the PM on global cellular processes at an earlier stage (44,45). Thus, we applied genome-wide 5hmC profiling in 16-week-old rCGG transgenic mice cerebella side-by-side with their wild-type age-matched littermates, noting that 16-week-old rCGG mice do not display noticeable FXTAS phenotypes at that age (21). We observed global reductions of 5hmC level in rCGG mice, mainly in gene bodies, and regions previously annotated as CGI. However, there were also gain-of-5hmC loci found in several repetitive elements, as well as cerebellum-specific enhancers. Genomic annotation and motif prediction using wild-type- and rCGG-specific differential 5-hydroxymethylated regions (DhMRs) reveals that they are highly correlated with genes and transcription factors that were important in neuronal development as indicated by functional pathway analysis. DhMR-associated genes partially overlapped with genes that were differentially associated with ribosomes in CGG mice. Our data offer important insights into the potential role of 5hmC in the early stages of FXTAS onset, and may point the way to possible therapeutic approaches in the future.
RESULTS
Global reduction of 5hmC in the cerebellum of a FXTAS mouse model
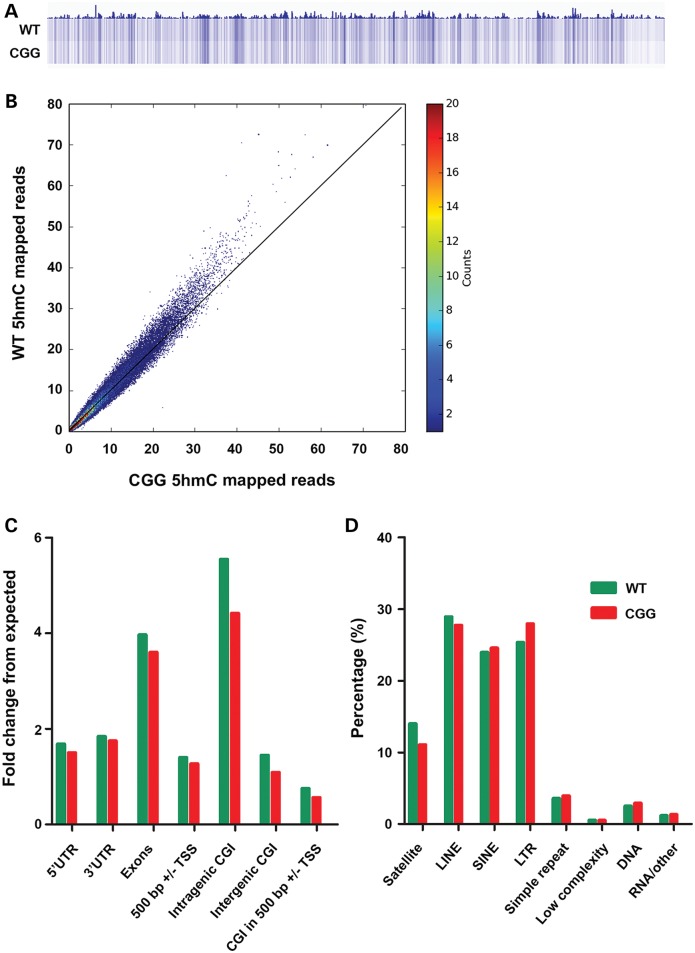
To determine the genome-wide 5hmC distribution in both FXTAS (rCGG) mice and their wild-type littermates, we employed a previously established chemical labeling and affinity purification method, coupled with high-throughput sequencing technology (36). Three rCGG mice at the age of 16 weeks were sacrificed as independent biological replicates, along with three age-matched littermates serving as controls. 5hmC-containing DNA sequences were enriched from cerebellar total DNA, and high-throughput sequencing resulted in a range of approximately 4.5 to 13.6 million non-duplicated reads from each biological replicate (Supplementary Material, Table S1). Chromosome-wide 5hmC read density mapping showed no visible differences between rCGG mice and wild-type controls (Supplementary Material, Fig. S1). 5hmC levels were depleted on the X chromosome, consistent with our previous observations (35). However, rCGG mice cerebella displayed distinguishable lower global 5hmC levels than wild-type controls, as shown in the global 5hmC heat map view (Fig. 1A). Genome-wide patterns of 5hmC were also evaluated by counting 5hmC mapped reads in each 10 kb bin from wild-type and rCGG mice genome and normalized to the total sequencing coverage. It is clearly shown that the majority of rCGG bins contained less 5hmC reads than wild-type, as the entire plot pattern moved toward the wild-type section (Fig. 1B). Figure 1C showed the overlapping features of normalized densities of 5hmC reads (per kilobase per million) in wild-type and rCGG mice with known genomic features on annotated gene bodies and defined CGI obtained from the UCSC Tables for NCBI37v1/mm9. Irrespective of phenotypic condition, 5hmC mapped reads were enriched mostly on intragenic CGI, with a 5.5-fold enrichment compared with its percent composition of the genome. In contrast, 5hmC reads were depleted (i.e. a fold change less than one) on CGIs within 500 bp of transcriptional start sites (TSS; Fig. 1C). Nonetheless, Figure 1C demonstrated an overall reduction of 5hmC levels in rCGG mice on these defined genomic features compared with wild-type controls.
Figure 1.
Global reduction of 5hmC in the cerebellum of a FXTAS mouse model. (A) Overall 5hmC levels were reduced in rCGG mice compared with wild-type. Global genomic heat maps were visualized by using the Integrated Genomics Viewer tools and browser. 5hmC reads were normalized to their coverage. (B) Genome-wide 5hmC reads density is reduced in the rCGG mouse model. Genome-wide 5hmC reads were counted within each 10 kb bin in wild-type and rCGG mice genome. 5hmC levels were higher in majority wild-type bin than rCGG. (C) Normalized 5hmC densities overlapping with various genomic features. Genomic coordinates were obtained from UCSC Tables for NCBI37v1/mm9 including 5′ UTR, exon, 3′ UTR, ±500 bp of TSS and CGI (±500 bp of CGI, Intergenic/Intragenic based on RefSeq Whole Gene). (D) Normalized 5hmC densities overlapping with repetitive elements. Aligned reads were assigned to repeat class annotations defined by RepeatMasker. 5hmC reads were counted in each repetitive class, and then divided by total aligned reads by Bowtie to achieve their percentage. WT, wild-type mice; CGG, rCGG transgenic mice. Pearson's Chi-squared test with Yates’ continuity correction were performed in (C) and (D) using their absolute mapped reads inside and outside of each genomic feature. P-values for these tests were significant (<2.2e−16).
DNA hypermethylation on repetitive elements is believed to play a critical role in maintaining genomic stability (46). To investigate the genome-wide distribution of 5hmC on repetitive elements, we aligned the total 5hmC reads to the RepeatMasker tracks of NCBI37v1/mm9. Although overall 5hmC levels were reduced in rCGG mice, notably several repetitive classes, including short interspersed nuclear elements (SINEs), long terminal repeats (LTRs) and simple repeats, showed specific acquisition of 5hmC in rCGG mice (Fig. 1D). This could imply that the specific gain of 5hmC on repetitive elements might correlate with the possible loss of 5mC in rCGG mice, thereby causing genomic instability and contributing to onset of FXTAS.
Differential 5hmC distribution in gene bodies, defined CGI, and enhancers in FXTAS mice
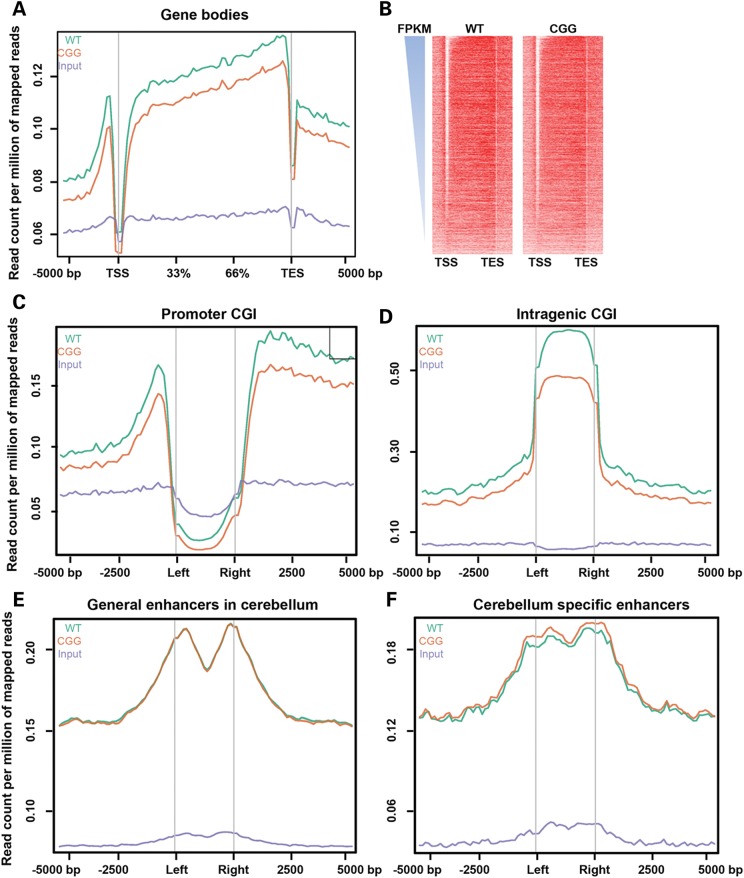
Since the overall 5hmC in rCGG mice was significantly reduced, we sought to visualize its distribution on defined genomic features. Figure 2A showed the 5hmC reads from wild-type and rCGG mice, and non-enriched DNA input mapped to 5 kb up- and downstream of gene bodies, TSS and transcription end sites (TES) by ngsplot (48), respectively. The mean and standard errors of 5hmC reads in each bin were obtained for group t-test analysis, and the average P-value was 0.0001. This indicates that the intragenic 5hmC is significantly different in wild-type and rCGG mice. The 5hmC levels in both wild-type and rCGG mice were largely depleted on TSS in cerebella, consistent with previous results (34,35). The distributions of 5hmC on TES showed the same depletion pattern as on TSS, but to a lesser extent. The wild-type intragenic 5hmC level was significantly higher in rCGG mice, in contrast to the mild differences on TSS or TES. This was particularly interesting since our previous data suggested that the intragenic 5hmC peaks are correlated with brain-specific gene expression (35). To further explore the correlation of 5hmC to gene expression, the aforementioned genes in wild-type and rCGG mice cerebella were ranked in descending order based on previously published wild-type mouse cerebellum RNA-seq data (see Materials and Methods). Interestingly, the strong 5hmC depletion, on TSS, and enrichment on the gene bodies were associated with genes with a higher expression profile in cerebellum. Thus, it is reasonable to speculate that the altered 5hmC on TSS, gene body and TES in rCGG mice may contribute to early onset of FXTAS by affecting gene with higher expression in the cerebellum.
Figure 2.
Differential 5hmC distribution in annotated RefSeq genes, defined CGI, and enhancers. (A) Normalized 5hmC read densities across TSS, TES and RefSeq gene bodies. Gene bodies were normalized to 0–100% as relative positions. 5 kb upstream and downstream of given genomic features were plotted. The read densities were normalized in millions. Group t-tests were performed using the mean and standard error of total 5hmC reads per bin from both wild-type and rCGG mice, and the average P-value is 0.0001033779. (B) Gene expression level-dependent 5hmC distributions. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) obtained from reference (47) were ranked in descending order. Both wild-type and rCGG 5hmC normalized read densities were measured in the regions shown in (A). Heat maps were visualized by Java Treeview. (C and D) Normalized 5hmC read densities on promoter or intragenic CGI. (E and F) Normalized 5hmC read densities on general enhancers in cerebellum or cerebellum-specific enhancers. Enhancer coordinates were obtained from reference (47).
Because DNA methylation occurs largely on CG dinucleotide regions termed CGI, we proceeded to examine the 5hmC distribution on defined CGI. As expected, the 5hmC levels in both wild-type and CGG mice were depleted on promoter CGI, but enriched on intragenic CGI (Fig. 2C and D), which correlates well with the observation in Figure 2A. However, a closer look at the 5hmC on promoters with differential CG abundance reveals that the depleted 5hmC was largely associated with high, but not intermediate or low, CpG promoters (HCP, ICP and LCP) as defined previously (49) (Supplementary Material, Fig. S1C and D). In addition, the rCGG mice overall 5hmC level was lower in all three promoter categories compared with wild-type controls (Supplementary Material, Fig. S1E). It had been suggested previously that 5hmC is located in cis-regulatory elements such as enhancers (41,50). Therefore, we plotted separately the wild-type and rCGG 5hmC in general enhancers and cerebellum-specific enhancers (47). Remarkably, the 5hmC level in rCGG mice was noticeably higher on cerebellum-specific enhancers than wild-type, but remained indistinguishable on general enhancers in the cerebellum (Fig. 2E and F), indicating explicit roles of 5hmC in the regulation of gene expression in the cerebellum, and loss of 5hmC in rCGG mice cerebellum-specific enhancer may cause selective mis-regulation of cerebellum-specific genes. Although the overall 5hmC level was lower in rCGG mice, gain of 5hmC on cerebellum-specific enhancers in rCGG mice could be a unique feature to account for their molecular abnormalities at early ages.
Identification and characterization of differentially hydroxymethylated regions (DhMRs) in a FXTAS mouse model
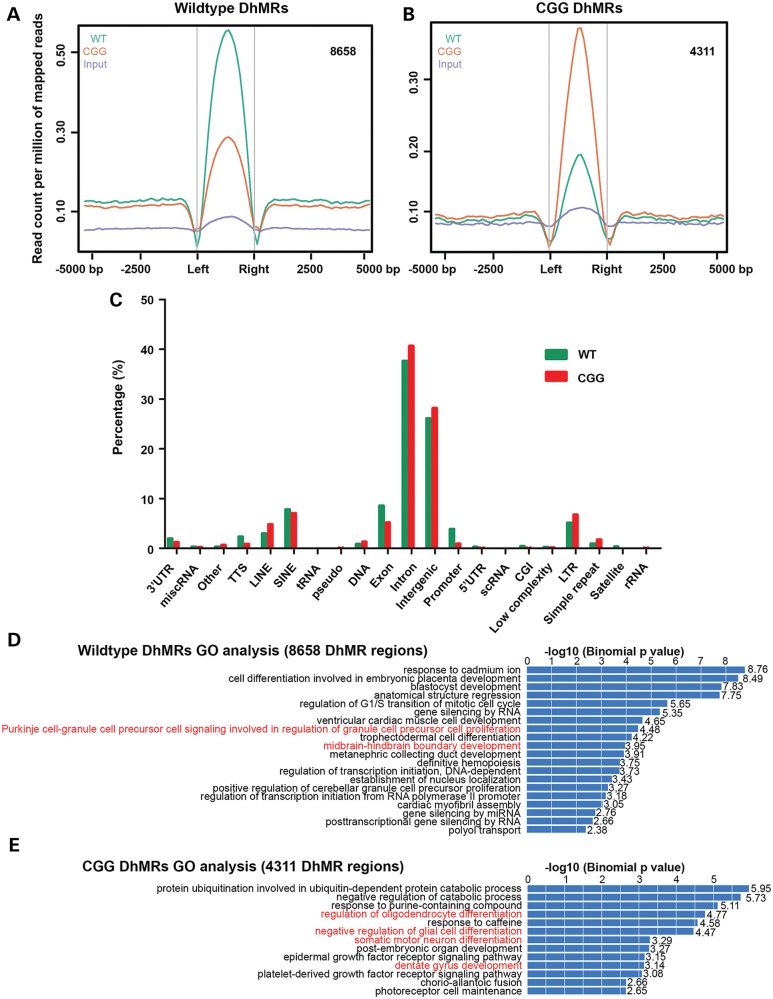
Since our present data suggested distinct 5hmC distribution patterns in wild-type and rCGG mice, we proceeded to identify and characterize the wild-type- and rCGG-specific DhMRs genome-wide, aiming to pinpoint specific loci that displayed altered 5hmC profiles between wild-type and rCGG mice (35,43). A total of 8658 wild-type-specific DhMRs and 4311 rCGG-specific DhMRs were identified (Fig. 3A, B and Supplementary Material, Tables S2 and S3). The lower number of rCGG-specific DhMRs was consistent with the global reduction of 5hmC in rCGG mice presented in Figure 1B. To investigate the defined genomic features associated with identified DhMRs, we annotated both wild-type- and rCGG-specific DhMRs of the mouse genome using HOMER (Hypergeometric Optimization of Motif EnRichment) software (51). HOMER annotations revealed that more wild-type-specific DhMRs were located on promoters and exons, whereas more rCGG-specific DhMRs were found on introns, repetitive elements and intergenic regions (Fig. 3C). To further explore the biological significance of these identified DhMRs, we used GREAT (Genomic Regions Enrichment of Annotations Tool) to perform gene ontology (GO) analyses for the wild-type- and rCGG-specific DhMRs (52). Remarkably, several GO biological processes associated with neuronal function and development in brain were illustrated (Fig. 3D and E). These included cerebellar Purkinje cell signaling in wild-type-specific DhMRs, as well as regulation of oligodendrocyte, negative regulation of glial cell and somatic motor neuron differentiation in rCGG-specific DhMRs (highlighted in red). In addition, mouse phenotype prediction also recapitulated several phenotypes in rCGG mice in a later disease stage, including the absence of Purkinje cell layers (Supplementary Material, Fig. S2).
Figure 3.
Identification and characterization of differentially hydroxymethylated regions (DhMRs) in a FXTAS mouse model. (A and B) Identification of wild-type- or rCGG-specific DhMRs. 8658 and 4311 wild-type- or rCGG-specific DhMRs were identified by directly comparing one to the other. The effective genome size = 1.87 × 109, tag size = 38, bandwidth = 200, P-value cutoff = 1.00 × 10−5. (C) Annotation of DhMRs to various genomic features. Identified wild-type and rCGG-specific DhMRs were annotated to various genomic regions and associated genes by HOMER software for a direct comparison (51). (D and E) GO analysis for defined DhMRs. Identified DhMRs were examined for their biological significance using the GREAT tool. GO biological processes associated with wild-type or rCGG DhMRs were shown.
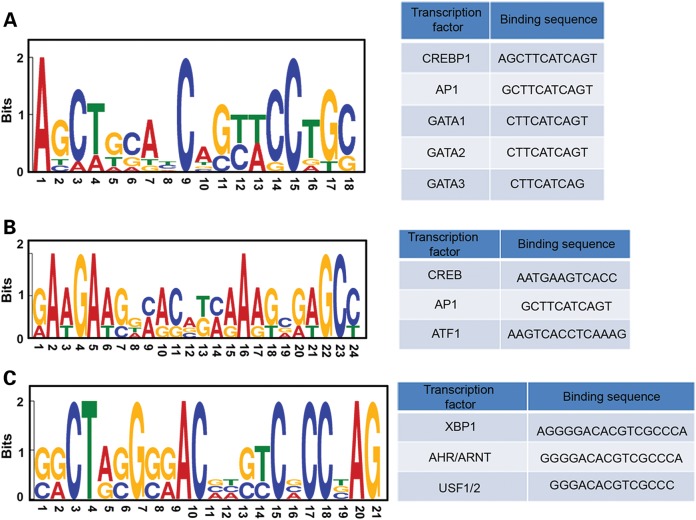
We thought it would be interesting to examine the common DNA sequence motifs associated with identified DhMRs, and eventually to predict altered DNA-binding protein occupancy, such as important transcriptional factors. To this end, we used the Multiple EM for Motif Elicitation (MEME) suite to predict the common motifs in wild-type- and rCGG-specific DhMRs (53). One common motif for wild-type-specific DhMRs and two major motifs for rCGG-specific DhMRs were predicted with relative certainty (Fig. 4A and B, left panel). The identified motifs were further analyzed by TFBIND, a software tool for searching transcription factor binding sites on given DNA motifs (54). Several putative transcriptional factors associated with either wild-type- or rCGG-specific DhMRs were identified (Fig. 4A and B, right panel). Many of these identified transcriptional factors, such as cyclic AMP response element binding protein 1 (CREBP1 or CREB2) (55,56), activator protein 1 (AP-1) (57,58), cAMP response element-binding protein (CREB) (59,60) and Aryl hydrocarbon receptor (AHR) (61,62), have been strongly linked to neurogenesis and neurological activities (see Discussion). These findings indicate that 5hmC-mediated epigenetic modulation plays a potential role in the onset of FXTAS.
Figure 4.
Identification of DhMR-associated common motifs and their putative binding factor. The MEME suite was used to predict the common motifs in the top 20% of wild-type (A) and rCGG-specific (B and C) DhMRs based on the P-value cutoff (53). Motifs with an E-value <10−3 were considered to be significant. Putative binding factors were predicted by TFBIND and listed in tables.
DhMR-associated genes partially overlap with altered ribosome-bound mRNAs in a FXTAS mouse model
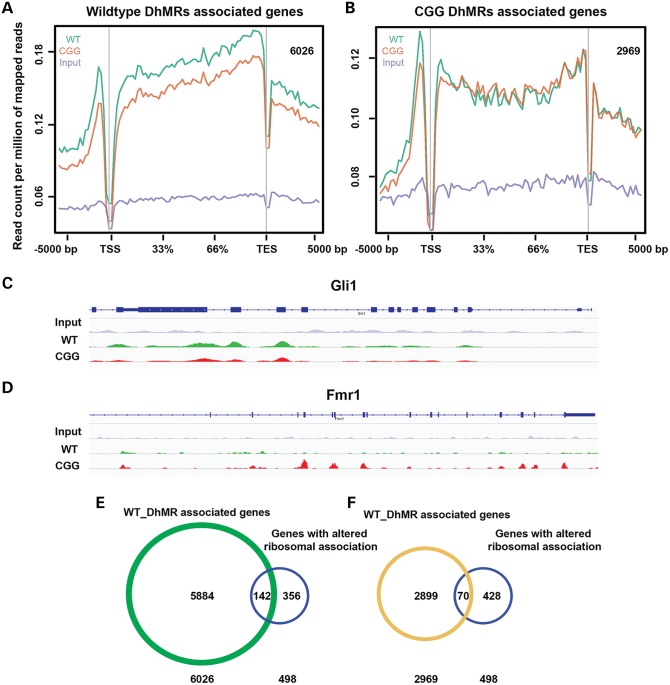
We investigated detailed lists of genes associated with identified DhMRs based on HOMER annotation results. Genes with DhMRs located on the UTRs, exons, introns and promoters were reported; genes with their closest TSSs to the intergenic DhMRs were also included. A total of 6026 and 2969 genes were found to be associated with wild-type- or rCGG-specific DhMRs, respectively (Supplementary Material, Tables S4 and S5). Plotting of 5hmC mapped reads on the two identified gene lists confirmed that the differential 5hmC distribution was primarily on the gene body (Fig. 5A and B). Many genes identified in these lists are known to play critical roles in neurological development and functions, among them Gli1 (63) and Fmr1 (5) (Fig. 5C and D, see Discussion). Taken together, we have identified lists of genes associated with differential 5hmC levels between rCGG and wild-type mice, suggesting their potential role in early FXTAS pathogenesis.
Figure 5.
DhMR-associated genes partially overlap with altered ribosome-bound mRNAs in a FXTAS mouse model. (A and B) DhMR-associated genes showed differential 5hmC distributions on gene bodies. 6026 wild-type- and 2969 rCGG-specific DhMR-associated genes were identified by HOMER annotation. Genes associated with wild-type DhMRs showed higher wild-type 5hmC plotting on their gene bodies, whereas genes associated with rCGG DhMRs showed higher rCGG 5hmC plotting. (C and D) Differential 5hmC distributions on gene bodies were visualized by IGV browser. Gli1 showed higher 5hmC in wild-type, while Fmr1 showed the opposite trend. (E and F) DhMR-associated genes partially overlapped with genes identified by TRAP. 142 or 70 genes overlapped with wild-type- or rCGG DhMR-associated genes, respectively. The P-values from Pearson's Chi-squared test with Yates’ continuity correction in (E) and (F) were 3.784e − 05 and 0.007686, respectively.
Beyond this, we also applied translating ribosome affinity purification (TRAP) to investigate the early translational changes in rCGG mice compared with wild-type littermates when they are 4 weeks of age (Galloway et al., co-submitted). A total of 498 genes were found to have altered ribosomal profiling on their mRNAs in rCGG mice. We compared our DhMR-associated gene lists to this ribosomal profiling. Interestingly, 142 out of 498 genes identified in TRAP experiments overlapped with wild-type DhMR-associated genes, and 70 out of 498 genes overlapped with rCGG DhMR-associated genes (Fig. 5E and F). Pearson's Chi-squared test with Yates’ continuity correction indicated these correlations were significant (P-values were 3.784e−05 and 0.007686, respectively). These data indicated that those genes that underwent translational changes could also be manipulated on a transcriptional level, possibly through the alteration of 5hmC levels.
DISCUSSION
In the present study, we have systematically characterized the cerebellar 5hmC profiles in Fmr1 transgenic mice bearing PM lengths of CGG repeats on their 5′UTR compared with wild-type littermates. We observed global reduction of 5hmC in our rCGG FXTAS mouse model, primarily on annotated gene bodies, including exons, introns, UTRs and promoters. However, we have also detected a gain of 5hmC on several repetitive classes, as well as cerebellum-specific enhancers. Aberrant distribution of DNA methylation has long been linked to various neurological diseases. For example, Wu et al. (64) found that DNA methyltransferase 3a (Dnmt3a) deletion leads to impaired postnatal neurogenesis. Mechanistically, Dnmt3a methylates intergenic regions and gene bodies flanking proximal promoters, with its knockout inducing the silencing of genes related to neurogenic activity. In FXS, the expanded CGG repeats can become hypermethylated and silence Fmr1 transcription (5). One recent report investigated the dynamic change of TET proteins and 5hmC in neurogenesis and found an increased 5hmC level during neuronal differentiation from neural stem cells (65). During the preparation of this article, Wang et al. (43) published data demonstrating the global loss of 5hmC in HD. Thus, it appears a genome-wide loss of 5hmC might be a signature for a spectrum of neurodegenerative diseases. Although global 5hmC loss has been seen in HD, there were no significant differences on RefSeq genes or repetitive elements (43). Our data reveal a unique FXTAS-specific 5hmC feature that distinguishes it from other neurodegenerative diseases. Substantial 5hmC enrichment in gene body CGI and depletion in TSS CGI in normal mouse cerebellum has been described in our earlier report (41). The present data confirm this trend in both wild-type and rCGG mice cerebellum. The 5hmC promoter depletion and gene body enrichment are associated with gene expression profiles and promoter CGI density, as shown in Figure 2 and Supplementary Material, Figure S1, implying a strong relationship between 5hmC and transcription state. Furthermore, we have also observed gain of 5hmC in specific genomic regions, such as cerebellum-specific enhancers, but not regular enhancers. 5hmC is known to occupy enhancer regions in both human and mouse embryonic stem cells (41,66,67). The 5hmC enrichment on cerebellum-specific enhancers indicates its unique roles in regulating cerebellum-specific gene expression, and a possible role in influencing key cerebellar gene aberrant expression in rCGG mice. In the meantime, our data indicate a gain of 5hmC on several repetitive classes in rCGG mice, including on SINEs, LTRs and simple repeats. DNA methylation on the repetitive elements was well known to maintain genomic stability (46); therefore, the increased 5hmC may be due to conversion from 5mC and make the rCGG genome vulnerable.
One key question remains to be resolved: the cause of the genome-wide 5hmC alterations from introducing extra copies of CGG repeats on the 5′UTR of Fmr1 transgene. Previous data from us and others pointed to a RNA gain-of-function model, based on the observations that a number of RNA-binding proteins are immobilized and sequestered by CGG repeats (5,68). Thus, it is reasonable to speculate that the target specificity or enzymatic activity of TET proteins or their co-factors could be ectopically affected in rCGG mice transcriptionally or post-transcriptionally. Proteomic analysis has revealed several TET-binding proteins, among them Ogt (69–71), Nanog (72), PARP1 (73) and SIN3A (74). Whether those proteins can be directly or indirectly sequestered by CGG repeats is not clear. Another possible cause of the loss of 5hmC in rCGG mice may be the altered availability of 5hmC or 5mC binding proteins. Overexpression of MeCP2 reduces global 5hmC levels (35), and MeCP2 was later shown to also bind to 5hmC (34). It is important to note that the maintenance of proper 5hmC homeostasis is critical for normal neuronal functions.
The identification of wild-type- and rCGG-specific DhMRs allow us to focus on unique genomic loci with altered 5hmC levels. Direct GO analysis using these loci reveal their strong correlation with various neuronal functions and brain development. Several transcriptional factors that recognize DhMR common motifs have been identified and found to play key roles in neurogenesis and neurological activities. For example, Lai et al. (55) showed CREBP1 localizes to distal dendrites of rodent hippocampal neurons and serves as cargo for importin alpha translocation from distal synaptic sites to the nucleus. CREBP1 is also known to be important for memory and plasticity (56). Moreover, AP-1 is required for axonal regeneration and is predicted to play a central role in neuronal gene regulatory networks (57,58). GATA1 is commonly known to play a role in erythroid development, with recent evidence also suggesting a role for it in neuronal activity. Kang et al. (75) showed that expression of GATA1 in prefrontal cortex neurons caused the loss of dendritic spines and dendrites, leading to depressive behavior in rat models of depression.
Other transcriptional factors, such as CREB, ATF, XBP1, AHR and USF1/2, are known to recognize rCGG-specific DhMR common motifs. Among those, CREB has been well characterized in neuronal plasticity and long-term memory formation (76), and changes in CREB function influenced specific populations of cells to achieve particular memory or learning activity (59,60). XBP1 is a key transcriptional factor for the unfolded protein response, and recent data suggested a function for it in normal brain development, as well as neurological diseases such as HD (77,78). AhR and Arnt proteins are present in mouse cerebellum from birth throughout postnatal development, and functional AhR can disrupt neural progenitor cell proliferation (61,62). Greenberg and colleagues (79) showed that upstream stimulatory factors (USF1/2) control gene expression in neurons, such as brain-derived neurotrophic factor, in a calcium-dependent manner. Overall, these transcriptional factors apparently participate in a wide range of neuronal activities and may lose or gain occupancy on these motifs when their binding sites are differentially hydroxymethylated, thus accounting for the FXTAS pathogenesis.
We have also generated detailed gene lists with DhMRs located on either the gene body or the nearest open reading frame. Many of the genes with differential 5hmC levels in rCGG mice are involved in critical neuronal functions. For example, the gene Gli1 shows reduced 5hmC on its gene body in rCGG mice compared with wild-type. This gene has been reported to play a critical role in modulating granule neuron precursor cell proliferation in the cerebellum by binding to hundreds of cerebellar gene promoters (63). In contrast, endogenous Fmr1 shows acquisition of 5hmC on the gene body in rCGG mice. Fmr1 mRNA is overexpressed in FXTAS patients and shows an increased ribosomal association in the TRAP experiments. The gain of 5hmC is likely to play a role in this process. The accurate correlation between 5hmC levels and gene expression needs to be studied further.
The TRAP methodology allows researchers to purify tagged ribosome, as well as its bound mRNA they are reading, for downstream high-throughput analysis (80). The Nelson laboratory has identified nearly 500 mRNA transcripts that showed altered ribosomal profiles in rCGG mice only 4 weeks old. Their data suggest an early abnormal translational profile, before any phenotypes developed. In line with their reported genes, we have found ∼30 and ∼14% overlap with wild-type- and rCGG-specific DhMR-associated genes. These data could imply the rCGG mice display abnormal gene expression on both transcription and translation stage, arguing that an ectopic condition has occurred, even before there is a noticeable phenotype. Recently, Vitamin C has been found to alter global 5hmC profile by controlling TET activity (81). Thus, 5hmC may make a good target for future drug development to treat PM carriers or even FXTAS patients.
In summary, our data presented here offer the first systematic 5hmC profile in a FXTAS mouse model, demonstrating a correlation of genome-wide 5hmC changes as a possible mechanism of FXTAS disease pathogenesis. Our research also sheds lights on possible future therapeutic interventions that can be used before the PM carriers develop FXTAS phenotypes.
MATERIALS AND METHODS
Transgenic animals
The FXTAS mouse model was described previously (21). Three 16-week-old rCGG transgene-positive mice were sacrificed at the Baylor College of Medicine, together with three age-matched wild-type littermates. Whole brains were dissected and flash-frozen in liquid nitrogen. Samples were transferred following the procedure of the Emory Incoming Material Transfer Agreement. Cerebella of each wild-type and rCGG mouse were dissected upon arrival and stored in −80° freezers. It has been previously reported that the rCGG mice start to show inclusions by 8 weeks of age and behavioral deficits at 20 weeks (21). We decided to investigate 5hmC distribution in the rCGG mice at 16 weeks of age without obvious phenotypes, to understand the impact of global 5hmC alteration before FXTAS onset.
Genomic DNA isolation and 5hmC-specific enrichment
Cerebella samples were homogenized on ice by 2 ml Dounce tissue grinder in lysis buffer [100 mm Tris–HCl (pH 8.5), 5 mm EDTA, 0.2% SDS (vol/vol), 200 mm NaCl]. Proteinase K was freshly added to the buffer, and samples were kept at 55°C overnight. An equal volume of phenol:chloroform:isoamyl alcohol [25:24:1, saturated with 10 mm Tris (pH 8.0) and 1 mm EDTA; P-3803, Sigma] was added at the second day, mixed completely and centrifuged at top speed on desktop centrifugation for 10 min. Supernatant was then mixed with an equal volume of isopropanol to precipitate DNA. DNA pellets were washed with 70% ethanol and eluted with 10 mm Tris–HCl (pH 8.0).
Chemical labeling-based 5hmC enrichment was described previously (36). Briefly, DNA was sonicated to 100–500 bp, and then mixed with 100 µl solution containing 50 mm HEPES buffer (pH 7.9), 25 mm MgCl2, 250 µm UDP-6-N3-Glu and 2.25 µm wild-type β-glucosyltransferase. Reactions were incubated for 1 h at 37°C. DNA substrates were purified via Qiagen DNA purification kit. 150 µm dibenzocyclooctyne modified biotin was then added to the purified DNA, and the labeling reaction was performed for 2 h at 37°C. The biotin-labeled DNA was enriched by Streptavidin-coupled Dynabeads (Dynabeads® MyOne™ Streptavidin T1, Life Technologies) and purified by Qiagen DNA purification kit.
Library preparation and high-throughput sequencing
5hmC-captured libraries were generated by the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina according to the manufacturer's protocol. In short, 25 ng of input genomic DNA or 5hmC-captured DNA were used. DNA fragments between 150 and 300 bp were gel-purified after the adapter ligation step. An Agilent 2100 BioAnalyzer was used to quantify the amplified DNA. 20-pM diluted libraries were eventually used for sequencing. We performed 38-cycle single-end sequencing using Version 4 Cluster Generation and Sequencing Kits (Part #15002739 and #15005236 respectively) and Version 7.0 recipes. Image processing and sequence extraction were done using the standard Illumina Pipeline.
Sequence alignment and mapped reads annotation
FASTQ sequence files from biological replicates were separately aligned to mouse NCBI37v1/mm9 references using Bowtie 0.12.9. Each unique mapped read (.bed files) with no more than two mismatches in the first 25 bp was concatenated to achieve combined wild-type and rCGG 5hmC sequence. Chromosome-wide densities were determined as reads per chromosome divided by the total number of reads in millions. Expected values were determined by dividing 106 by the total genome length and multiplying by chromosome length. Association of mapped reads with genomic features was performed by overlapping reads files with known genomic features obtained from UCSC Tables for NCBI37v1/mm9, including 5′ UTR, exon, 3′ UTR, ±500 bp of TSS, CGI (±500 bp of CGI, Intergenic/Intragenic based on RefSeq Whole Gene).
Repeat-associated 5hmC
Unique mapped reads were aligned to the RepeatMasker track of NCBI37v1/mm9 using Bowtie–q–best parameters, allowing no more than two mismatches across the entire 38 bp read. Aligned reads were assigned to repeat class annotations defined by RepeatMasker. 5hmC reads were counted in each repetitive class, and then divided by total aligned reads by Bowtie to achieve their percentage.
Mapped reads plotting to various genomic features and heat map generation
Unique 5hmC mapped reads were plotted to various genomic regions using an R program package termed ngsplot (48). Five kilobase upstream and downstream of given genomic features were divided into 100 bins; unique mapped reads were calculated in each bin and then normalized to reads per millions. The gene bodies were defined between TSS and TES. Relative gene positions were calculated due to the variable gene length. The various genomic regions were defined by ngsplot database or downloaded from mm9 table browser. Cerebellum RNA-seq data, cerebellum-specific enhancers and cerebellum general enhancers were downloaded from the ENCODE project (47). Mouse cerebellum RNA-seq data were also obtained from reference (47), and the 5hmC reads mapped to each gene were ranked in descending order based on their FPKM value (Fragments Per Kilobase of transcript per Million mapped reads) from the RNA-seq data. Heat maps were generated and visualized by ngsplot or java treeview.
DhMR identification, annotation and motif analysis
We used Model-based Analysis of ChIP-Seq (MACS) software (82) to identify DhMRs between wild-type and rCGG mice by directly comparing one to the other, rather than comparing to the input. The effective genome size = 1.87 × 109, tag size = 38, bandwidth = 200, P-value cutoff = 1.00 × 10−5. Identified wild-type- and rCGG-specific DhMRs were annotated to various genomic regions and associated genes by HOMER software (51). DhMRs were also analyzed by GREAT for GO analysis. The MEME suite was used to predict the common motifs in the top 20% of wild-type- and rCGG-specific DhMRs based on the P-value cutoff (53). Motifs with an E-value of <10−3 were considered to be significant. Identified motifs were input to the TFBIND, a software tool for searching transcription factor binding sites on given DNA motifs (54).
Read coverage and visualization
Genomic views of read coverage were generated using the Integrated Genomics Viewer tools and browser (IGV 1.4.2, http://www.broadinstitute.org/igv/), with a window size of 25 and a read extend of 200.
Statistics
Pearson's χ2 test with Yates’ continuity correction and group t-tests were performed by using R software (http://www.r-project.org/).
ACCESSION NUMBER
Sequencing data have been deposited to GEO with accession number GSE49463.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by the FRAXA Research Foundation and NIH grants NS051630 (P.J. and D.L.N.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank C. Strauss for critical reading of the article.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Warren S.T., Nelson D.L. Advances in molecular analysis of fragile X syndrome. JAMA. 1994;271:536–542. [PubMed] [Google Scholar]
- 2.Till S.M. The developmental roles of FMRP. Biochem. Soc. Trans. 2010;38:507–510. doi: 10.1042/BST0380507. [DOI] [PubMed] [Google Scholar]
- 3.Kwan K.Y., Lam M.M., Johnson M.B., Dube U., Shim S., Rasin M.R., Sousa A.M., Fertuzinhos S., Chen J.G., Arellano J.I., et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin P., Warren S.T. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet. 2000;9:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 5.Tan H., Li H., Jin P. RNA-mediated pathogenesis in fragile X-associated disorders. Neurosci. Lett. 2009;466:103–108. doi: 10.1016/j.neulet.2009.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S.T., Schlessinger D., Sutherland G.R., Richards R.I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 7.Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J.C., Warren S.T., Schlessinger D., et al. Fragile X genotype characterized by an unstable region of DNA. Science. 1991;252:1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- 8.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 9.Oberle I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boue J., Bertheas M.F., Mandel J.L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 10.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurol. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 11.Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W., et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- 12.Greco C.M., Berman R.F., Martin R.M., Tassone F., Schwartz P.H., Chang A., Trapp B.D., Iwahashi C., Brunberg J., Grigsby J., et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 13.Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Gen. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 15.Sellier C., Rau F., Liu Y., Tassone F., Hukema R.K., Gattoni R., Schneider A., Richard S., Willemsen R., Elliott D.J., et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin P., Duan R., Qurashi A., Qin Y., Tian D., Rosser T.C., Liu H., Feng Y., Warren S.T. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sofola O.A., Jin P., Qin Y., Duan R., Liu H., de Haro M., Nelson D.L., Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qurashi A., Li W., Zhou J.Y., Peng J., Jin P. Nuclear accumulation of stress response mRNAs contributes to the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2011;7:e1002102. doi: 10.1371/journal.pgen.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bontekoe C.J., Bakker C.E., Nieuwenhuizen I.M., van der Linde H., Lans H., de Lange D., Hirst M.C., Oostra B.A. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum. Mol. Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 20.Entezam A., Biacsi R., Orrison B., Saha T., Hoffman G.E., Grabczyk E., Nussbaum R.L., Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashem V., Galloway J.N., Mori M., Willemsen R., Oostra B.A., Paylor R., Nelson D.L. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum. Mol. Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 23.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 24.Gopalakrishnan S., Van Emburgh B.O., Robertson K.D. DNA methylation in development and human disease. Mutat. Res. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams K., Christensen J., Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan L., Shi Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Branco M.R., Ficz G., Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 28.Cimmino L., Abdel-Wahab O., Levine R.L., Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaffeneder T., Hackner B., Truss M., Munzel M., Muller M., Deiml C.A., Hagemeier C., Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Cehm. Int. Ed. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G.L., Luo C., Jiang H., He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellen M., Ayata P., Dewell S., Kriaucionis S., Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I., et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X., et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munzel M., Globisch D., Bruckl T., Wagner M., Welzmiller V., Michalakis S., Muller M., Biel M., Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Cehm. Int. Ed. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 39.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M., McLoughlin E.M., Brudno Y., Mahapatra S., Kapranov P., et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szulwach K.E., Li X., Li Y., Song C.X., Han J.W., Kim S., Namburi S., Hermetz K., Kim J.J., Rudd M.K., et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F., Yang Y., Lin X., Wang J.Q., Wu Y.S., Xie W., Wang D., Zhu S., Liao Y.Q., Sun Q., et al. Genome-wide loss of 5-hmC is a novel epigenetic feature of huntington's disease. Hum. Mol. Genet. 2013;22:3641–3653. doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- 44.Cornish K.M., Hocking D.R., Moss S.A., Kogan C.S. Selective executive markers of at-risk profiles associated with the fragile X premutation. Neurology. 2011;77:618–622. doi: 10.1212/WNL.0b013e3182299e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry-Kravis E., Hall D.A. Executive dysfunction in young FMR1 premutation carriers: forme fruste of FXTAS or new phenotype? Neurology. 2011;77:612–613. doi: 10.1212/WNL.0b013e3182299f98. [DOI] [PubMed] [Google Scholar]
- 46.Coufal N.G., Garcia-Perez J.L., Peng G.E., Yeo G.W., Mu Y., Lovci M.T., Morell M., O'Shea K.S., Moran J.V., Gage F.H. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L., Shao N., Liu X., Nestler E. Ngs.plot: an easy-to-use global visualization tool for next-generation sequencing data. 2013 https://code.google.com/p/ngsplot/ [Google Scholar]
- 49.Ku M., Koche R.P., Rheinbay E., Mendenhall E.M., Endoh M., Mikkelsen T.S., Presser A., Nusbaum C., Xie X., Chi A.S., et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 51.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsunoda T., Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 55.Lai K.O., Zhao Y., Ch'ng T.H., Martin K.C. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc. Natl Acad. Sci. USA. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu R.Y., Shah S., Cleary L.J., Byrne J.H. Serotonin- and training-induced dynamic regulation of CREB2 in Aplysia. Learn. Mem. 2011;18:245–249. doi: 10.1101/lm.2112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raivich G., Bohatschek M., Da Costa C., Iwata O., Galiano M., Hristova M., Nateri A.S., Makwana M., Riera-Sans L., Wolfer D.P., et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Miller G.M., Ogunnaike B.A., Schwaber J.S., Vadigepalli R. Robust dynamic balance of AP-1 transcription factors in a neuronal gene regulatory network. BMC Syst. Biol. 2010;4:171. doi: 10.1186/1752-0509-4-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y., Won J., Karlsson M.G., Zhou M., Rogerson T., Balaji J., Neve R., Poirazi P., Silva A.J. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat. Neurosci. 2009;12:1438–1443. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han J.H., Kushner S.A., Yiu A.P., Cole C.J., Matynia A., Brown R.A., Neve R.L., Guzowski J.F., Silva A.J., Josselyn S.A. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 61.Williamson M.A., Gasiewicz T.A., Opanashuk L.A. Aryl hydrocarbon receptor expression and activity in cerebellar granule neuroblasts: implications for development and dioxin neurotoxicity. Toxicol. Sci. 2005;83:340–348. doi: 10.1093/toxsci/kfi031. [DOI] [PubMed] [Google Scholar]
- 62.Latchney S.E., Lioy D.T., Henry E.C., Gasiewicz T.A., Strathmann F.G., Mayer-Proschel M., Opanashuk L.A. Neural precursor cell proliferation is disrupted through activation of the aryl hydrocarbon receptor by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Stem Cells Dev. 2011;20:313–326. doi: 10.1089/scd.2009.0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee E.Y., Ji H., Ouyang Z., Zhou B., Ma W., Vokes S.A., McMahon A.P., Wong W.H., Scott M.P. Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc. Natl Acad. Sci. USA. 2010;107:9736–9741. doi: 10.1073/pnas.1004602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hahn M.A., Qiu R., Wu X., Li A.X., Zhang H., Wang J., Jui J., Jin S.G., Jiang Y., Pfeifer G.P., et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H., D'Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stroud H., Feng S., Morey Kinney S., Pradhan S., Jacobsen S.E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kapeli K., Yeo G.W. Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: implications for neurological diseases. FNINS. 2012;6:144. doi: 10.3389/fnins.2012.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 70.Deplus R., Delatte B., Schwinn M.K., Defrance M., Mendez J., Murphy N., Dawson M.A., Volkmar M., Putmans P., Calonne E., et al. TET2 And TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Q., Chen Y., Bian C., Fujiki R., Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2012;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A., et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang H.J., Voleti B., Hajszan T., Rajkowska G., Stockmeier C.A., Licznerski P., Lepack A., Majik M.S., Jeong L.S., Banasr M., et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat. Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silva A.J., Kogan J.H., Frankland P.W., Kida S. CREB and memory. Annu. Rev. Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 77.Hayashi A., Kasahara T., Iwamoto K., Ishiwata M., Kametani M., Kakiuchi C., Furuichi T., Kato T. The role of brain-derived neurotrophic factor (BDNF)-induced XBP1 splicing during brain development. J. Biol. Chem. 2007;282:34525–34534. doi: 10.1074/jbc.M704300200. [DOI] [PubMed] [Google Scholar]
- 78.Vidal R.L., Figueroa A., Court F.A., Thielen P., Molina C., Wirth C., Caballero B., Kiffin R., Segura-Aguilar J., Cuervo A.M., et al. Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum. Mol. Genet. 2012;21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen W.G., West A.E., Tao X., Corfas G., Szentirmay M.N., Sawadogo M., Vinson C., Greenberg M.E. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J. Neurosci. 2003;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suarez-Farinas M., Schwarz C., Stephan D.A., Surmeier D.J., et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A., et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.