Abstract
The biomedical literature presents a uniquely challenging text mining problem. Sentences are long and complex, the subject matter is highly specialized with a distinct vocabulary, and producing annotated training data for this domain is time consuming and expensive. In this environment, unsupervised text mining methods that do not rely on annotated training data are valuable. Here we investigate the use of random indexing, an automated method for producing vector-space semantic representations of words from large, unlabeled corpora, to address the problem of term normalization in sentences describing drugs and genes. We show that random indexing produces similarity scores that capture some of the structure of PHARE, a manually curated ontology of pharmacogenomics concepts. We further show that random indexing can be used to identify likely word candidates for inclusion in the ontology, and can help localize these new labels among classes and roles within the ontology.
Introduction
Biomedical text mining algorithms typically require normalization: mapping the diversity of natural language to a smaller set of canonical concepts or “features”. This feature reduction process is critical for prediction, since the risk of overfitting to a particular training set goes up as the number of features increases.
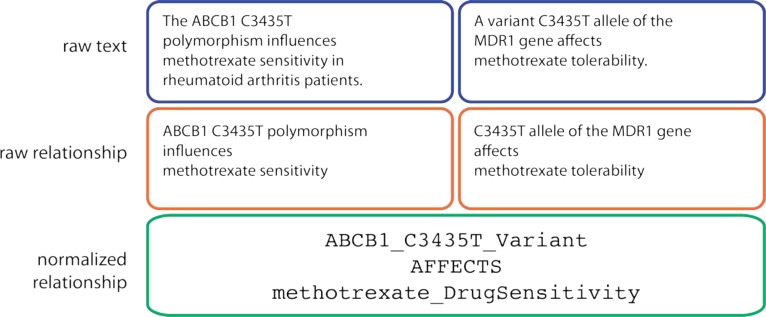
In many biomedical applications, terms are normalized using a manually-constructed ontology. For example, the PHARE (PHArmacogenomic RElationship) ontology normalizes pharmacogenomic relationships observed in text (Figure 1) [1]. PHARE includes rules for recognizing relationships in sentences; it extracts pharmacogenomic relations with 80% precision. Coulet et al used PHARE to extract and normalize over 40,000 relationships among drugs, genes and phenotypes [2]. Later work used PHARE-normalized gene-drug relations to predict drug-drug interactions [3].
Figure 1:
An example of relation normalization using PHARE. Here two sentences that look very different on the surface are mapped to the same normalized “fact”.
Ontology-based normalization works well for many purposes. However, as the volume of the scientific literature grows, and especially as biomedical text mining enters new domains like clinical text, patient forums on the Internet, and the patent literature, it becomes increasingly costly to construct domain-specific ontologies like PHARE. At the same time, unsupervised algorithms that can assess word and phrase similarity automatically based on usage patterns in large corpora become increasingly attractive. In particular, there has been much recent interest in the use of automated natural language processing methods to learn the structure of biomedical ontologies [4].
Here we compare the structure of PHARE to the structure predicted using a popular method for unsupervised word similarity assessment called random indexing. We show that the word pair similarities predicted by random indexing correlate significantly with the words’ relative positions within PHARE. We further examine the degree to which random indexing could be expected to “reproduce” PHARE; that is, to assign PHARE’s word labels to the appropriate concepts and roles within the ontology. Although random indexing, at least as it was applied here, is not sufficient to fully reproduce the PHARE ontology, we conclude that it shows promise for identifying candidate terms for inclusion in future versions of the ontology. Future work will also explore the intriguing possibility that applications where normalization is critical, such as biomedical sentence alignment, might benefit from the use of distributional methods rather than rules-based approaches like domain-specific ontologies.
Background
The PHARE ontology
The (PHARE) ontology was created in 2010 by Adrien Coulet and colleagues at Stanford University. The researchers extracted approximately 40,000 raw relationships (verbs and nominalized verbs) among 3007 drugs, 41 genes and 4202 phenotypes from biomedical sentences and identified the 200 most frequent relationship types from within this set. They then manually merged similar relationship types into conceptual “roles” and organized these roles in a hierarchy [1]. They repeated this process for the nouns most often modified by drug and gene entities, such as “expression” and “polymorphism”, creating a hierarchy of modifier “concepts”. Finally, they defined a set of rules for application of the roles and concepts to drug, gene, and phenotype terms found in real English sentences. In particular, they limited the application of certain roles and concepts to certain classes of entities. (“Polymorphism”, for example, was only permitted to modify gene names, not drug or phenotype names.) The English words that map to each concept and role are called “labels”. The final version of PHARE consists of (a) a hierarchy of roles, (b) a hierarchy of concepts, and (c) a set of labels associated with each role or concept.
Recently, we investigated the degree to which pharmacogenomic relationships of interest described in PubMed sentences conformed to the grammatical structures PHARE is able to recognize. We found that although PHARE is excellent at extracting relationships of that form (nearly 100% sensitivity), its recall on interesting pharmacogenomic relationships as a whole is quite low. Of 72 sentences describing an inhibitory relationship between itraconazole and CYP3A4, for example, PHARE was able to extract only 2 relations. We have estimated PHARE’s overall recall at approximately 30%, though this number has high variance depending on the specific nature of the drug-gene relationship involved. We concluded that to extract all useful pharmacogenomic relationships from Medline sentences, we would need to account for greater variability in sentence structure and phrasing than PHARE currently supports. As a first step in expanding PHARE’s coverage, we decided to experiment with automated techniques for identifying other potential labels and their likely locations within PHARE.
Random indexing
Vector space models of semantics have gained prominence in the text mining community as a way to teach computers the “meaning” of words and phrases. They represent each word as a vector that is constructed based on how the word is used in context; there are endless variations for how best to determine and construct these context vectors, each of which captures a slightly different aspect of word meaning [5, 6]. This work dates back to the 1990s, when some of the earliest methods – Latent Semantic Analysis (LSA) [7] and Hyperspace Analogue to Language (HAL) [8] – were invented. One popular approach that has emerged more recently is random indexing, which builds similar vector space representations to LSA and HAL, but is more computationally efficient [9].
In random indexing, each word in a corpus is assigned a random, sparse “elemental” vector. The “dimension” of this vector is its length, and the “seed length” is how many of the terms in the vector are nonzero; typical values for dimension and seed length are 100–1000 and 5–20. An elemental vector is built by initializing all of its elements to zero and then randomly assigning s/2 “+1” elements and s/2 “−1” elements, where s is the seed length. After the elemental vectors are assigned, a “context vector” is built for a particular target word by adding together the elemental vectors from words that occur within some pre-specified radius (“window width”) of the target word. It is important at this stage to distinguish the elemental from the context vectors: elemental vectors are randomly assigned, and context vectors are built for each word using the elemental vectors of the other words that surround it. It is the context vectors that will be used to compare word meanings.
The process for building the context vectors is simple: one moves through the corpus with a bin of width 2w+1, where w is the window width, and adds elemental vectors for all words within the bin to the context vector for the word in the middle. These added elemental vectors may additionally be weighted according to some predefined metric, such as their corresponding words’ overall frequencies in the corpus. Finally, the context vectors are normalized to unit length. To evaluate the similarity of two words, one calculates the cosine similarity of the context vectors corresponding to those two words. The cosine similarity is a unitless metric between −1 and 1; more similar words have cosine similarities closer to 1.
Encoding word order
Different variants of random indexing encode word order for surrounding terms in context vectors in different ways. The most basic version ignores it completely; elemental vectors for all words within the bin are added directly to the context vector for the word in the middle. More elaborate versions use convolution [10] or permutations [11] to encode word order.
Methods
Data set construction
We extracted all sentences from Medline 2012 that mentioned a drug and a gene and were between 4 and 50 words in length (approximately 95% of all sentences in Medline fell within this range). Drug and gene mentions were established using simple string matching and lexicons of drug and gene terms from PharmGKB [12]. We included only single-word drug and gene names for simplicity. We manually removed several common words that were accidentally included in the lexicons and were not actually drugs or genes (such as “enzymes”, “glycine”, and “vaccines” for drugs; “dehydrogenase”, “protease”, and “murine” for genes). The final lexicons included 1470 unique drug strings and 37,922 unique gene strings. Our final corpus consisted of 494,804 sentences.
The Semantic Vectors package
We used the Java-based Semantic Vectors package [13] to construct vector representations of all words occurring at least three times in our corpus. Semantic Vectors is a convenient implementation of random indexing based on Apache Lucene. We varied the window size, vector dimension and seed length to evaluate how much these parameters affected our representations, and to find the combination that created the optimal vectors for our task. We also evaluated the means by which word order was encoded: “basic” vectors did not encode word order, “drxn” vectors encoded only the direction associated with a context word (before or after the target word), and “perm” vectors used permutations to encode the relative position of each context word relative to the target word. The degree of semantic similarity between two [unit-normalized] vectors was calculated using cosine similarity as described above.
Calculating concordance with the PHARE ontology
We wanted to see how well similarity scores for word pairs calculated using random indexing corresponded to those words’ semantic relatedness within PHARE. Because we could not calculate the semantic relatedness of PHARE’s concepts and roles directly using random indexing (since they are not English words), we instead calculated pairwise similarity scores between all concept labels, and independently, all role labels, in PHARE. We also wanted to determine whether a particular formulation of the semantic vectors we generated (such as a particular window width, dimension, or seed length) optimized the vectors’ concordance with the structure of the PHARE ontology. We tested all combinations of the following: window widths 1, 3, and 5, vector dimensions 50, 100, 150, 300, 500, and 1000, word order encodings “basic”, “drxn”, and “perm”, and seed lengths 4, 10, and 20.
We hypothesized that high similarity scores would correspond to close ontological relationships, meaning larger numbers of common ontological parents. For each label pair, we measured (a) the cosine similarity of its two labels’ context vectors and (b) the number of common ontological parents for the labels in that pair (traversing the ontology upward until we reached the root node). We then repeated these measurements for all concept label pairs and, separately, all role label pairs in the ontology. We used the Kendall-Tau nonparametric correlation coefficient, specifically the implementation in R’s “stats” package, to test the correlation between cosine similarity and number of common ontological parents separately for both concepts and roles. Unfortunately, the algorithm for calculating the Kendall-Tau coefficient is O(n2); because the number of data points in our experiments was so large and the number of ties so high, and because we performed many different trials with different parameter values for our semantic vectors, calculating the full Kendall-Tau coefficient for each trial took too long. We therefore used 1000-point bootstrap samples of our data and repeated the calculation of the Kendall-Tau coefficient 100 times for each sample; here we report the medians of those results. For all subsequent analyses, we used the best performing vectors, the specific formulation of which differed for roles and concepts.
Reassigning labels within the ontology
Next, we evaluated how well random indexing could localize labels within the ontology. We removed each concept or role label from the ontology, one at a time. (Call the removed label L, and call its corresponding context vector VL.) We then evaluated VL’s (a) mean and (b) maximum cosine similarity with the vectors for the remaining labels from each ontological group (a concept, if L was a label for a concept, or a role, if L was a label for a role). We ranked the groups according to their label vectors’ similarity with VL to ascertain which concepts or roles L was most likely to belong to. The result was a ranked list of candidate concepts or roles for each L. Ideally, the correct concept/role assignment for each label would rise to the top of its ranked list of candidates.
There are 228 concepts and 77 roles in the PHARE ontology. However, if a role was the passive-voice version of another role (“isInducedBy”, rather than “induced”) it was excluded from our analysis and its labels added to the active form version of the role. We therefore evaluated our performance on 54 of the 77 original roles.
Identifying new word candidates for inclusion in PHARE
Finally, we wanted to see if our semantic vectors could be used to efficiently augment the PHARE ontology. PHARE only incudes a few hundred of the most common role and concept labels found in Medline; since its precision is only 80%, there are likely other reasonable labels that it missed. We wanted to see which other words might logically be added as labels to each concept and role. As a preliminary investigation of this possibility, we compared the vectors for each non-ontology term to all known label vectors from the ontology. For each concept or role label within the ontology, we found the top non-ontology term whose semantic vector best matched its own. This led to a ranked list of possible ontology candidates, ordered by their similarity to a current label in the ontology. For role labels, we restricted our list to verbs or nominalized verbs. For concept labels, we restricted our list to nouns (nominalized verbs, like “identification”, were also acceptable here). We then manually reviewed the lists for the most likely “ontology augmentation candidates”.
Results
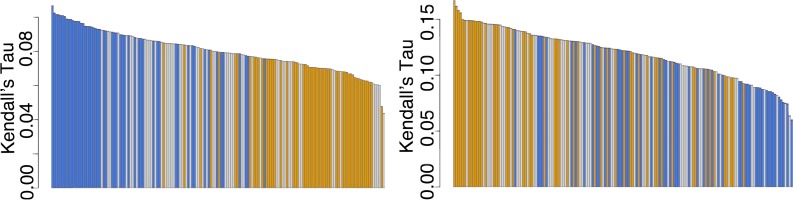
Figure 2 shows the results of our initial experiments to ascertain which type of semantic vector, generated by random indexing, best captured the structure of the PHARE ontology. As described in the Methods, we evaluated a variety of different vector types (window widths, dimensions, word order encodings and seed lengths) to see which led to the highest Kendall-Tau correlation between X = cosine similarity of label vectors and Y = number of common parents for those labels within the ontology. No matter what type of semantic vector we constructed, the correlation between X and Y was significant at the 95% confidence level; the best performing vectors had median correlations of 0.108 (p = 0.00121; concepts) and 0.165 (p < 0.0001; roles). Interestingly, the window widths associated with the best-performing vectors differed between concept and role labels. Concept labels correlated most highly with ontology position when a window width of 5 was used, while role labels were just the opposite; the correlation was highest with a window width of 1. Intuitively, this makes sense; concepts are nouns and roles are verbs, so one might speculate that most of the information about verbs is contained within the words immediately preceding and following them, while nouns’ meaning depends on the more general “theme” of the sentence.
Figure 2.
Bar plots of correlations between number of common parents in ontology and distributional similarity scores for (left) concepts and (right) roles. Each bar represents a different type of semantic vector. Orange bars represent vectors with width 1, gray width 3, and blue width 5.
Some examples of highly similar concept and role labels, where similarity was assessed using the cosine similarity of the respective words’ vectors, are shown in Tables 1 (concepts) and 2 (roles). The semantic relatedness of most of these word pairs is obvious. However, we do notice one peculiarity of the random indexing approach, which is that antonyms are not separated; in fact, antonyms have a high similarity score. This makes sense when one considers the nature of random indexing’s context vector assembly process; there is no context where “downregulation” occurs in which “upregulation” could not also occur. However, it does raise a red flag in terms of random indexing’s ability to reproduce the structure of the PHARE ontology; in the “role” portion of the ontology, for example, “induces” and “inhibits” live on separate branches. Random indexing could potentially localize them only to within the same parent branch, “regulates”.
Table 1.
Examples of high-ranking pairs of concept labels from drug-gene sentences, ordered by cosine similarity.
| Concept Label 1 | Concept Label 2 | Cosine Similarity Score |
|---|---|---|
| inhibition | suppression | 0.983 |
| downregulation | upregulation | 0.982 |
| incidence | prevalence | 0.981 |
| assessment | evaluation | 0.977 |
| pharmacokinetics | disposition | 0.974 |
| association | interaction | 0.973 |
| inactivation | inhibition | 0.973 |
| tolerability | safety | 0.972 |
Table 2.
Examples of high-ranking pairs of role labels from drug-gene sentences, ordered by cosine similarity.
| Role Label 1 | Role Label 2 | Cosine Similarity Score |
|---|---|---|
| investigate | examine | 0.999 |
| assess | evaluate | 0.999 |
| suggest | indicate | 0.997 |
| alter | affect | 0.996 |
| modulation | inhibition | 0.992 |
| suppress | stimulate | 0.990 |
| inhibit | prevent | 0.988 |
| catalyzed | catalysed | 0.986 |
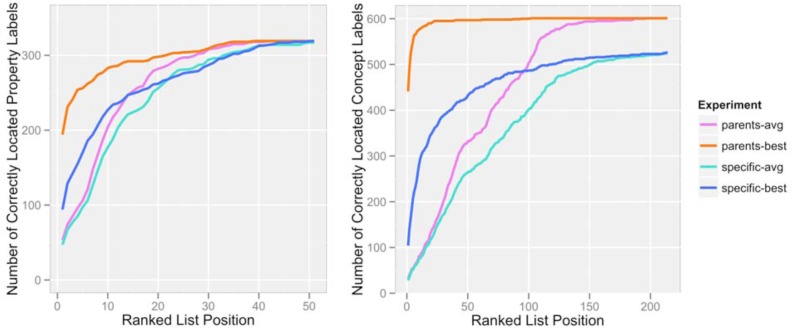
Our results for the “label reassignment” portion of our assessment are shown in Figure 3. The graphs display four lines: “specific-avg” and “specific-best” contain the number of correct concept/role assignments for labels that occurred within the top k items on their ranked lists (where k is the “Ranked List Position” value on the horizontal axis). The avg/best designation refers to the way in which the concept/role assignments were ranked; in the “avg” case, we calculated the test label’s similarity to all labels within a concept/role and took the mean of those values as our match score for that concept/role. In the “best” case, we took the maximum of those values. Practically speaking, this means that if a test label was highly similar to only one member label of a concept/role, that concept/role would be ranked highly in the “best” case but not in the “avg” case.
Figure 3.
Correct concepts/roles found, by position in the ranked list. Separate graphs are shown for (left) roles and (right) concepts. The total number of concepts included here was 228 and the total number of roles was 54.
The “specific” vs. “parents” designation in Figure 3 refers to what we counted as a “hit”. In the “specific” case, a concept/role label was considered correctly classified by position k only if its most specific matching concept/role appeared on the ranked list by that point. In the “parents” case, the most specific concept/role or one of its parent concepts/roles in the ontology could appear. We simply wanted to see whether some of our missed assignments were the result of the test label’s being assigned to a more general super-class of the correct concept/role, which would be less of a problem than if it were assigned to an entirely incorrect part of the hierarchy.
Of the 602 concept labels we examined, 104 (17.3%) were correctly classified (i.e. the correct role was first on the ranked list) when the “best” method was used to assign the matches, and 17 (2.8%) were correctly classified when the “avg” method was used. This seems to indicate that often a label will be distributionally similar to some, but not all, other labels within its concept/role. Of the 319 role labels we examined, 94 (29.5%) were correctly classified when the “best” method was used and 25 (7.8%) were correctly classified when the “avg” method was used. If we relax our restriction on the concept/role assignment such that a parent of a given concept/role is also acceptable, 443 (73.6%) of concept labels are assigned correctly for “best” and 20 (3.3%) for “avg”, and 194 (60.8%) of role labels are assigned correctly for “best” and 31 (9.7%) for “avg”.
In addition, performance increases if we consider assignments beyond rank position #1. Considering only the “best” assignment methods, since those seem to outperform “avg” at every turn, 234 (73.4%) of correct role labels and 420 (69.8%) of correct concept labels occur in the top 20% of the labels’ ranked lists.
The final part of our analysis sought to identify those terms, not currently part of the ontology, that would make good candidates for inclusion as labels, and to localize those new labels within the ontology. Tables 3 and 4 show the best candidates, evaluated in terms of the criteria described in the Methods. Some of these terms, such as “tumors” and “combinations” in Table 4, were minor variants of other words that were already present in the ontology. In the case of both “tumors” and “combinations”, their respective singular forms (“tumor”, “combination”) were already present as labels within the concepts assigned to them using random indexing. Findings like this boosted our confidence in random indexing considerably. Many of our findings from Tables 3 and 4 are already under review for possible inclusion in future versions of PHARE. However, so as not to over-sell this method to the reader, we have also included some errors in Tables 3 and 4. “Capillary” was the highest-similarity word to “gel”, for example, probably due to their common proximity to the relatively uncommon word “electrophoresis”, but “gel”’s corresponding concept in the ontology is “TopicalFormulation”. Similarly, because “treated” is often used to describe chemical treatment of cell cultures in our corpus, it matched closely with “pretreated”, while in PHARE “treated” is only permitted to describe a drug’s treatment of a disease. A similar problem occurs for “incubation” and “treatment”.
Table 3.
Top 15 ontology augmentation candidates for roles. Errors are denoted by a gray background.
| Candidate Label | Matching Role Label | Cosine Similarity | Role (Active Form) | Candidate Label’s Occurrences in Corpus |
|---|---|---|---|---|
| suppression | inhibition | 0.985 | inhibits | 2809 |
| ascertain | determine | 0.972 | demonstrates | 145 |
| abrogated | abolished | 0.960 | suppresses | 517 |
| impact | influence | 0.940 | influences | 1292 |
| infused | injected | 0.935 | administers | 911 |
| given | administered | 0.928 | administers | 5345 |
| uptake | transport | 0.926 | transports | 4813 |
| formed | generated | 0.881 | produces | 1128 |
| utilizing | using | 0.871 | uses | 325 |
| display | exhibit | 0.866 | has | 378 |
| underwent | received | 0.839 | accepts | 897 |
| verified | confirmed | 0.835 | demonstrates | 148 |
| documented | established | 0.794 | demonstrates | 567 |
| devised | developed | 0.755 | produces | 48 |
| maintain | sustain | 0.749 | demonstrates | 417 |
| pretreated | treated | 0.971 | treats | 1621 |
| incubation | treatment | 0.923 | treats | 2527 |
Table 4.
Top 15 ontology augmentation candidates for concepts. We include one example of a concept associated with the given concept label; there could have been more than one in the ontology, since labels are not unique for concepts. Errors are denoted by a gray background.
| Candidate Label | Matching Concept Label | Cosine Similarity | Concept | Candidate Label’s Occurrences in Corpus |
|---|---|---|---|---|
| participation | involvement | 0.984 | GeneProductFunction | 246 |
| enhancement | augmentation | 0.97 | Overexpression | 1349 |
| tumors | neoplasms | 0.96 | Cancer | 3430 |
| utility | usefulness | 0.958 | DrugEfficacy | 371 |
| combinations | coadministration | 0.952 | DrugTreatment | 962 |
| estimation | measurement | 0.952 | GeneAnalysis | 221 |
| superfusion | perfusion | 0.949 | DrugTreatment | 150 |
| identification | detection | 0.943 | PhenotypeAnalysis | 575 |
| comparable | similar | 0.935 | DrugAnalog | 1472 |
| assembly | formation | 0.913 | Synthesis | 270 |
| cascade | pathway | 0.907 | GenePathway | 434 |
| protocol | regimen | 0.862 | DrugTreatment | 776 |
| perturbation | modification | 0.856 | ChemicalModification | 78 |
| chronic | acute | 0.822 | DiseaseSeverity | 8493 |
| reactivation | recurrence | 0.802 | DiseaseRelapse | 204 |
| capillary | gel | 0.621 | TopicalFormulation | 529 |
| summary | conclusion | 0.988 | DrugEffect | 494 |
It is interesting to note that training semantic vectors on domain-specific corpora like our ∼500,000 drug-gene sentences seems to yield an increase in the specificity with which word senses are represented. For example, a context vector for the word “given” trained on text from the Wall Street Journal probably would not share much similarity with one for the verb “administered”. However, because of the specific contextual cues found in drug-gene sentences, “given”’s closest vector neighbor is indeed “administered”. This is because, in drug-gene sentences, to “give” something (a rat, a human) a drug is to administer that drug. There are not many other contexts within these sentences in which “given” is used. The same argument is probably also true for “cascade” and “pathway” (Table 4) and “uptake” and “transport” (Table 3).
Discussion
Distributional semantics methods, specifically vector space representations of word and phrase meanings, have gained popularity in recent years as a scalable alternative to rules-based approaches to term and sentence normalization [4, 6, 14]. Here, in an attempt to evaluate the degree to which these techniques could augment or even replace our lab’s current ontology-based approach to pharmacogenomics sentence normalization, we examined how closely semantic word vectors generated using random indexing captured the overall conceptual hierarchy of the manually-generated PHARE ontology. We discovered that word pairs’ semantic vectors became increasingly similar as the words shared more common parents within the ontology. We also discovered that words could be assigned to reasonable concepts and roles within the ontology if we scored them based on their maximum similarity with other word labels within a given concept or role. We expanded this approach to assign some new word candidates that are not currently in the ontology to their most likely ontological locations.
Correlation strength and its interpretation
The relatively weak correlation between the proximity of word labels within the PHARE ontology and their vector space similarities is a strong indication that there is more information in the ontology than can be captured purely by looking at how words are used in context. For example, several of the ontological concepts and roles contained labels that were common terms, like “find”, that gained additional specificity by the rules PHARE provides on how they are to be applied to real biomedical sentences. Our investigations here take none of these “word sense” factors into account, aside from our selection of a training corpus in which the word senses in question are limited. To our semantic vectors, “established” (as in “established methods”) is the same as “established” (as in “established a new technique for”). This is a major limitation of the distributional approach used here; ambiguities like this were one reason PHARE was created.
However, it is interesting to consider the degree to which these imperfections matter for real biomedical applications. For example, consider the two dependency parses shown in Figure 3. (A dependency parse is one technique for representing the deep grammatical structure of a sentence.) These parses are for the two example sentences shown in the normalization example in Figure 1. Noted biomedical relation extraction algorithms like RelEx [15] already use dependency parses in their analysis, but they apply manually-generated rules to them to extract relations of interest. (PHARE was also inspired by Coulet et al’s observation of common structural “motifs” in dependency-parsed biomedical sentences.) We immediately notice that the sentences in this figure are structurally similar, and that we might conceive of aligning the two dependency graphs and using vector space representations of word meanings to compare the quality of these alignments. This assessment of the sentences’ similarity would perform a task akin to normalization. In this case, even if “arthritis” and “tolerability” somehow ended up with similar distributional representations, it wouldn’t matter for the purposes of assigning the alignment score because they exist in different grammatical “places” within the two graphs. Alignment-based approaches like this are already common in the computer science literature; for example, in automatic essay grading [16] and entailment recognition [17]. So, practically speaking, even a weak distributional “signal” might be enough for some interesting applications.
Additional limitations of our approach
Our approach suffers from a few additional limitations that are worth mentioning. First, our corpus consisted of individual Medline sentences containing drug and gene names; we did not consider additional contextual cues from the rest of the abstracts. We did this in the interest of building semantic vectors that were as domain specific as possible; however, the lack of additional domain cues probably hurt us, especially with respect to concept assignments (which, as we observed, preferred wider bin widths).
Second, as briefly alluded to earlier and as lamented frequently in the distributional semantics literature, our techniques did not capture the opposing nature of antonyms. As far as we know, there is no way to reliably distinguish antonyms using distributional means.
In addition, our evaluation of word similarities, and our assignment of word labels to concepts and roles within the PHARE ontology, ignored much of the ontology’s deeper structure. For example, some concepts are only permitted to modify phenotypes, while others are only permitted to modify genes. We ignored this structure and compared the labels from these different concepts directly. This was done in the interest of quick exploration and simplicity, but restricting our comparisons to labels from specific concepts/roles could very well have improved our performance reassigning labels to PHARE. However, since the point of our study was to see how much the structure of PHARE could be captured without human intervention, we did not choose to restrict our analysis in this way.
And finally, label assignments within PHARE are unique for roles but not concepts. This meant that a given label could have more than one concept associated with it, and it probably explains the huge increase in performance we experienced when we included parent concepts in our analysis in Figure 2.
Conclusion
Random indexing produces vector representations of words that correlate significantly with these words’ positions within a biomedical ontology. Although these representations do not capture all of the information contained in the ontology, they have several advantages. First, they are quick and easy to produce, and can easily be adapted to different corpora (Medline, other biomedical text, or specialized subsets of Medline such as the drug-gene sentences we examined here). Second, they seem to capture much of the semantic meaning of individual words, at least as those words are represented within the PHARE ontology, and they can be used to quickly and easily “bootstrap” connections to other words in the corpus that could be suitable for inclusion in the ontology. They can also provide a rough sense of where those words should be located within the ontology. And finally, and most importantly, construction and evaluation of these vectors requires no manual rule-making or annotation; the vectors are learned in an unsupervised manner from unlabeled text corpora. Although our explorations here are preliminary and much work remains to be done to fully establish the role of distributional semantics methods within biomedical text mining, increasing interest in this field within the biomedical community could lead to exciting new applications in the areas of named entity recognition, concept normalization, and specialized ontology building within bioinformatics.
Figure 4.
Dependency parses for the two example sentences shown in Figure 1. Because the structure of these sentences is so similar, one could conceive of using distributional semantics methods to establish an alignment between them, thus performing a task akin to normalization without the use of an ontology.
Acknowledgments
This work was supported by a grant from Oracle Corporation, NIH GM61374 and MH094267. We used computational resources supplied by NSF award CNS-0619926.
References
- 1.Coulet A, Shah NH, Garten Y, Musen M, Altman RB. Using text to build semantic networks for pharmacogenomics. Journal of biomedical informatics. 2010;43(6):1009–1019. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coulet A, Garten Y, Dumontier M, Altman RB, Musen MA, Shah NH. Integration and publication of heterogeneous text-mined relationships on the Semantic Web. J Biomed Semantics. 2011;2(Suppl 2):S10. doi: 10.1186/2041-1480-2-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Percha B, Garten Y, Altman RB. Discovery and explanation of drug-drug interactions via text mining. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing; 2012. p. 410. [PMC free article] [PubMed] [Google Scholar]
- 4.Liu K, Hogan WR, Crowley RS. Natural language processing methods and systems for biomedical ontology learning. Journal of Biomedical Informatics. 2011;44:163–179. doi: 10.1016/j.jbi.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turney PD, Pantel P. From frequency to meaning: Vector space models of semantics. Journal of Artificial Intelligence Research. 2010;37(1):141–188. [Google Scholar]
- 6.Cohen T, Widdows D. Empirical distributional semantics: Methods and biomedical applications. Journal of biomedical informatics. 2009;42(2):390. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landauer TK, Foltz PW, Laham D. An introduction to latent semantic analysis. Discourse processes. 1998;25(2–3):259–284. [Google Scholar]
- 8.Lund K, Burgess C. Producing high-dimensional semantic spaces from lexical co-occurrence. Behavior Research Methods, Instruments, & Computers. 1996;28(2):203–208. [Google Scholar]
- 9.Sahlgren M. An introduction to random indexing. Methods and Applications of Semantic Indexing Workshop at the 7th International Conference on Terminology and Knowledge Engineering, TKE; 2005. [Google Scholar]
- 10.Jones MN, Mewhort DJ. Representing word meaning and order information in a composite holographic lexicon. Psychological Review; Psychological Review. 2007;114(1):1. doi: 10.1037/0033-295X.114.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Sahlgren M, Holst A, Kanerva P. Permutations as a Means to Encode Order in Word Space. Proceedings of the 30th Annual Meeting of the Cognitive Science Society (CogSci'08); July 23–26; Washington D.C., USA. 2008. [Google Scholar]
- 12.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widdows D, Ferraro K. Semantic vectors: a scalable open source package and online technology management application. Proceedings of the Sixth International Language Resources and Evaluation (LREC’08); 2008. pp. 1183–1190. [Google Scholar]
- 14.Cohen T, Schvaneveldt R, Widdows D. Reflective Random Indexing and indirect inference: A scalable method for discovery of implicit connections. Journal of Biomedical Informatics. 2010;43(2):240–256. doi: 10.1016/j.jbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Fundel K, Küffner R, Zimmer R. RelEx—Relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 16.Mohler M, Bunescu R, Mihalcea R. Learning to grade short answer questions using semantic similarity measures and dependency graph alignments. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies; 2011. pp. 752–762. [Google Scholar]
- 17.Herrera J, Penas A, Verdejo F. Textual entailment recognition based on dependency analysis and wordnet. Machine Learning Challenges. Evaluating Predictive Uncertainty, Visual Object Classification, and Recognising Tectual Entailment. 2006. pp. 231–239.