Abstract
Temporal abstraction, a method for specifying and detecting temporal patterns in clinical databases, is very expressive and performs well, but it is difficult for clinical investigators and data analysts to understand. Such patterns are critical in phenotyping patients using their medical records in research and quality improvement. We have previously developed the Analytic Information Warehouse (AIW), which computes such phenotypes using temporal abstraction but requires software engineers to use. We have extended the AIW’s web user interface, Eureka! Clinical Analytics, to support specifying phenotypes using an alternative model that we developed with clinical stakeholders. The software converts phenotypes from this model to that of temporal abstraction prior to data processing. The model can represent all phenotypes in a quality improvement project and a growing set of phenotypes in a multi-site research study. Phenotyping that is accessible to investigators and IT personnel may enable its broader adoption.
Introduction
Healthcare quality improvement relies on electronic health record (EHR) data to identify and characterize patient populations whose care is suboptimal.1,2 Clinical research increasingly leverages EHR data to understand real-world performance of interventions and reduce enrollment and data collection costs. Much information exists in EHRs implicitly as patterns in billing codes, clinical events and observations, and concepts embedded in text reports. These data are hospital-specific, temporal and have high dimensionality, thus application of traditional statistical analysis and machine learning algorithms is difficult. Abstracting away the hospital-specific, temporal and high dimensionality features of these data could enhance greatly the application of EHR data for secondary use.3
Clinical phenotypes are derived variables computed from EHR data that signify disease, treatment and therapeutic response.4 Clinical phenotyping infers such information using temporal pattern finding, in addition to concept extraction from text and other techniques.5 Temporal abstraction,6 a method for finding frequency, sequence and overlap temporal patterns in structured time-stamped clinical and administrative data, allows specifying and computing a broad range of phenotypes in quality improvement7 and research.8 Our previously developed open source clinical phenotyping software, the Analytic Information Warehouse (AIW),7 employs temporal abstraction and has a web-based user interface for controlling data processing, called Eureka! Clinical Analytics.9 Software engineers and informatics researchers have had to specify phenotypes in a temporal abstraction ontology7 prior to data processing. The need to edit this ontology has limited the adoption of AIW and Eureka!.
We report on an extension to Eureka! to support a broad range of users in specifying phenotypes without having to learn temporal abstraction or perform ontology editing. We have created a high-level model of temporal patterns and relations for this purpose and a user interface for specifying phenotypes in terms of the model. The Eureka! software converts specified phenotypes into AIW’s existing temporal abstraction representation for data processing. The model is based on how phenotypes have been expressed to us by stakeholders of two projects: 1) an analysis of factors leading to hospital readmission within 30 days; and 2) a multi-site study of minority hypertension, the Minority Health Grid project. We aim to determine whether the model and our implementation are sufficiently expressive to specify these projects’ phenotypes.
Background
The AIW software is a Java framework that is callable through defined Application Programming Interfaces (APIs). Phenotypes are specified in a temporal abstraction ontology that is an extension of that provided in the RESUME and IDAN temporal abstraction systems.6 These earlier systems10 were designed for application in clinical decision support and guideline monitoring. Their ontology captures clinically relevant thresholds in numerical observations, hierarchies of clinical events and observations, sequence and overlap temporal patterns in events and observations, and the disease states and other contexts in which specified thresholds and patterns apply. Temporal abstraction data processing algorithms compute intervals with a start time and a stop time that represent instances of the thresholds and patterns that are specified in the ontology.
AIW extends this ontology and corresponding data processing algorithms for application in large-scale retrospective analyses of population data (see Methods). A database-agnostic data model represents entities found in EHR datasets, their attributes (called properties in AIW) and associations to each other, and concepts from the temporal abstraction ontology that they represent. Data retrieval from a source system is enabled by two kinds of mappings. The first are from the thresholds and patterns in the ontology to the entities in the data model containing the data from which they are computed. The second are from those entities to their physical schema in the source database. These two kinds of mappings allow AIW to generate SQL to retrieve the raw data needed to compute thresholds and patterns of interest. AIW’s data processing is initiated by passing the ontology, data model and mappings, database connection information, names of the phenotypes and raw data of interest, and a date range as input. AIW identifies and outputs intervals and raw data of interest either in delimited files or into a specified i2b2 project. AIW is a scalable and hardened implementation of an earlier proof-of-concept system, PROTEMPA.11 AIW has been shown to process tens of millions of patients efficiently.7 Its architecture is described in greater detail elsewhere.7
AIW’s initial application was in identifying patient features associated with 30-day readmissions. We specified in the ontology more than 100 phenotypes as patterns in laboratory test results, medication orders, diagnosis and procedure codes, admissions, discharges, geographic data and bills.7 Software engineers specified phenotypes using the Protégé ontology editor.12 We processed five years of clinical and administrative data from our institution’s clinical data warehouse and a national database extracted from 200 hospitals associated with academic health systems (UHC Clinical Database13). We found chronic disease conditions and exacerbations that have statistically significant association with an elevated readmission rate and can be predictive.3 We have since applied the AIW in research. The AIW is a part of the CardioVascular Research Grid, in which it supports creating i2b2 data marts containing raw data and phenotypes in cardiovascular studies.14 In the Minority Health Grid, we are creating such a data mart containing EHR and case report form data to characterize hypertension-related clinical history, treatment and co-morbidities in whole exome and whole genome studies. A data mart created using AIW supports lung cancer studies for our cancer center, and another data mart for lymphoma studies is under development.
Eureka!, AIW’s user interface, is implemented in Java using standard web application technologies (CSS, jQuery, JPA, Google Guice, Hibernate). Its initial version supported processing clinical and administrative data loaded from an Excel spreadsheet that conforms to a specified structure and semantic data representation.9 Users uploaded a spreadsheet when prompted, and Eureka! scanned the data in it for a predefined set of phenotypes representing co-morbidity and readmissions-related conditions and patterns. The software subsequently loaded the spreadsheet data and found phenotypes into a specified i2b2 project. These phenotypes were represented in a temporal abstraction ontology provided with the software. The phenotype editor described below is an extension of that user interface.
Methods
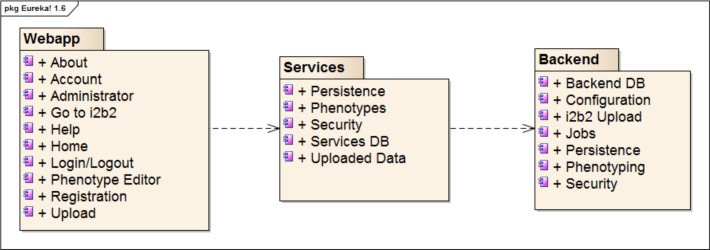
Eureka! has a three-tiered architecture, shown in Figure 1, with web application (user interface), services and backend layers that communicate via RESTful APIs. The backend layer contains the AIW temporal abstraction implementation, and the webapp and services layers implement the user interface, spreadsheet upload, security and phenotype editor storage. Eureka’s architecture, minus the phenotype editor, has been described in detail elsewhere.9
Figure 1:
Package diagram of the Eureka architecture.
AIW Temporal Abstraction Ontology
AIW’s temporal abstraction ontology is frames-based and may be edited with the Protégé editor. Category abstractions group events, observations or abstractions. They are useful for specifying hierarchies of procedure and diagnosis codes, medication orders, laboratory test results, and other data. Low-level abstractions allow specifying clinically significant thresholds on the values of a numerical observation or on the slope of sequential values of such observations, such as High systolic blood pressure. Compound low-level abstractions allow specifying clinically significant contemporaneous low-level abstractions, for example, High blood pressure specified as High systolic blood pressure or high diastolic blood pressure. Temporal pattern abstractions allow specifying temporal relationships between pairs of abstractions, events or observations, for example, High blood pressure with prior hypertension diagnosis. Quantitative temporal relationships are supported such as within 6 months before. Slice abstractions allow specifying the first, second, third etc. interval of an event, observation or abstraction on a timeline. They are useful in specifying frequencies when only the first instances of some event, observation or abstraction are of interest (e.g., the first two high blood pressure readings). Constraints may be specified on the minimum and maximum time duration of an abstraction or event specified in an abstraction definition (e.g., encounters at least 1 week in duration). Abstraction definitions may specify a gap function (causes non-adjacent intervals within a specified time distance of each other to be combined into a single longer interval), a concatenable flag (whether adjacent intervals will be combined into a single longer interval) and a solid flag (whether overlapping intervals will be combined into a single interval). Each type of abstraction has an associated mechanism for computing it, implemented in the AIW software, that outputs intervals with a start time and end time corresponding to the temporal extent of one or more input raw data values or intervals. Intervals and data values also may have different temporal granularities that are resolved automatically and propagated to output abstractions. Eureka’s open source distribution comes with a temporal abstraction ontology containing temporal patterns and relationships specifying chronic disease and hospital readmission phenotypes. This ontology also contains hierarchies of standard diagnosis and procedure codes (ICD9, CPT), and custom hierarchies of laboratory test results, medication orders and vital signs. These phenotypes and raw data types are accessible to the GUI phenotype editor as described below.
Phenotype Editor
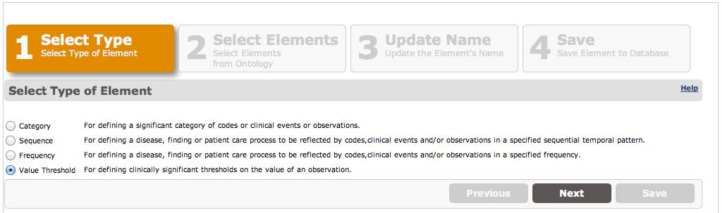
The editor supports creating four types of user-defined data element definitions (Category, Sequence, Frequency, and Value Threshold) that may be combined to build up higher-level phenotypes. A wizard-style interface guides users through creating these definitions. In the first step of the wizard, shown in Figure 2, the user selects one of the four data element types. In the second step, described in detail below, the user populates a form that is specific to the type of element selected in step 1. Each of these forms has on its left-hand side two tabs, called System and User Defined, containing a browser of the hierarchy of raw data types and abstractions specified in the ontology, and data elements the user has previously created, respectively. The user selects these data elements by dragging and dropping them from these tabs into drop boxes in the forms. In step 3, the user selects a name and a description for the data element (not shown). In step 4, the user reviews the data element definition and saves it (not shown). A similar user interface allows editing existing user-defined phenotypes. A data element list interface, not shown, allows the user to select data elements for editing or deletion. We selected these four types of data element definitions because we believe them to support the most commonly needed capabilities of temporal abstraction.
Figure 2:
Data element editing wizard showing the four types of data elements that may be created. A value threshold type has been selected.
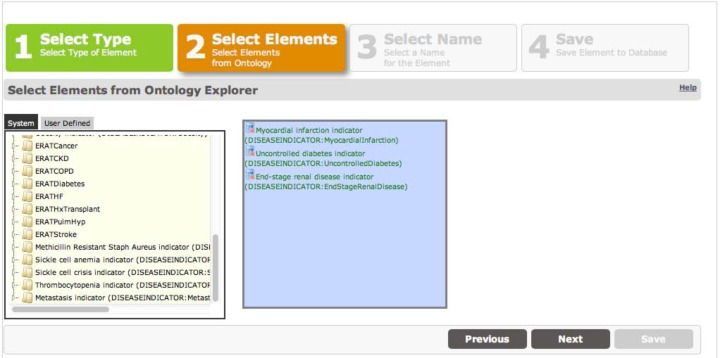
Category data elements allow specifying clinically significant groupings of other data elements. The user interface is shown in Figure 3. We have used category elements to specify custom groupings of billing diagnosis codes and procedure codes that signify co-morbidities of interest in hospital readmissions analyses. During data processing, intervals will be created that correspond to the timestamps of the category’s members that are found.
Figure 3:
Category data element editing screen showing a custom grouping of data elements found in the System tab that represent disease conditions.
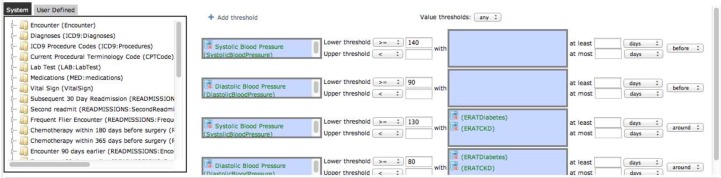
Value threshold data elements allow specifying lower and/or upper limits on one or more numerical observations such as laboratory test results or vital signs. The user interface is shown in Figure 4. A context may be specified on each observation. A context is one or more data elements that must be present in order for the specified limits to apply. Contextual information may be required to be within a specified time distance before or after the observation being thresholded. Contexts allow specifying multiple thresholds on the same observations that apply depending on what conditions and interventions the patient has had. For example, for the hypertension study we have specified two thresholds for high blood pressure: a 140/90 threshold, and a lower threshold of 130/80 if the patient has diabetes or chronic kidney disease as indicated by diagnosis codes. The combo box near the top right of Figure 4 allows selecting whether all of the specified thresholds need to be satisfied or just any of the thresholds. Intervals will be created with start and stop times equal to the timestamps of the observations that satisfy the specified thresholds.
Figure 4:
Data element editing screen showing a value threshold data element being created: high blood pressure, defined as systolic blood pressure >= 140 or diastolic blood pressure >= 90, or systolic blood pressure >= 130 or diastolic blood pressure >= 80 with a diabetes (ERATDiabetes) or chronic kidney disease (ERATCKD) discharge diagnosis code.
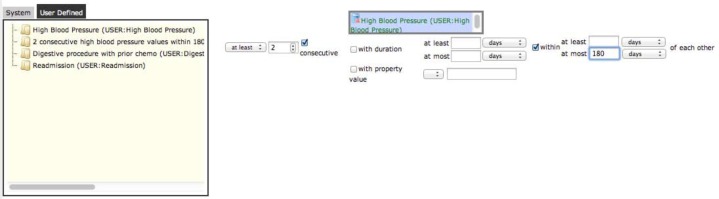
Frequency data elements allow specifying the number of times a specified data element must be present in or computed from a patient’s data. The user interface is shown in Figure 5. The left-most combo box in the user interface allows selecting whether to find any interval during which the specified number of values is present (select At least) or only the first interval (select First). After specifying the frequency count, the consecutive checkbox, which is visible only if a value threshold data element has been dragged into the drop box to the checkbox’s right, specifies that only consecutive values that meet the specified value thresholds will cause the frequency data element to be computed. For example, the at least two consecutive high blood pressure readings within 180 days data element shown in Figure 5 will be computed only if there are no intervening not-high blood pressure readings. This example also shows how users may specify that values be within a certain amount of time of each other (at most 180 days in this example). Intervals will be created that span the temporal extent of all of the intervals or data that satisfy the frequency threshold.
Figure 5:
Data element editing screen showing a frequency data element being created: 2 consecutive high blood pressure values within 180 days of each other (consecutive means with no intervening not-high values).
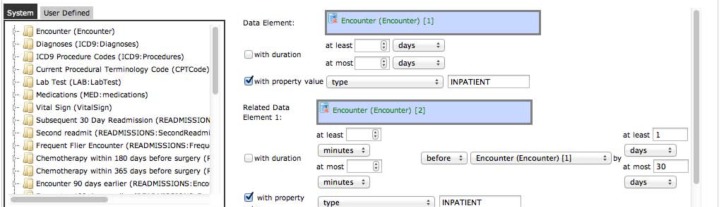
Sequence data elements allow specifying two or more data elements that must occur in a specified temporal order. The user interface is shown in Figure 6. Each data element that is specified may be constrained with a minimum and/or maximum temporal duration and a property value. The distance between pairs of data elements may be constrained to minimum and maximum time values. Intervals will be created as the temporal extent of intervals of the top-most specified data element. An example is shown in Figure 6.
Figure 6:
Data element screen showing a sequence data element being created: two inpatient encounters with the second encounter before the first by at most 30 days.
Converting data elements to abstraction definitions
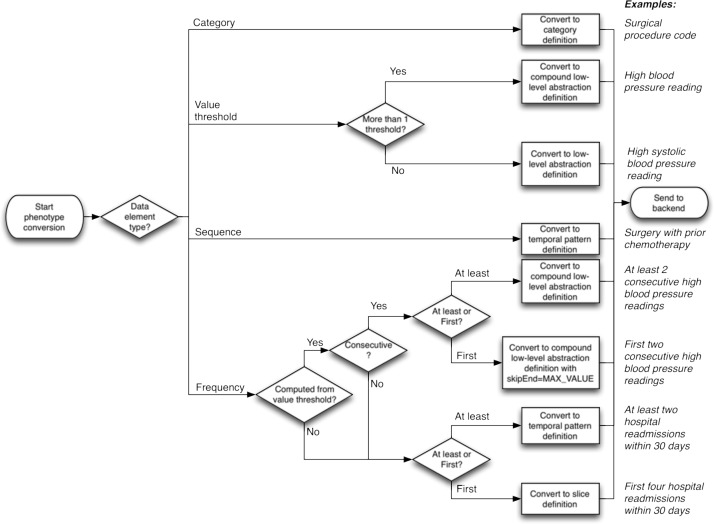
The process of converting Eureka! data element definitions into abstraction definitions is illustrated in Figure 7. Category and sequence data elements are converted directly into category and temporal pattern abstraction definitions, respectively. The corresponding temporal pattern abstraction definitions are configured with a gap function of 0 (meaning non-adjacent intervals will not be combined), and set to be concatenable and solid.
Figure 7:
Flow chart showing the process of converting Eureka! data elements into temporal abstraction definitions. This occurs in the services layer (Phenotypes component in Figure 1). Example phenotypes and how they would go through this workflow are illustrated on the right.
The conversions of value threshold and frequency data elements are more complex. Value threshold data elements in which one observation is specified are converted into low-level abstraction definitions that detect value thresholds, a gap function of 0, and the concatenable and solid properties set to true. Value threshold data elements with multiple specified observations are converted into compound low-level abstraction definitions with a gap function of 0, and the concatenable and solid properties set to true.
Frequency data elements are converted into abstraction definitions according to the following algorithm:
- If the frequency is computed from a value threshold, then:
- If the consecutive checkbox (Figure 5) is checked, then:
- If At least is selected (Figure 5), then convert to a compound low-level abstraction definition with a gap function of 0 and the concatenable and solid properties set to true.
- If First is selected (Figure 5), then convert to a compound low-level abstraction definition with a gap function of 0, the concatenable and solid properties set to true, and the skipEnd property set to MAX_VALUE. The latter property will cause data processing to stop after the first interval that matches the definitions’ criteria is found.
- If the consecutive checkbox is not checked, then:
- If At least is selected, then convert to a temporal pattern abstraction definition with a gap function of 0 and the concatenable and solid properties set to true.
- If First is selected, then convert to a slice abstraction definition with a gap function of 0 (the concatenable and solid properties always are false for slice abstractions).
If the frequency is not computed from a value threshold, then convert as in 1b above.
Data Processing
Users select phenotype definitions for processing. The selected phenotype definitions are converted to temporal abstractions. These abstractions, abstractions specified in the ontology, a spreadsheet and a date range are passed into the Eureka! backend layer (Figure 1) for data processing. The data in the spreadsheet, computed intervals, and the hierarchies and threshold definitions and temporal pattern definitions from the phenotype editor and the ontology are loaded into i2b2’s data and metadata tables. The hierarchies specified in the ontology are visible in i2b2’s concept hierarchy for use in queries in i2b2’s web-based Query and Analysis Tool. The prepackaged readmissions and comorbidity phenotypes (see Background) are visible in Hospital Readmissions and Comorbidities folders in the hierarchy, respectively. The phenotypes specified in the Eureka! phenotype editor are listed in a User-defined Data Element folder in the hierarchy.
Results
We have implemented our simplified phenotype model as part of the Eureka! services layer’s persistence component, which stores specified phenotypes in a relational database (Oracle Corp., Redwood Shores, CA). The phenotype editor user interface is integrated into the existing Eureka! website9 and may be accessed after user login by clicking a hyperlink in the site’s main button bar. The editor is available starting in Eureka! version 1.6. Eureka! is available as open source under the Apache 2 license from http://aiw.sourceforge.net.
Selected temporal and value threshold phenotypes from the readmissions project (see Background) are shown in Table 1. All phenotypes from that project that were specified in the ontology can be specified in the user interface of version 1.7 of Eureka!, which was released in Spring, 2013. For the Minority Health Grid project, we currently are adding support for representing phenotypes from the eMERGE project15 for diabetes, chronic kidney disease and hypertension. The portions of those phenotypes that are supported by Eureka! currently are shown in Table 2.
Table 1:
Temporal and value threshold phenotypes used in the readmissions analysis.
| Name | Definition | Data element(s) |
|---|---|---|
| Hospital readmission within 30 days | An inpatient encounter with another inpatient encounter within 30 days before. | Sequence |
| First four 30-day readmissions | The first four intervals of Hospital readmission within 30 days | Frequency |
| Frequent-flier encounter | Any encounter that is after the First four 30-day readmissions | Sequence |
| Suggest heart failure from BNP (B-type natriuretic peptide) | BNP test result >= 300 pg/ml | Value threshold |
| Hospital encounter in last 90 days | An inpatient encounter with another inpatient encounter that ends at most 90 days before | Sequence |
| Surgical procedure with prior chemotherapy | Any surgical procedure with a chemotherapy encounter that ends within 365 days earlier | Sequence (surgical procedure is a category of ICD9 procedure codes, and chemotherapy encounter is a category of ICD9 V-codes) |
| At least two myocardial infarctions | At least two myocardial infarction events across all encounters for a patient | Frequency (Myocardial infarction is a category of ICD-9 diagnosis codes) |
Table 2:
Supported phenotypes of interest in the Minority Health Grid study.
| Name | Definition | Data element(s) |
|---|---|---|
| Diabetes from codes – outpatient encounter | Outpatient encounter that ends with a discharge diagnosis of diabetes | Sequence (diabetes is defined as a category of discharge diagnoses) |
| Diabetes from codes – outpatient encounter | At least two Diabetes from codes – outpatient encounter (intermediate) | Frequency |
| Diabetes from codes – inpatient encounter | Inpatient encounter that ends with a discharge diagnosis of diabetes | Sequence (diabetes is defined as a category of discharge diagnoses) |
| Diabetes from codes | Diabetes from codes – outpatient encounter or Diabetes from codes – inpatient encounter | Category |
| Diabetes from prescriptions | Dispense of any anti-diabetic medication | Category containing anti-diabetic medications |
| Elevated fasting blood glucose measurements | Fasting blood glucose > 126 mg/dl | Value threshold |
| Diabetes from fasting blood glucose | At least 2 Elevated fasting blood glucose measurements | Frequency |
| Diabetes from hemoglobin A1c | A hemoglobin A1c measurement > 7% | Value threshold |
| Low estimated GFR (eGFR) using the modifications of diet in renal disease study group equation | eGFR < 60 mL/min | Value threshold |
| Elevated serum creatinine (sCr) with low eGFR | Serum creatinine > 1.5 in context of Low estimated GFR within 3 months | Value threshold |
| Chronic kidney disease from lab tests | At least two Elevated serum creatinine observations | Frequency |
| Chronic kidney disease from codes | Any chronic kidney disease discharge diagnosis code | Category of ICD-9 codes |
| Elevated blood pressure | Systolic blood pressure (SBP) >= 140 or diastolic blood pressure (DBP) >= 90; or SBP >= 130 or DBP >= 80 in the context of Diabetes from codes, Diabetes from prescriptions, Diabetes from fasting blood glucose, Diabetes from HbA1c, Chronic kidney disease from lab tests, or Chronic kidney disease from codes | Value threshold |
| Hypertension from blood pressure readings | At least two Elevated blood pressure measurements at least 1 day apart | Frequency |
| Hypertension from codes | At least two hypertension discharge diagnosis codes at least 1 day apart | Frequency (using a Category of hypertension ICD-9 codes) |
| Antihypertensive medication | A pharmacy dispensing for an antihypertensive medication | A category of hypertension medication dispenses |
| Hypertension from meds | An Antihypertensive medication and a Hypertension from codes within 6 months of each other | 2 sequences (Antihypertensive medication before Hypertension from codes by 0 to 6 months; and Antihypertensive medication after Hypertension from codes by 1 to 6 months) |
Eureka! is available in demonstration form at https://eureka.cci.emory.edu. Anyone may request an account, create phenotypes using the phenotype editor, and load data and phenotypes into an i2b2 project hosted on the demonstration site. As of August 16, 2013, there are 28 users of the demonstration site who have created 34 phenotypes using the phenotype editor. Users have included software engineers, informatics and clinical research faculty, and IT and clinical research staff.
Discussion
Eureka! aims to provide user interfaces for all of the functionality of the AIW, including phenotype editing, configuring access to relational databases, and delimited file and i2b2 output. We have implemented user interfaces for phenotype editing, spreadsheet file upload, data processing job control, and loading data and phenotypes into i2b2. While our current deployments restrict access to the services and backend layers (Figure 1) to the web application only, ultimately we plan to publish stable RESTful APIs for other applications that have a need for temporal pattern finding. The AIW project has the broad goal of enabling integrated management of clinical data and metadata describing images, EKG and other data so as to enable investigators to construct datasets consisting of, e.g., image metadata in patients meeting specified phenotypic criteria, or conversely, clinical variables in patients with specific image metadata values.
AIW employs temporal abstraction algorithms for its data processing that have well-understood performance.11 Converting phenotypes specified in our simplified model to temporal abstraction definitions for processing has allowed us to leverage AIW’s highly optimized temporal abstraction implementation that can process data from tens of millions of patients efficiently. We expect that the expressiveness of temporal abstraction will accommodate growing our model and user interfaces to meet the demands of new use cases with only limited additions to AIW’s temporal abstraction implementation. A potential limitation of the conversion from the simplified model to temporal abstraction definitions is that it is one-way in some cases. For example, a temporal pattern definition may correspond to a sequence or frequency data element (see Figure 7). Thus, we cannot visualize the contents of the temporal abstraction ontology in the Eureka! user interface using the language of the simplified model in all cases. We expect that, over time, we will move most if not all phenotype definitions to the simplified model and storage, thus substantially mitigating this concern.
By creating a simplified model of temporal patterns and relationships, and a graphical user interface for specifying phenotypes using the model, we hope to enable clinical investigators and their staff to specify phenotypes directly in many cases. The model and user interface allow us to specify the broad array of phenotypes shown in Table 1 and Table 2. To keep the user interface as simple as possible, we have purposefully omitted capabilities of temporal abstraction that we have not needed. The model’s four data element types are sufficient to cover the basic types of abstractions in the temporal abstraction ontology. We expect it to be possible to add back most features as configuration options for those four basic types. For more complex needs, the temporal abstraction ontology may still be edited in the Protégé editor as described above. There have been several attempts at graphical user interfaces for specifying temporal abstractions,16 but none to our knowledge have been deployed in production settings. While others have proposed simplified temporal query models and processing algorithms,17 none satisfy all of the phenotyping requirements of our projects, and their data processing performance has not been demonstrated. Now that we have a model in place, we expect our user interface to undergo substantial refinement in response to user feedback. Formal usability studies with clinical investigators and IT analyst personnel are needed.
The use cases that have driven our designs and implementation have a broad range of phenotyping requirements using structured data. While concept extraction from clinical documents would be valuable in understanding a patient’s past medical and social histories in these projects, we do not support it at this time. We believe that AIW’s architecture is compatible with such support, though how to integrate concepts extracted from documents into a patient’s overall clinical data timeline is an unsolved challenge in general. We hope that the open source availability of our software will lead external investigators to try using our tools in unexpected ways that will help the informatics community arrive at a reasonably complete feature set for phenotype editing.
Conclusion
Clinical phenotyping that is accessible to investigators and IT personnel would enable its broader adoption. Temporal abstraction provides a powerful representation of clinical phenotypes but needs accessible user interfaces. Eureka’s phenotype editor can represent a broad array of phenotypes specified in terms of temporal patterns and relationships in EHR data. User feedback, usability studies, and the requirements of publicly available phenotypes such as those found in the PheKB clinical phenotype knowledge base4 will inform future development of the system.
Acknowledgments
This work was supported in part by NHLBI grant R24 HL085343; PHS Grant UL1 RR025008, KL2 RR025009 and TL1 RR025010 from the CTSA program, NIH, NCRR; NIMHD Grant RC4MD005964; Emory Healthcare; and Emory Winship Cancer Institute.
Footnotes
Disclosure: Dr. Saltz is on the Scientific Advisory Board of a company called Appistry.
References
- 1.Blumenthal D. Implementation of the federal health information technology initiative. N Engl J Med. 2011;365(25):2426–31. doi: 10.1056/NEJMsr1112158. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal D. Wiring the health system--origins and provisions of a new federal program. N Engl J Med. 2011;365(24):2323–9. doi: 10.1056/NEJMsr1110507. [DOI] [PubMed] [Google Scholar]
- 3.Cholleti S, Post A, Gao J, Lin X, Bornstein W, Cantrell D, et al. Leveraging Derived Data Elements in Data Analytic Models for Understanding and Predicting Hospital Readmissions. Proc AMIA Annu Fall Symp. 2012:103–11. [PMC free article] [PubMed] [Google Scholar]
- 4.PheKB. Vanderbilt University; 2012. [cited 2013 Mar 13]; Available from: http://phekb.org. [Google Scholar]
- 5.Conway M, Berg RL, Carrell D, Denny JC, Kho AN, Kullo IJ, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc. 2011:274–83. [PMC free article] [PubMed] [Google Scholar]
- 6.Shahar Y. A framework for knowledge-based temporal abstraction. Artif Intell. 1997;90:79–133. doi: 10.1016/0933-3657(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 7.Post AR, Kurc T, Cholleti S, Gao J, Lin X, Bornstein W, et al. The Analytic Information Warehouse (AIW): A platform for analytics using electronic health record data. J Biomed Inform. 2013 doi: 10.1016/j.jbi.2013.01.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Post A, Kurc T, Overcash M, Cantrell D, Morris T, Eckerson K, et al. A Temporal Abstraction-based Extract, Transform and Load Process for Creating Registry Databases for Research. AMIA Summits Transl Sci Proc. 2011:46–50. [PMC free article] [PubMed] [Google Scholar]
- 9.Post A, Kurc T, Rathod H, Agravat S, Mansour M, Torian W, et al. Semantic ETL into i2b2 with Eureka! AMIA Summits Transl Sci Proc. 2013 accepted. [PMC free article] [PubMed] [Google Scholar]
- 10.Stacey M, McGregor C. Temporal abstraction in intelligent clinical data analysis: A survey. Artif Intell Med. 2007;39(1):1–24. doi: 10.1016/j.artmed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Post AR, Harrison JH., Jr PROTEMPA: A Method for Specifying and Identifying Temporal Sequences in Retrospective Data for Patient Selection. J Am Med Inform Assoc. 2007;14:674–83. doi: 10.1197/jamia.M2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanford Medical Informatics The Protege Ontology Editor and Knowledge Acquisition System. 2012. [cited 2012 December 11]; Available from: http://protege.stanford.edu/
- 13.Clinical Data Base/Resource Manager UHC. 2012. [cited 2012 May 3]; Available from: http://www.uhc.edu/11536.htm.
- 14.Winslow RL, Saltz J, Foster I, Carr JJ, Ge Y, Miller MI, et al. The CardioVascular Research (CVRG) Grid. Proceedings of the AMIA Summit on Translational Bioinformatics. 2011:77–81. [Google Scholar]
- 15.Pathak J, Wang J, Kashyap S, Basford M, Li R, Masys DR, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc. 2011;18(4):376–86. doi: 10.1136/amiajnl-2010-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Combi C, Oliboni B. Visually defining and querying consistent multi-granular clinical temporal abstractions. Artif Intell Med. 2012;54(2):75–101. doi: 10.1016/j.artmed.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Plaisant C, Lam S, Shneiderman B, Smith MS, Roseman D, Marchand G, et al. Searching electronic health records for temporal patterns in patient histories: a case study with Microsoft Amalga. AMIA Annu Symp Proc. 2008:601–5. [PMC free article] [PubMed] [Google Scholar]