Abstract
Knowledge about maternal history is critical for guiding certain aspects of newborn clinical care as well as for research on neonatal issues. However, often the only maternal history available in the newborn record is in the clinical notes. We are using data from the MIMIC-II database for a clinical study on newborns admitted to the intensive care unit. Important maternal data were only available in the newborn notes, so we developed a simple algorithm to extract those data. We manually derived patterns for maternal age, gravida/para status, and laboratory results by reviewing a small set of notes. Using regular expressions and specific filters for notes and results, we extracted maternal data with recall of 0.91–0.99 and precision of 0.95–1.0 for the 289 infants in our study. Our methods could be used with other research datasets and with clinical documentation systems to extract maternal data into a more useful, structured format.
Introduction
Newborn infants are unique in that they have no true “past medical history” and that their initial newborn course is directly linked to their mothers’ general medical and pregnancy history. Certain aspects of the prenatal history are especially relevant to the immediate newborn period, including maternal blood type and antibody status, vaginal Group B Strep culture, and Hepatitis B status, all of which directly affect the newborn’s medical management during the first 24 to 48 hours of life. For example, if a mother’s Hepatitis B surface antigen result is positive or unknown at the time of delivery, her newborn should receive hepatitis B immune globulin within 12 hours of birth (in addition to the hepatitis B vaccine) to reduce the risk of perinatal hepatitis B transmission.1
One challenge for newborn medical records is transferring maternal data to the newborn’s chart. In the days of paper charts pediatric providers would transcribe the relevant data from the mother’s record to the newborn’s chart. In theory, with electronic health records (EHRs) maternal data should flow into the newborn’s record. However, many barriers to that process exist, including prohibitions on transferring maternal data electronically to her newborn’s chart,2 lack of structured fields to record maternal history in the newborn’s chart, and lack of data connections between birth hospitals and stand-alone children’s hospitals. As recently as June 2012, the National Institute of Standards and Technology recommended that maternal EHR data be linked to the newborn record,3 which clearly suggests that this linkage is still uncommon. Instead, maternal data are still transcribed by the pediatric provider, usually into a free-text portion of the neonatal admission note, for both healthy newborns as well as those in the neonatal intensive care unit (NICU).
Extraction of data from clinical notes remains an active and practical issue for clinical natural language processing (NLP). Clinical NLP presents unique problems in addition to the common NLP challenges.4 For example, hyphens, slashes and plus signs are often ignored in processing English text in other domains, but these tokens play important roles in clinical notes and often are ambiguous. Other challenges of clinical text include the abundance of ambiguous abbreviations, misspellings, lists and Table-like structures intermixed with properly formed sentences, as well as formatting imposed by some EHR software. Many of these challenges are addressed in the currently available clinical NLP systems,5 however we could not identify a freely available system that would address our specific needs and extract maternal medical history from the neonatal clinical notes.
Background
Given that prenatal information is directly relevant to a newborn’s health and clinical care, it is not surprising that these same data are important for research on neonatal diseases. However, obtaining the relevant data can be difficult when maternal and newborn records are disconnected. On occasion, the mother’s data may be accessible to the pediatric researcher either electronically, directly from interviewing the mother, or via manual chart review. However, researchers using large de-identified data collections (which are increasingly used for clinical research given the amount of data available and relative time- and cost-savings) do not have any access to prenatal information except what is documented in the newborn record, most often in unstructured notes.
We are using the Multiparameter Intelligent Monitoring in Intensive Care (MIMIC-II) database to answer a variety of current clinical research questions.6 MIMIC-II contains de-identified data on over 30,000 critically ill patients including almost 8,000 babies admitted to the neonatal intensive care unit (NICU). The database is maintained by the Laboratory for Computational Physiology at the Massachusetts Institute of Technology (MIT) and contains data from patients hospitalized in an ICU at Beth Israel Deaconess Medical Center from 2001 to 2008. The database is freely available in that any researcher who accepts the data use agreement and has completed “protecting human subjects” training can apply for permission to access the data.7 The MIT research team de-identified the data according to the Health Insurance Portability and Accountability Act Privacy Rules.8 The de-identification process included random date shifting that preserved the temporal relationship within a given patient but not across patients, which effectively removed any links that may have existed between the maternal and newborn records.
One study we are working on using the MIMIC-II data relates to blood transfusions given to newborns in the NICU, and we want to include several maternal factors as variables. Given that no structured maternal data are available, we decided to use NLP to extract the information from the NICU notes. As all other clinical notes, NICU notes vary significantly in their structure, content and style. Clinicians enter maternal data in several types of notes at various points in time, as needed for the newborn’s clinical care. The language and syntax for describing maternal health ranges from grammatically correct sentences with the clinical variables of interest fully spelled out to table-like structures presenting the results of laboratory measurements in shorthand that is easily understood in the context of the note by other clinicians, but ambiguous otherwise.
We did not find any previously published literature on this topic. The only publication we found describing NLP methods for a pediatric topic was by Mendonça and colleagues, who adapted an existing application that identified adult pneumonia cases using MedLEE9 for surveillance of healthcare-related pneumonia in newborns.10 The most relevant related work we found was by Garvin and colleagues and described an NLP system for extracting ejection fraction (EF) data from echocardiogram (echo) reports.11 Similar to maternal history in neonatal records, EF results are important for patient management and most often are only found in echo reports, not as structured data. Garvin et al were able to develop a system that accurately classified reports as either reporting an EF < 40% or an EF >= 40%.11 However, they did not attempt to extract the actual EF percentage.
The maternal data we extracted are important for our current study as well as future NICU studies, and we plan to give the data to the database administrators to include in the next release of MIMIC-II so that other researchers will also have ready access to this information. The methods and patterns we used can easily be adapted for use with other research datasets that include neonatal patients. In addition, given that the problem of maternal history only being available in newborn notes is not limited to research data sets, our methods could potentially be used in a clinical application that would extract prenatal data from a newborn’s note into a structured format that could more easily be used for clinical care.
Methods
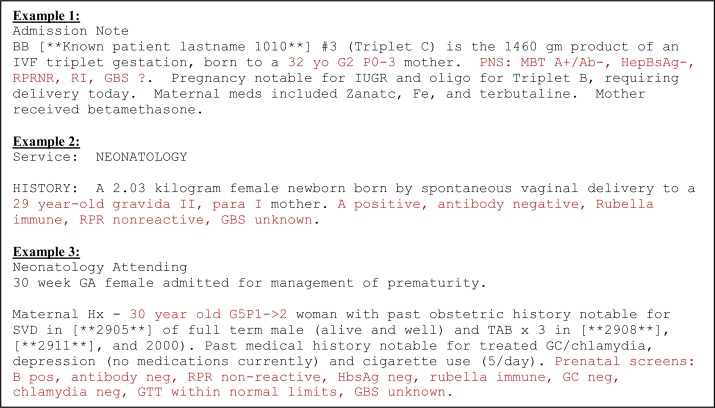
We chose a specific set of maternal variables that are typically found in a newborn’s history: 1) age; 2) gravida/para*; 3) blood type∞; 4) antibody (Ab) status§; 5) vaginal group B strep (GBS) culture; 6) hepatitis B (hepB) immune status; 7) rubella immune status; and 8) syphilis screen (also called RPR). Pediatric providers typically record all of these results in a compact format using typical shorthand (see Figure 1 for examples).
Figure 1:
Excerpts from newborn notes with maternal history recorded in typical shorthand (in red).
We focused on the 289 newborns we had previously selected for our clinical study as the target population for our initial maternal data extraction efforts. We first looked at different types of notes to see which ones contained maternal history. Both the NICU admission notes and discharge summaries had maternal data, so in the first iteration we looked at 25 of these notes to find representative phrases and develop patterns for all of the variables. The gravida/para information almost always immediately followed the maternal age, so we created one pattern for age + gravida/para and six other patterns for the laboratory variables. We extracted maternal age + gravida/para information for 92 babies with the first set of patterns as well as at least 3 out of the 6 other components for 262 babies. We went through two more iterations of reviewing ten notes for the newborns who were missing maternal age + gravida/para information and ten others for those missing lab data and added to the existing patterns. Examples of cases we did not detect in the first 25 notes but found in subsequent iterations were the use of Roman numerals for gravida/para (e.g., “GIV PII”) and the use of a “0” instead of an “O” for the blood group (“0+” instead of “O+”).
We implemented the patterns and the file filtering module in Java. We had four filters in the filtering module. The first two filters limited the number of notes that the algorithm searched over based on certain inclusion criteria. The 289 newborns in our study had 89,896 provider notes and radiology reports associated with their NICU stays, most of which did not contain any data relevant to our study. Because we only wanted maternal data and not newborn data for variables that they had in common (for example, blood type and antibody status), we originally planned to search for the maternal data patterns in a subset of the notes consisting of admission and discharge summaries. However, although the discharge summaries had a “DISCHARGE_SUMMARY” note type, the admission notes did not have any specific designation or titles that consistently indicated they were admission notes. Some clearly stated “Admission note” but others had generic titles such as “Service: NEONATOLOGY.” Our baseline filter only allowed notes with the note type “DISCHARGE_SUMMARY” or with a note title that included the words “admission note,” “admit note,” “attending admission,” “attending admit,” “neonatology attending,” “attending note,” “neonatology,” or “newborn med attending.” Even with this filter we still had 14,709 remaining notes. Most of the notes we had looked at that contained maternal lab data also included maternal age and gravida/para information, so we decided to use the presence of the first pattern (maternal age + gravida/para) as the second filter, which determined whether to further evaluate a newborn’s note for the remaining maternal variables.
The remaining two filters removed results that were most likely irrelevant based on specific exclusion criteria. Many of the notes passed through the first two filters based only on the appropriate note type or title and the presence of a potential age and upper case G followed by a number or an upper case I. For example, one progress note contained the text “Lytes - 125 5.2 89 21 [new line] GI - no evidence of feeding intolerance” which was interpreted as maternal age 21, gravida status I. Therefore, if a particular note only had age and gravida information and another note for that newborn had more information (i.e., para status), we excluded the former. Finally, even after passing through the first three filters we found a large number of notes with “0−“ or “B−“ falsely extracted as blood type without any other lab results, so we excluded those from the final results.
One of the authors (SA) manually checked all of the results that were extracted for accuracy. Because each newborn often had more than one note with maternal data, multiple data points were frequently extracted for the same maternal variable for a given newborn. We evaluated the data both on the individual note level to see if the data that were extracted were relevant as well as on the newborn level to see if any correct values were extracted for a given variable for each newborn.
Results
As we were creating the patterns, we found that although the general format is similar between notes, the order in which the lab results are recorded as well as other results included with the typical set of labs are quite variable (see Figure 1). Therefore, we had to extract each lab variable independently and not as a single group. Table 1 contains the patterns we developed for each variable and Table 2 gives a detailed interpretation of the age + gravida/para pattern.
Table 1.
Java regular expressions for maternal age + gravida/para status and individual laboratory variables.
| Variable | Regular expression |
|---|---|
| Age + gravida/para | Pattern.compile(“(\\d{2})\\W*[yearhs]*\\W*[oldp]*\\W*(Gravida|gravida|G|primparous|primiparous|primigravida|multiparous|primip)\\W*([0–9IVX]*)\\W*(now)*\\W*(para|Para|P|p)*\\W*([0–9IVX]*) \\W*(now)*\\W*([0–9IVX]*)”); |
| Blood type | Pattern.compile(“(?:pns|prenatal screen|mat labs|serologies)?.*?(?:blood type|MBT|BT)? [^hepatis]*\\W+(O|A|B|AB|0)\\W*(\\+|pos|positive|neg|negative|\\−)”); |
| Antibody status (Ab) | Pattern.compile(“(?:antibody|Ab|DAT)\\W*(\\+|pos|positive|neg|negative|\\−)”, Pattern.CASE_INSENSITIVE); |
| Group beta streptococcus (GBS) | Pattern.compile(“(?:GBS|Group Beta|Streptococcus status|strep status|Group B Streptococcus|Group B strep|Group Streptococcus|Group beta strep|group Beta strep status)\\W* (\\+|pos|positive|neg|negative|unk|unknown|was unknown|\\?|\\−)”,Pattern.CASE_INSENSITIVE); |
| Hepatitis B (HepB) surface antigen | Pattern.compile(“(?:HBsAg|hep b|HepBsAg|hepatitis surface antigen|hep b status|hepatitis B surface antigen|HBS antigen|hep)\\W*(\\+|pos|positive|neg|negative|unk|unknown|not reported|\\?|\\−)”, |
| Rubella immune status | Pattern.compile(“(?:rubella|rub|\\W+R)\\W*(immune|unknown|unk|nonimmune|non\\−immune|im|I) \\W+”,Pattern.CASE_INSENSITIVE); |
| Syphilis (RPR) result | Pattern.compile(“(?:RPR)\\W*(\\+|pos|positive|neg|negative|unk|unknown|not reported|NR|nonreactive|non\\−reactive|\\?|\\−)”,Pattern.CASE_INSENSITIVE); |
Table 2.
Step-by-step interpretation of the Age + gravida/para pattern.
| Java regular expression component | Translation (meaning of the regular expression component => translation in context of age + gravida/para status) | Example 1: 32-yo G1 now P2 | Example 2: 24 years old gravida IV para I now II |
|---|---|---|---|
| (\\d{2}) | Exactly 2 numbers => maternal age digits | 32 | 24 |
| \\W* (only defined in this table once for brevity) | (Same translation for every instance of \\W*) Non-letter/underscore => a space or punctuation separating the other age components | - | |
| [yearhs]* | Any 0 or more of the letters in the brackets => representation of the word “year” in the maternal age (e.g., “year” or the “y” part of “yo”); the extra h is not a typo in the pattern but rather represents a typo in the note text that we wanted to capture | y | years |
| [oldp]* | Any 0 or more letters in the brackets => representation of the word “old” in the maternal age (e.g., “old” or the “o” part of “yo”); again, the extra p is a typo in the note text that we want to capture | o | old |
| (Gravida|gravida|G| primparous|primiparous| primigravida| multiparous|primip) | Any of the terms separated by the vertical bar => representation of “gravida” (the number of pregnancies to date), “primiparous” (having one live birth), or “multiparous” (having more than one live birth) | G | gravida |
| [0–9IVX]* | Any of the digits 0–9 or upper case letters I, V or X => representation of the gravida value, either with Arabic or Roman numerals | 1 | IV |
| (now)* | The word “now” => indicates the change in para information from before the current delivery to after the current delivery | now | |
| (para|Para|P|p)* | Any of the terms separated by the vertical bar => representation of “para” (i.e., the number of live births) | P | para |
| ([0–9IVX]*) | See above | 2 | I |
| (now)* | See above | now | |
| ([0–9IVX]*) | See above | II |
A star (*) in the pattern means that the pattern can occur 0 or more times. We had to account for 0 occurrences because every note was missing at least one component of the pattern.
Out of the nearly 90,000 notes for the newborns in our data set, 563 passed through the first three filters, meaning they contained maternal age + gravida/para information. We searched these 563 notes for the maternal lab result patterns, and a total of 482 notes passed through the fourth and final filter. Table 3 summarizes our results at the note level. Out of 482 notes, 414 to 479 contained results for the maternal laboratory variables. Other than maternal antibody status, which had a recall of 0.88, the remaining variables had a recall of 0.90 or above. Other than maternal para and antibody status, which had a precision of 0.925 and 0.947, respectively, precision by individual note ranged from 0.975 to 1.0, indicating that nearly all of the results extracted were accurate. We could not calculate the recall for maternal age because the presence of maternal age was one of our filters, and it would have been impossible to manually determine the recall denominator for age (i.e., how many of the remaining 89,000+ notes contained a maternal age).
Table 3.
Recall and precision for each maternal variable by individual note.
| Maternal variable | Number of notes (Total N = 563) | Number of notes from which values were extracted | Number of notes from which values were extracted correctly | Recall | Precision |
|---|---|---|---|---|---|
| Age | § | 563 | 562 | § | 0.998 |
| Gravida | 563 | 559 | 557 | 0.989 | 0.996 |
| Para | 528 | 519 | 480 | 0.909 | 0.925 |
| Blood type | 479 | 471 | 459 | 0.958 | 0.975 |
| Ab | 466 | 433 | 410 | 0.880 | 0.947 |
| GBS | 431 | 407 | 406 | 0.942 | 0.998 |
| HepB | 431 | 389 | 389 | 0.903 | 1.000 |
| RPR | 451 | 426 | 426 | 0.945 | 1.000 |
| Rubella | 414 | 406 | 406 | 0.981 | 1.000 |
We could not calculate the recall for maternal age because the presence of maternal age was one of our filters, and it would have been impossible to manually determine the recall denominator for age (i.e., how many of the 89,000+ notes contained a maternal age).
We also calculated the results by individual newborn, because these results are more clinically relevant and necessary for our clinical study (see Table 4). Out of 289 newborns, 284 had notes with maternal age. One additional newborn had maternal gravida/para information but not the mother’s age (“…mother is a G12P8…”), so the gravida/para values were not extracted even though they were present in the note. Maternal lab data were available for 247 to 276 of the 289 newborns. To calculate recall and precision for each variable at the newborn level, we counted the number of newborns for whom at least one of the extracted values was correct. For example, if the value for mother’s blood type was extracted as O+ from one note and B− from another note, if either one of those was correct we counted that newborn as having a relevant result for maternal blood type. The incorrect blood type extraction was still reflected in the results on the individual note level given in Table 3. Given that we accepted even one correct answer per newborn, it makes sense that our recall and precision numbers were better by newborn than by note. Recall ranged from 0.912 for antibody status to 0.996 for maternal age, and precision was 0.950 or greater for all of the variables.
Table 4.
Recall and precision for each maternal variable by newborn.
| Maternal variable | Number of newborns (Total N = 289) | Number of newborns for which values were extracted | Number of newborns for which at least one value was correct | Recall | Precision |
|---|---|---|---|---|---|
| Age | 284 | 283 | 283 | 0.996 | 1.000 |
| Gravida | 285 | 283 | 282 | 0.989 | 0.996 |
| Para | 285 | 283 | 271 | 0.951 | 0.958 |
| Blood type | 276 | 269 | 259 | 0.938 | 0.963 |
| Ab | 273 | 262 | 249 | 0.912 | 0.950 |
| GBS | 259 | 249 | 248 | 0.958 | 0.996 |
| HepB | 261 | 246 | 246 | 0.943 | 1.000 |
| RPR | 267 | 256 | 256 | 0.959 | 1.000 |
| Rubella | 247 | 242 | 242 | 0.980 | 1.000 |
Discussion
Overall, by using pattern-based extraction for retrieving maternal history data from newborn NICU notes, we got results that were sufficiently accurate to use in our clinical study. Maternal data are important for newborn care as well as research on neonatal issues. Given that these data are typically not available in a structured format in the newborn record and in general, maternal and newborn records are not linked electronically either in clinical EHRs or de-identified data collections, our method is a feasible option for accessing this information.
We discovered several issues while we were developing the patterns. One, we only reviewed a small subset of notes to develop the patterns, so it was impossible to capture all of the patterns that existed in the larger set. Maternal para information had the least consistency in how it was reported and the subset of notes we reviewed during pattern development did not include two important ones: 1) para x TO y and 2) Px…Py, where x and y stand for the number of live births prior to the current delivery and the number of live births including the current delivery, respectively. In the first case, the previous and current para numbers were connected by a “to” and in the second, the current para number was preceded by a second “P.” Other examples that we did not discover during pattern development were “Unknown GBS status” in which the result “unknown” was given before the variable “GBS status,” RPR spelled out as “rapid plasma reagin,” and “hepatitis negative” without any indication of which type of hepatitis. We plan to fix these patterns and re-evaluate our results.
The second major issue was case sensitivity. At first, we did not use case-sensitive pattern matching, but we retrieved a large number of false positive results, primarily for the maternal blood type. For example, admission notes usually include information about the mother’s obstetrician, and if a note said “Ob− Dr. X” the “b−“ portion of that phrase was extracted as blood type B−. By restricting the blood type results A, B, AB, and O to upper-case matching only, the number of false positives was greatly reduced and recall was not affected.
The third issue was the relationship between blood type and antibody results, and overlap between the blood type AB and the abbreviation for antibody “Ab.” Initially, we assumed that maternal blood type was always followed by her antibody status and therefore had a single pattern for blood type + antibody; however, after the first iteration of pattern development we discovered that while this holds true in the majority of cases, we found several instances in which other labs were interpolated between the blood type and antibody results and/or the antibody result appeared before the blood type. Therefore, we searched for blood type and antibody status independently, which created the new problem described next.
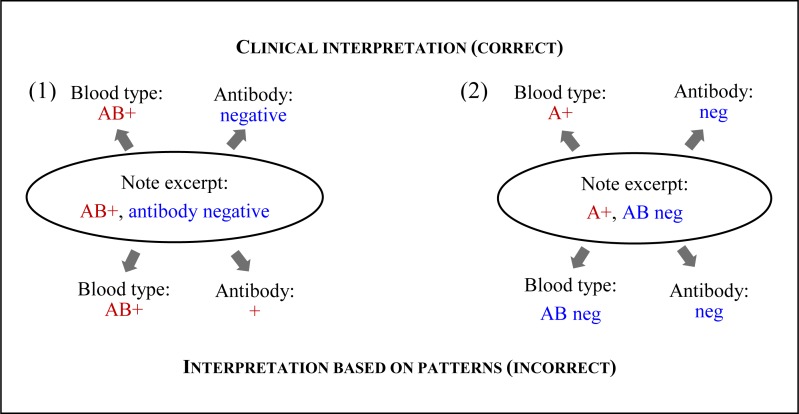
When we searched for the two variables together, once a portion of text was extracted for the blood type (which came first in our pattern), that same text could not be re-used to match a later part of the pattern. However, when searching independently, the same text could be re-used if it matched the pattern for each independent variable. We had not considered the similarity between the blood type and antibody patterns. As a result, in several cases the blood group AB was interpreted as the antibody abbreviation with the Rh status interpreted as antibody result as well as the converse, where the antibody abbreviation “AB” was interpreted as the blood group and the antibody result as the Rh status (see Figure 3). Fortunately, we had already restricted blood type results to upper case so that the antibody abbreviations “Ab” and “ab” were not also extracted as blood type AB.
In a few cases, during the manual result validation we found examples where the problem was not an incomplete pattern but rather incorrect representations of the variable in the note. One example is “hepatitis C surface antigen,” which clearly was meant to indicate hepatitis B (there is no such thing as hepatitis C surface antigen) but was (correctly) not picked up by the pattern due to the “C.” In other cases, de-identification was the issue, such as “Hepatitis [**Name2 (NI) **] negative” and “para [**1–31**].” In both of these cases we counted the result for that specific variable as missing and did not include them when we calculated recall at both the note- and newborn-level.
We used regular expressions instead of more linguistically-grounded methods (such as syntactic parsing) because maternal information was entered in table-like format in the first 25 notes that we reviewed prior to implementing the extraction module. However, during the evaluation of the extraction results, we encountered several examples where parsing likely would have improved results. For example, one note contained the phrase “HBsAg, RPR, Rubella and GBS status unknown at the time of delivery” and based purely on pattern matching, no results were extracted for HepB, RPR or rubella. In another instance, although the phrase “DAT negative on … but positive today” clearly means that the current antibody status is positive, based on the antibody pattern we implemented, the antibody result was extracted as negative. Additionally, more formal NLP may have ameliorated the results for maternal blood type AB and antibody status abbreviated “AB” as well as cases where the newborn’s blood type or antibody status was incorrectly extracted as a maternal result.
Our next step is to use the small set of 563 manually annotated notes from this study to train supervised machine learning algorithms and extract the same maternal data for the remaining 7,000+ newborns in MIMIC-II. If we are successful, our work could be used to develop applications to extract maternal history from newborn notes both in a clinical setting in order to streamline and potentially improve the quality of clinical care, as well as in a research setting to facilitate maternal data gathering for neonatal research.
Conclusion
Maternal information is very important for a newborn’s clinical care but is often difficult to obtain due to the disconnect between the maternal and newborn records. We have presented a simple algorithm that uses a few regular expressions to extract maternal history from newborn notes. Although some of the pattern components were ambiguous, when we combined them with filtering for both the notes and the extraction results, we obtained good quality results from a small set of notes. With a small manual review effort, we achieved recall between 0.91 and 0.99 and precision between 0.95 and 1.0. If pediatric researchers and clinicians are comfortable with this level of accuracy, our algorithm is freely available for developers to adapt for use with other research data sets or in a clinical setting.
Figure 2.
Pattern interpretation errors based on: (1) blood group AB, which was interpreted as the antibody variable; and (2) antibody abbreviation “AB,” which was interpreted as the blood group AB.
Funding/Support Acknowledgment
This research was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health, under NIH IRB exemption.
Footnotes
Gravida means the number of pregnancies, and para is the number of live births. Typically, at the time of delivery, a woman’s gravida number includes her current pregnancy, the first para number is how many live births she had in the past, and the second para number is how many live births she has now. For example, G1P0−>1 indicates that a woman has been pregnant once (the current pregnancy), previously had no children, and now has one living newborn. G2P1−>3 indicates that a woman has been pregnant twice, previously delivered one live newborn, and now has delivered twins (for a total of three live births).
Blood type includes the blood group (A, B, AB, or O) and Rh antibody (positive or negative), which together create blood types such as O+ and B−.
Antibody status is the presence of non-Rh antibodies. The Rh status is included in blood type as explained above.
References
- 1.Advisory Committee on Immunization Practices Recommendation of the Immunization Practices Advisory Committee (ACIP) Postexposure Prophylaxis of Hepatitis B. MMWR Morb Mortal Wkly Rep. 1984;33:285–290. [PubMed] [Google Scholar]
- 2.Pediatric Practice Action Group and Task Force on Medical Informatics Privacy protection of health information: patient rights and pediatrician responsibilities. Pediatrics. 1999;104:973–977. [PubMed] [Google Scholar]
- 3.Lowry SZ, Quinn MT, Ramaiah M, et al. A Human Factors Guide to Enhance EHR Usability of Critical User Interactions when Supporting Pediatric Patient Care. Gaithersburg (MD): National Institute of Standards and Technology (US); 2012. Jun 28, pp. 27–28. Report No.: NISTIR 7865. Chapter 4, Section VII, Detailed guidance for critical user interactions: Newborn care. [Google Scholar]
- 4.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011 Sep-Oct;18(5):544–51. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009 Oct;42(5):760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeed M, Lieu C, Raber G, Mark RG. MIMIC II: A Massive Temporal ICU Patient Database to Support Research in Intelligent Patient Monitoring. Comput Cardiol. 2002;29:641–644. [ http://mimic.physionet.org/] [PubMed] [Google Scholar]
- 7.Saeed M, Villarroel M, Reisner AT, et al. Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC–II): A public-access intensive care unit database. Crit Care Med. 2011 Jan 28;39:952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Standards for Privacy of Individually Identifiable Health Information 2002. Final Rule, 45 CFR Parts 160 and 164. [ http://www.hhs.gov/ocr/hipaa/] [PubMed]
- 9.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1161(2):74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendonça EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform. 2005;38:314–321. doi: 10.1016/j.jbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc. 2012;19:859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]